Abstract
The oncogenic MUC1-C subunit is aberrantly overexpressed in most human breast cancers by mechanisms that are not well understood. The present studies demonstrate that stimulation of non-malignant MCF-10A cells with epidermal growth factor (EGF) or heregulin (HRG) results in marked upregulation of MUC1-C translation. Growth factor-induced MUC1-C translation was found to be mediated by PI3K->AKT, and not MEK->ERK1/2, signaling. We also show that activation of the mTORC1->S6K1 pathway decreases PDCD4, an inhibitor of the eIF4A RNA helicase, and contributes to the induction of MUC1-C translation. In concert with these results, treatment of growth factor-stimulated MCF-10A cells with the eIF4A RNA helicase inhibitors, silvestrol and CR-1-31-B, blocked increases in MUC1-C abundance. The functional significance of the increase in MUC1-C translation is supported by the demonstration that MUC1-C, in turn, forms complexes with EGFR and promotes EGFR-mediated activation of the PI3K->AKT pathway and the induction of growth. Compared to MCF-10A cells, constitutive overexpression of MUC1-C in breast cancer cells was unaffected by EGF stimulation, but was blocked by inhibiting PI3K->AKT signaling. The overexpression of MUC1-C in breast cancer cells was also inhibited by blocking eIF4A RNA helicase activity with silvestrol and CR-1-31-B. These findings indicate that EGF-induced MUC1-C expression is mediated by the PI3K->AKT pathway and the eIF4A RNA helicase, and that this response promotes EGFR signaling in an autoinductive loop. The findings also indicate that targeting the eIF4A RNA helicase is a novel approach for blocking MUC1-C overexpression in breast cancer cells.
Keywords: MUC1-C, translation, AKT, mTOR, PDCD4, silvestrol
Introduction
Protein synthesis is a tightly regulated process that is limited by translation initiation, a step controlled by the eIF4F complex at the level of ribosomal recruitment (1). The eIF4F complex is formed by binding of eIF4E to the 5′ cap structure of mRNAs and thereby recruitment of eIF4G and eIF4A. Overexpression of eIF4E has been documented in diverse human cancers and linked to transformation (2). eIF4E contributes to the malignant phenotype by selectively promoting the translation of certain oncoproteins, such as cyclin D1, MYC and MCL1, that are involved in growth and survival (2; 3). The PI3K->AKT pathway is a major regulator of protein synthesis that is upstream to the mammalian target of rapamycin complex 1 (mTORC1) (4). mTORC1 regulates eIF4E activity by phosphorylation and thereby inactivation of the inhibitory eIF4E binding proteins (4E-BPs). mTORC1 also contributes to cap-dependent translation by activating 40S ribosomal protein S6 kinases (S6Ks) that, in turn, enhance the eIF4A RNA helicase activity (1). S6K induces degradation of the tumor suppressor programmed cell death protein 4 (PDCD4), which is an eIF4A inhibitor (5). eIF4A initiates translation by unwinding highly structured 5′ untranslated regions (UTRs) in mRNAs, such as those encoding cyclin D1 and MYC (6). In this way, cancer cells can modulate translation in response to growth signals through mTORC1-induced (i) binding of eIF4E to the 5′ cap structure and (ii) activation of the eIF4A RNA helicase function. Dysregulation of translation in malignant cells has supported the development of agents that target eIF4E (7) and eIF4A (8; 9). For example, the natural product silvestrol is a potent inhibitor of the eIF4A RNA helicase function that blocks cap-dependent translation and decreases production of cyclin D1, MYC and MCL1 (9; 10). Silvestrol has also been shown to be active against cancer cells growing in vitro and in animal models (9; 8; 10). These findings have indicated that constitutive activation of PI3K->AKT->mTORC1 signaling in cancer cells can be blocked in part by targeting downstream effectors of translation.
Mucin 1 (MUC1) is an oncoprotein that is aberrantly overexpressed in human cancers by mechanisms that are not clearly understood (11). MUC1 consists of two subunits that form a non-covalent complex at the cell membrane (11). The MUC1 N-terminal (MUC1-N) ectodomain is the mucin component of the heterodimer that contains glycosylated tandem repeats. The transmembrane MUC1 C-terminal subunit (MUC1-C) has a 58 amino acid (aa) extracellular domain that interacts with the epidermal growth factor receptor (EGFR) and other receptor tyrosines (12; 11). Overexpression of MUC1 in transgenic mouse models is associated with binding to EGFR in mammary glands and the induction of breast tumors (13; 14). The interaction between MUC1 and EGFR increases EGFR internalization and recycling at the cell membrane (15). Other studies have shown that MUC1-C contributes to EGFR-mediated activation of the PI3K->AKT pathway (16). In this context, the 72 aa MUC1-C cytoplasmic domain binds to PI3K and contributes to activation of the PI3K->AKT pathway (17; 16). Overexpression of the MUC1-C subunit, as found in diverse human cancers, is sufficient to induce anchorage-independent growth and tumorigenicity (18; 19; 11). Upregulation of MUC1-C also attenuates the induction of cell death in response to genotoxic, oxidative and hypoxic stress (20–22). MUC1-C localizes to the nucleus, where it associates with transcription factors, such as NF-κB RelA and STAT3, and promotes activation of their target genes, including MUC1 itself (23; 24). Thus, MUC1-C contributes, at least in part, to its own overexpression through autoinductive regulatory loops (11). Based on these findings, MUC1-C has emerged as an attractive target for cancer treatment using approaches that block its function and thereby overexpression. For example, cell-penetrating peptides and small molecules that inhibit the MUC1-C cytoplasmic domain attenuate localization of MUC1-C to the nucleus of cancer cells and downregulate its overexpression (25–27). There is, however, no available information about whether MUC1-C can be targeted in cancer cells by blocking its expression at the level of translation.
The present results demonstrate that growth factor stimulation of non-malignant MCF-10A breast epithelial cells is associated with activation of the PI3K->AKT->mTORC1 pathway and thereby induction of MUC1-C translation. In concert with involvement of the eIF4A RNA helicase, growth factor-induced MUC1-C translation in MCF-10A cells was inhibited by silvestrol and another eIF4A inhibitor, designated CR-1-31-B. The results also show that treatment of human breast cancer cells with eIF4A inhibitors is associated with downregulation of MUC1-C expression.
Results
Growth factor stimulation induces MUC1-C expression
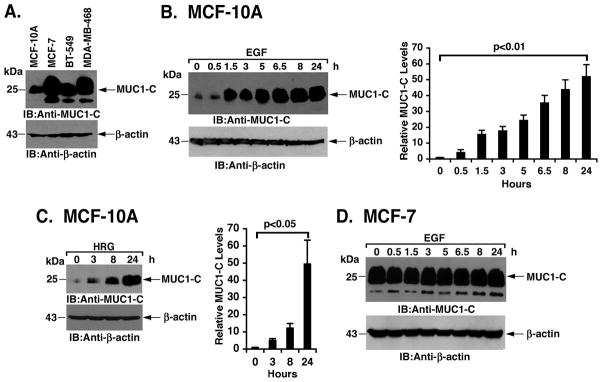
Abundance of the ~25 kDa MUC1-C protein is relatively lower in non-malignant MCF-10A breast epithelial cells as compared to that MCF-7, BT-549 and MDA-MB-468 breast cancer cells (Fig. 1A). Consequently, we reasoned that MCF-10A cells might represent a potential model to study mechanisms responsible for the overexpression of MUC1-C in breast cancer cells. In this context, we found that stimulation of MCF-10A cells with EGF is associated with marked upregulation of MUC1-C expression with an increase of over 50-fold at 24 h compared to baseline levels (Fig. 1B, left). Densitometric scanning of the signals from repetitive experiments further demonstrated a time-dependent increase in MUC1-C abundance (Fig. 1B, right). Treatment of MCF-10A cells with heregulin (HRG) was similarly associated with a substantial increase in MUC1-C abundance (~50-fold at 24 h compared to baseline) (Figs. 1C, left and right). By contrast, EGF had no apparent effect on MUC1-C levels in MCF-7 breast cancer cells (Fig. 1D). Stimulation of MCF-7 cells with HRG also had no effect on MUC1-C abundance (data not shown), indicating that MUC1-C expression is inducible by growth factors in MCF-10A, but not MCF-7, cells.
Figure 1. Stimulation of non-malignant MCF-10A breast epithelial cells with EGF or HRG induces MUC1 expression.
A. Lysates from MCF-10A cells and the indicated breast cancer cells were immunoblotted with anti-MUC1-C and anti-β-actin. B. MCF-10A cells were stimulated with 100 ng/ml EGF for the indicated times. Lysates were immunoblotted with anti-MUC1-C and anti-β-actin (left). Intensity of the MUC1-C signals was determined by densitometric scanning. The results (mean±SD of three replicates) are expressed as relative MUC1-C levels compared to that obtained for the untreated control (assigned a value of 1) (right). C. MCF-10A cells were stimulated with 10 ng/ml HRG for the indicated times. Lysates were immunoblotted with the indicated antbodies (left). The results (mean±SD of three replicates) are expressed as relative MUC1-C levels compared to that obtained for the untreated control (assigned a value of 1) (right). D. MCF-7 cells were stimulated 100 ng/ml EGF for the indicated times. Lysates were immunoblotted with the indicated antibodies.
MUC1-C translation is induced by the PI3K->AKT->mTOR pathway
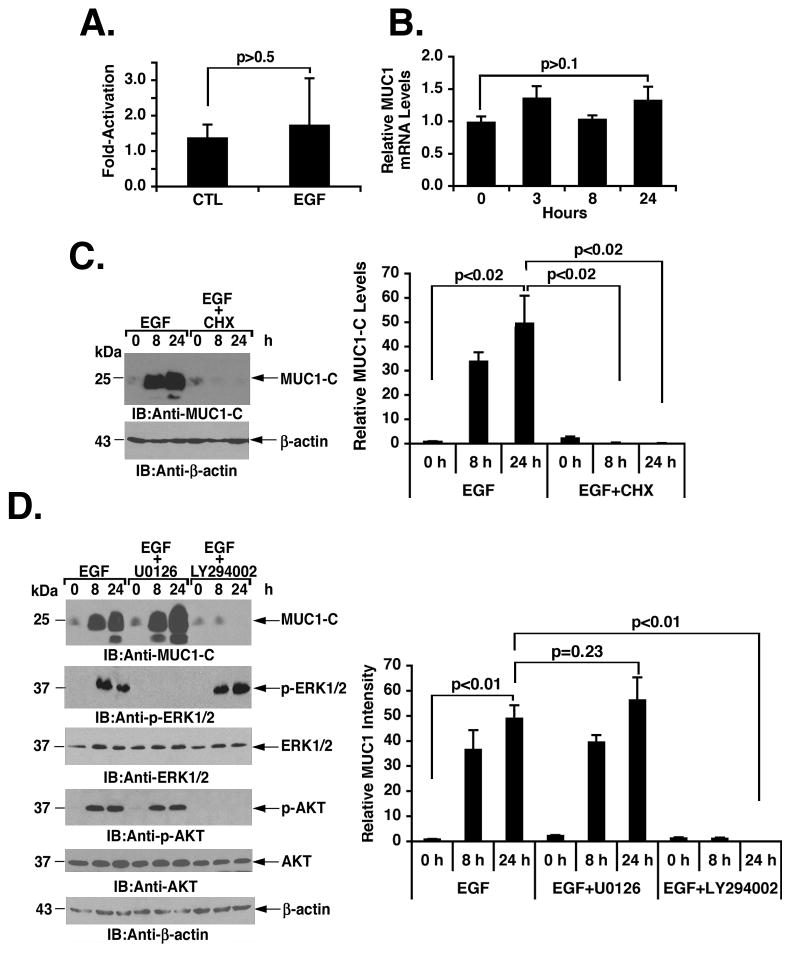
To define the basis for growth factor-induced increases in MUC1-C expression, we asked if the upregulation in levels is mediated by transcriptional and/or post-transcriptional mechanisms. EGF stimulation of MCF-10A cells had no significant effect on activation of the MUC1 gene promoter in MCF-10A cells (Fig. 2A). Moreover, EGF had no apparent effect on MUC1 mRNA levels as determined by qRT-PCR, indicating that the increase in MUC1-C protein is regulated at the post-transcriptional level (Fig. 2B). As a control, inhibition of protein synthesis with cycloheximide (CHX) blocked EGF-induced increases in MUC1-C abundance (Figs. 2C, left and right), confirming that translation of MUC1-C is upregulated in the response to growth factor stimulation. Of note, the basal levels of MUC1-C in MCF-10A cells varies among experiments as a result of differences in exposure times used for detection of the signals. Certain signaling pathways, such as MEK->ERK1/2 and PI3K->AKT, have been linked to the activation of protein translation (1). To assess potential involvement of ERK1/2 and/or PI3K in the regulation of MUC1-C translation, EGF-stimulated MCF-10A cells were treated with the dual ERK1/2 inhibitor, U0126 (28), or the PI3K inhibitor, LY294002 (29). Inhibition of PI3K, but not ERK1/2, blocked EGF-mediated induction of MUC1-C expression (Figs. 2D, left and right).
Figure 2. Activation of the PI3K pathway induces MUC1-C translation.
A. MCF-10A cells were transfected with the control pGL3 (CTL) or pMUC1-Luc in the presence of Renilla plasmid for 24 h. The cells were then stimulated with EGF for 5 h and then assayed for luciferase activity. The results are expressed as the fold-activation (mean±SD of three determinations) relative to that obtained for the pGL3 plasmid. B. MCF-10A cells were stimulated with EGF for the indicated times. MUC1 mRNA levels (mean±SD of three determinations) were assayed by qRT-PCR. C. MCF-10A cells were stimulated with EGF in the absence and presence of 10 ng/ml CHX for the indicated times. Lysates were immunoblotted with anti-MUC1-C and anti-β-actin (left). The results (mean±SD of three replicates) are expressed as relative MUC1-C levels compared to that obtained for the untreated control (assigned a value of 1) (right). D. MCF-10A cells were stimulated with EGF in the presence of 10 μM U0126 or 50 μM LY294002 for 8 and 24 h. Lysates were immunoblotted with the indicated antibodies (left). The results (mean±SD of three replicates) are expressed as relative MUC1-C levels compared to that obtained for the untreated control (assigned a value of 1) (right).
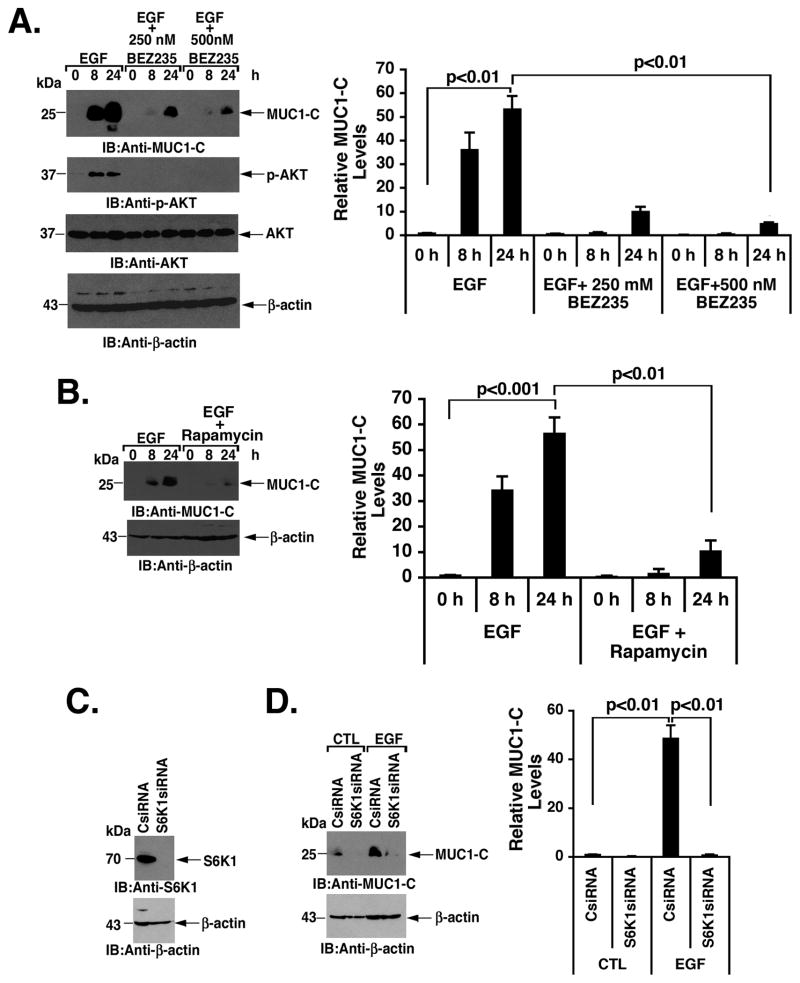
To extend this analysis, experiments were performed with BEZ235, an inhibitor of PI3K and mTOR (30). As found with LY294002, BEZ235 blocked EGF-induced increases in MUC1-C abundance (Figs. 3A, left and right). In concert with these results, treatment of EGF-stimulated MCF-10A cells with rapamycin, an allosteric inhibitor of mTOR (1), was also associated with a block in the induction of MUC1-C expression (Figs. 3B, left and right). mTOR is part of the mTORC1 complex, which phosphorylates the ribosomal protein S6 kinase 1 (S6K1) and thereby contributes to the initiation of translation (1). The demonstration that silencing S6K1 (Fig. 3C) inhibits EGF-induced upregulation of MUC1-C levels (Figs. 3D, left and right) provided further support for involvement of the PI3K->AKT->mTORC1->S6K1 pathway in the activation of MUC1-C translation.
Figure 3. MUC1-C translation is induced by PI3K->mTOR->S6K1 signaling.
A. MCF-10A cells were stimulated with EGF in the presence of 250 or 500 nM BEZ235 for the indicated times. Lysates were immunoblotted with the indicated antibodies (left). The results (mean±SD of three replicates) are expressed as relative MUC1-C levels compared to that obtained for the untreated control (assigned a value of 1) (right). B. MCF-10A cells were stimulated with EGF in the presence of 100 nM rapamycin for the indicated times. Lysates were immunoblotted with anti-MUC1-C and anti-β-actin (left). The results (mean±SD of three replicates) are expressed as relative MUC1-C levels compared to that obtained for the untreated control (assigned a value of 1) (right). C. MCF-10A cells were transfected to express a control siRNA or a S6K1 siRNA pool. Lysates from the transfected cells were immunoblotted with the indicated antibodies. D. MCF-10A cells transfected with the CsiRNA or S6K1siRNA were left untreated (CTL) or stimulated with EGF for 24 h. Lysates were immunoblotted with the indicated antibodies (left). The results (mean±SD of three replicates) are expressed as relative MUC1-C levels compared to that obtained for the control (assigned a value of 1) (right).
Inhibiting cap-dependent translation blocks growth factor-induced increases in MUC1-C abundance
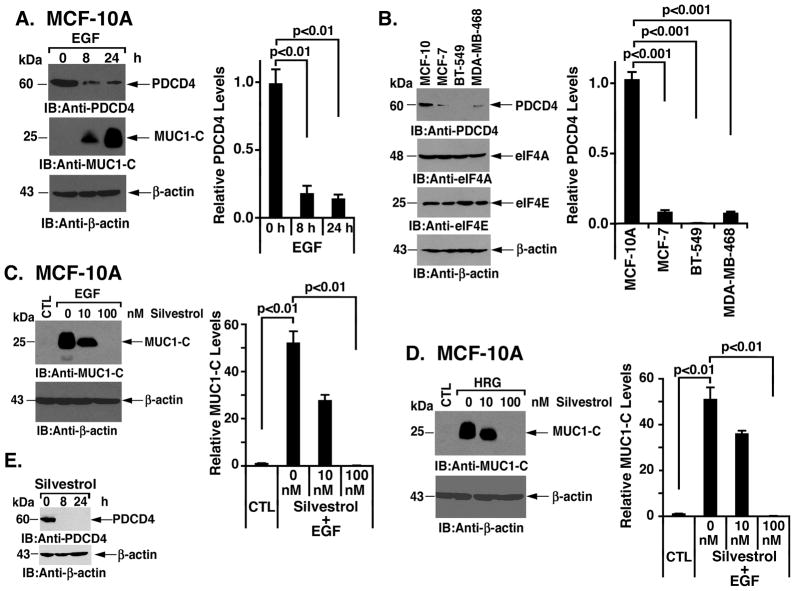
S6K1-mediated activation of the eIF4A RNA helicase is essential for unwinding of certain 5′UTRs and induction of translation (4). To address the potential role of eIF4A, we first examined expression of the tumor suppressor programmed cell death protein 4 (PDCD4), which inhibits the eIF4A RNA helicase activity (31; 32). Stimulation of MCF-10A cells with EGF resulted in downregulation of PDCD4 levels in association with increases in MUC1-C abundance (Figs. 4A, left and right). Moreover and in concert with the constitutive upregulation of MUC1-C in breast cancer cells, PDCD4 was low to undetectable in MCF-7, BT-549 and MDA-MB-468 cells (Figs. 4B, left and right). In contrast to PDCD4, there was little difference among these cells in terms of eIF4A and eIF4E expression (Fig. 4B, left). Whereas PDCD4 inhibits the eIF4A RNA helicase, EGF-stimulated MCF-10A cells were treated with silvestrol, an inhibitor of eIF4A RNA helicase activity (8). In this context, there is presently no assay for monitoring eIF4A activity in cells and silvestrol is used to assess dependence on this helicase (9; 10). Notably, silvestrol blocked EGF-mediated activation of MUC1-C translation in a dose-dependent manner (Figs. 4C, left and right). Similar dose-dependent inhibitory effects were obtained when HRG-stimulated MCF-10A cells were treated with silvestrol (Figs. 4D, left and right). MCF-10A cells were also treated with silvestrol and monitored for effects on PDCD4 expression. The results demonstrate that silvestrol treatment is associated with a marked decrease in PDCD4 abundance (Fig. 4E). These findings indicate that (i) growth factor-induced MUC1 translation is associated with degradation of PDCD4 and activation of the eIF4A RNA helicase, and (ii) silvestrol blocks growth-factor-induced MUC1 translation by inhibiting eIF4A activity. Silvestrol also decreased PDCD4 expression in a potential feedback response to the inhibition of eIF4A activity.
Figure 4. Growth factor-induced MUC1-C expression is regulated by cap-dependent translation.
A. MCF-10A cells were stimulated with EGF for 8 and 24 h. Lysates were immunoblotted with the indicated antibodies (left). The results (mean±SD of three replicates) are expressed as relative PDCD4 levels compared to that obtained for the untreated control (assigned a value of 1) (right). B. Lysates from MCF-10A, MCF-7, BT-549 and MDA-MB-468 cells were immunoblotted with the indicated antibodies (left). The results (mean±SD of three replicates) are expressed as relative PDCD4 levels compared to that obtained for MCF-10A cells (assigned a value of 1) (right). C and D. MCF-10A cells were left untreated (CTL) and stimulated with EGF (C) or HRG (D) in the presence of the indicated concentrations of silvestrol for 24 h. Lysates were immunoblotted with anti-MUC1-C and anti-β-actin (left). The results (mean±SD of three replicates) are expressed as relative MUC1-C levels compared to that obtained for the control (assigned a value of 1) (right). E. MCF-10A cells were treated with 100 nM silvestrol for the indicated times. Lysates were immunoblotted with anti-PDCD4 and anti-β-actin.
Upregulation of MUC1-C expression contributes to EGFR-mediated signaling
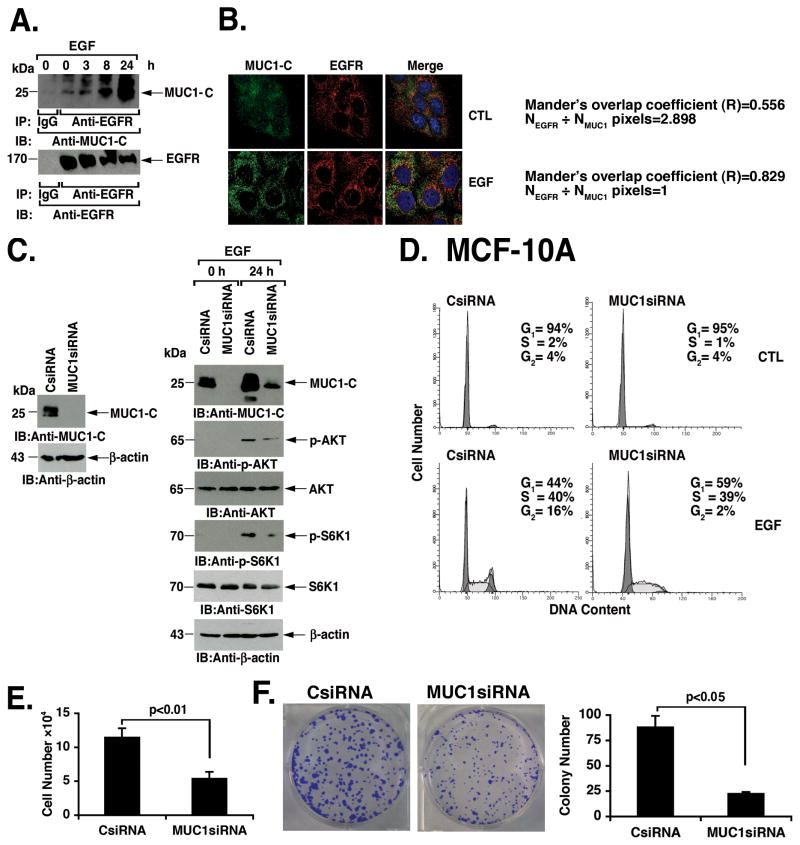
MUC1-C forms complexes with EGFR at the cell membrane of breast cancer cells (12). Coimmunoprecipitation studies were therefore performed to determine whether the upregulation of MUC1-C expression affects the formation of EGFR/MUC1-C complexes. Analysis of anti-EGFR precipitates demonstrated a time-dependent increase in the association of EGFR and MUC1-C (Fig. 5A). In addition, the increases in EGFR-MUC1-C complexes were associated with partial downregulation of EGFR levels observed in the response to EGF stimulation (Fig. 5A) (33; 34). These results were extended with confocal microscopy studies of EGF-stimulated MCF-10A cells demonstrating increased colocalization of EGFR and MUC1-C (Fig. 5B, left). Analysis of the images using Image J (35) confirmed a significant increase in EGFR and MUC1-C colocalization after EGF stimulation as supported by an enhanced Mander’s overlap coefficient with a NEGFR/NMUC1-C pixels=1. To further assess the functional role of MUC1-C, we generated MCF-10A cells that were stably silenced for MUC1-C expression (Fig. 5C, left). Whereas MUC1-C contributes to EGFR-mediated activation of the PI3K->AKT pathway (16), studies were performed to assess the effects of MUC1-C silencing on EGF-induced PI3K->AKT signaling (Fig. 5C, right). Notably, EGF-induced increases in p-AKT and p-S6K1 were suppressed in association with the silencing of MUC1-C (Fig. 5C, right). In concert with these results, EGF-induced cell cycle progression was attenuated as a result of MUC1-C silencing with increases in G1 phase and decreases in G2 phase (Fig. 5D). In addition, decreases in MUC1-C abundance attenuated EGF-stimulated MCF-10A cell growth (Fig. 5E) and colony formation (Fig. 5F, left and right). These findings indicate that EGF stimulates MUC1-C expression and, in turn, MUC1-C promotes EGFR-induced PI3K->AKT signaling and cell growth.
Figure 5. MUC1-C contributes to EGFR-mediated signaling and cell growth.
A. MCF-10A cells were stimulated with EGF for the indicated times. Lysates were precipitated with a control IgG or anti-EGFR. The precipitates were immunoblotted with anti-MUC1-C or anti-EGFR. B. MCF-10A cells were left untreated (CTL) or stimulated with EGF for 24 h. Cells were stained with anti-MUC1-C and anti-EGFR, and analyzed by confocal microscopy (left). The images were analyzed by Image J (35) to confirm increased colocalization of EGFR and MUC1-C in the response to EGF stimulation (right). C. Lysates from MCF-10A cells stably transfected to express a control siRNA or a MUC1 siRNA were immunoblotted with the indicated antibodies (left). The MCF-10A/CsiRNA and MCF-10A/MUC1siRNA cells were left untreated or stimulated with EGF for 24 h (right). Lysates were immunblotted with the indicated antibodies. D. MCF-10A/CsiRNA and MCF-10A/MUC1siRNA cells were stimulated with EGF for 24 h. Control (CTL) and EGF-treated cells were stained with PI and analyzed for cell cycle distribution by flow cytometry. The percentage of cells in G1, S and G2 phase are included in the panels. E. MCF-10A/CsiRNA and MCF-10A/MUC1siRNA cells were stimulated with EGF for 24 h, reseeded and then counted at 48 h. The results are expressed as cell number (mean±SD of three determinations). F. MCF-10A/CsiRNA and MCF-10A/MUC1siRNA cells were stimulated with EGF for 24 h and reseeded into 6-well plates (1000 cells per well). Colonies were stained with crystal violet, photographed (left) and counted (right) on day 7. The results are expressed as colony number (mean±SD of three determinations) (right).
eIF4A RNA helicase activity confers translation of MUC1-C in growth factor-stimulated MCF-10A cells
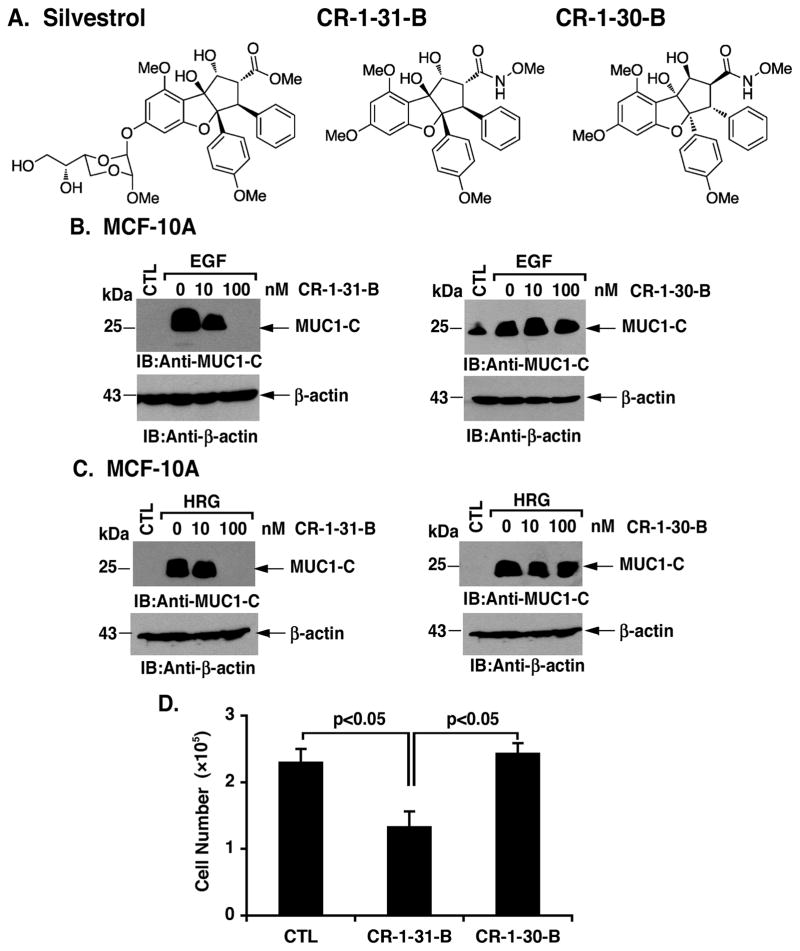
To confirm the notion that eIF4A RNA helicase activity induces MUC1-C translation, we assessed the effects of a silvestrol analog, designated CR-1-31-B, and its enantiomer CR-1-30-B, which is inactive against eIF4A (36) (Fig. 6A). As found with silvestrol, CR-1-31-B treatment of EGF-stimulated MCF-10A cells was associated with a dose-dependent decrease in MUC1-C abundance (Fig. 6B, left). By contrast, the inactive CR-1-30-B had no effect on EGF-induced MUC1-C expression (Fig. 6B, right). Treatment of MCF-10A cells with CR-1-31-B, but not the inactive CR-1-30-B, also blocked HRG-induced increases in MUC1-C abundance (Fig. 6C, left and right). In concert with the effects of inhibiting the eIF4A RNA helicase on the cap-dependent translation of multiple oncoproteins, growth of MCF-10A cells in response to EGF was attenuated by CR-1-31-B and not CR-1-30-B (Fig. 6D).
Figure 6. MUC1-C translation is inhibited by CR-1-31-B in MCF-10A cells.
A. Structures of the indicated compounds. B and C. MCF-10A cells were left untreated (CTL) and stimulated with EGF (B) or HRG (C) in the presence of the indicated concentrations of CR-1-31-B (left) or inactive CR-1-30-B (right) for 24 h. D. MCF-10A cells were stimulated with EGF in the absence (CTL) or presence of 100 nM CR-1-31-B or CR-1-30-B for 24 h, reseeded and then counted at 48 h. Viable cell number (mean±SD of three determinations) was determined by trypan blue exclusion.
PI3K->AKT pathway and the eIF4A RNA helicase contribute to MUC1-C translation in breast cancer cells
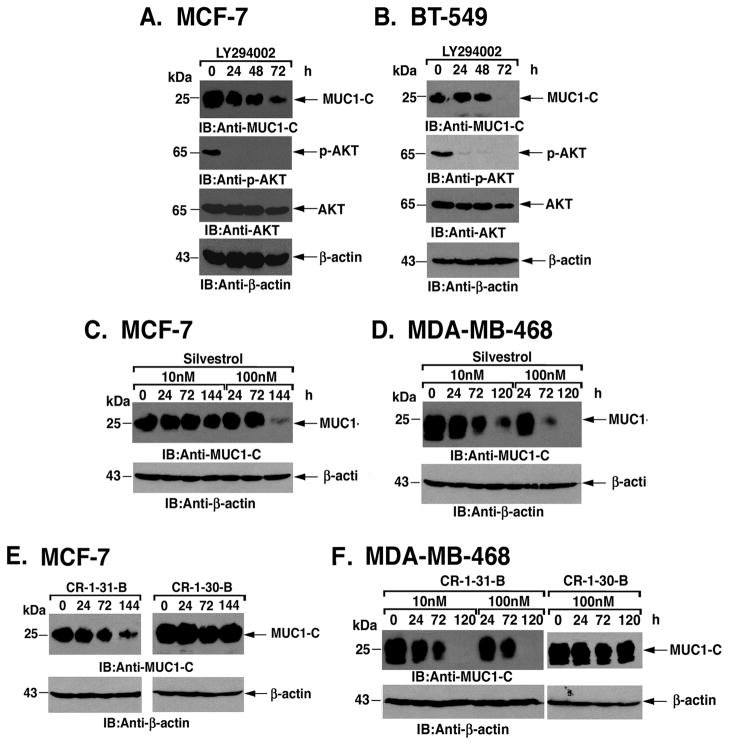
Based on the results obtained in MCF-10A cells, we asked if PI3K->AKT-induced activation of MUC1-C translation contributes to the constitutive overexpression of MUC1-C in breast cancer cells. Accordingly, treatment of MCF-7 cells with LY294002 was associated with progressive decreases in MUC1-C abundance that corresponded with inhibition of p-AKT (Fig. 7A). In addition, downregulation of MUC1-C protein levels by LY294002 occurred in the absence of a detectable effect on MUC1 mRNA levels (data not shown). Similar results were obtained with LY294002-treated BT-549 breast cancer cells (Fig. 7B), indicating that PI3K->AKT signaling contributes to MUC1-C overexpression. To extend these observations to the regulation of MUC1-C translation, we treated breast cancer cells with silvestrol. Exposure of MCF-7 cells to 10 nM silvestrol had a limited effect on MUC1-C levels (Fig. 7C). Moreover, treatment with 100 nM silvestrol was associated with a more pronounced decrease in MUC1-C abundance (Fig. 7C). Treatment of BT-549 cells resulted in a similar dose-dependent effect of silvestrol on MUC1-C levels (Supplemental Fig. S1A). Treatment of MDA-MB-468 breast cancer cells also demonstrated decreases in MUC1-C that were clearly detectable in response to 10 and 100 nM silvestrol (Fig. 7D). Treatment of MCF-7 cells with 100 nM CR-1-31-B was associated with downregulation of MUC1-C abundance (Fig. 7E, left). By contrast, the inactive CR-1-30-B enantiomer had no apparent effect (Fig. 7E, right). We also found that 100 nM CR-1-31-B, but not the inactive CR-1-30-B, decreases MUC1-C expression in BT-549 cells (Supplemental Fig. S1B, left and right). In addition, treatment of MDA-MB-468 breast cancer cells with 10 and 100 nM CR-1-31-B was associated with decreases in MUC1-C abundance (Fig. 7F). These findings collectively indicated that PI3K->AKT signaling activates eIF4A RNA helicase-mediated translation of MUC1-C in breast cancer cells.
Figure 7. MUC1-C translation is regulated by PI3K->AKT signaling and eIF4A in breast cancer cells.
A and B. MCF-7 (A) and BT-549 (B) cells were treated with LY294002 for the indicated times. Lysates were immunoblotted with the indicated antibodies. C–D. MCF-7 (C) and MDA-MB-468 (D) cells were treated with 10 or 100 nM silvestrol for the indicated times. Lysates were immunoblotted with anti-MUC1-C and anti-β-actin. E and F. MCF-7 (E) and MDA-MB-468 (F) cells were treated with 100 nM CR-1-31-B or inactive CR-1-30-B for the indicated times. Lysates were immunoblotted with anti-MUC1-C and anti-β-actin.
Discussion
Growth factor-induced PI3K->AKT signaling induces MUC1-C translation
The MUC1 heterodimer is localized at the apical border of normal epithelial cells and is thus sequestered from EGFR and other RTKs that reside at the basal-lateral borders (11). In the response to stress, epithelial cells lose apical-basal polarity in association with activation of a proliferation and survival program (37). Under these circumstances, the MUC1-C subunit is now repositioned to form complexes with RTKs, such as EGFR, and promote their activation of downstream growth and survival signals (Fig. 8) (11). The present studies demonstrate that stimulation of non-malignant MCF-10A mammary epithelial cells with EGF results in pronounced increases in MUC1-C translation. Similar effects were observed with HRG, an activator of ErbB2, indicating that this increase in MUC1-C levels is not restricted to EGFR stimulation. Indeed, our results do not exclude the possibility that activation of non-ErbB RTKs similarly induces MUC1-C expression. We also found that EGF- and HRG-induced increases in MUC1-C abundance are suppressed by inhibitors of the PI3K->AKT pathway and not those that block MEK->ERK1/2 signaling (Fig. 8). AKT controls protein synthesis at multiple levels, including ribosome biogenesis, translation initiation and elongation, leading to changes in translation of select mRNAs (38). The present results demonstrate that like other oncoproteins, for example cyclin D1, MYC and MCL1 (4; 1), translation of MUC1-C is selectively induced by growth factor stimulation and activation of AKT signaling. The functional significance of upregulating MUC1-C translation is supported by the findings that MUC1-C in turn forms complexes with EGFR and promotes EGFR-mediated signaling (Fig. 8). In this capacity, previous work has shown that EGFR phosphorylates the MUC1-C cytoplasmic domain and that this domain binds to PI3K and contributes to the activation of PI3K->AKT signaling (Fig. 8) (39; 17; 12; 16). Accordingly, MUC1-C silencing in EGF-stimulated MCF-10A cells attenuated activation of AKT and the induction of a proliferative response. These findings support a model in which EGF-induced MUC1-C rtranslation activates an autoinductive loop in which MUC1-C in turn contributes to EGFR signaling (Fig. 8).
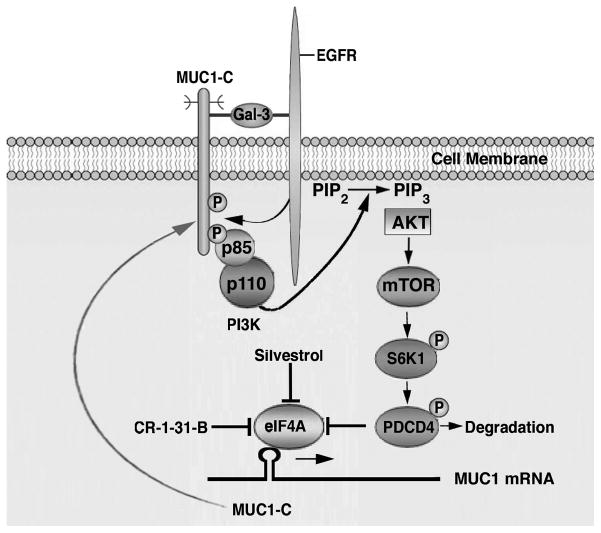
Figure 8. Proposed autoinductive loop in which MUC1-C contributes to activation of the EGFR->PI3K->AKT->mTOR pathway and thereby increased translation of the MUC1-C protein.
MUC1-C forms complexes with EGFR at the cell membrane that are mediated by extracellular galectin-3 bridges. Stimulation of EGFR with EGF induces phosphorylation of the MUC1-C cytoplasmic domain, promotes binding of the PI3K SH2 domains and thereby activation of the PI3K->AKT->mTOR pathway. mTOR-mediated phosphorylation and activation of S6K1 induces degradation of PDCD4, an inhibitor of the eIF4A RNA helicase. Derepression of eIF4A activity stimulates MUC1-C translation with marked increases in MUC1-C protein and, in turn, the formation of EGFR/MUC1-C complexes. This autoinductive loop is constitutively activated in breast cancer cells and disrupted by the eIF4A inhibitors, silvestrol and CR-1-31-B.
MUC1-C translation is induced by the eIF4A RNA helicase
AKT controls translation in part through the activation of mTORC1, which results in the phosphorylation of several substrates, including S6K (Fig. 8) (4). In turn, S6K phosphorylates and thereby induces the degradation of PDCD4, an inhibitor of eIF4A RNA helicase activity that regulates translation of proteins, such as p53, that are involved in growth and survival (Fig. 8) (5; 40). S6K also phosphorylates eIF4B, which interacts with eIF4A and contributes to eIF4A activation (41). In our studies with MCF-10A cells, EGF-induced increases in MUC1-C translation were blocked by silencing S6K1. Moreover, EGF stimulation was associated with downregulation of PDCD4, suggesting that the induction of MUC1-C translation could be mediated by the eIF4A RNA helicase. To directly address this possibility, we found that EGF-and HRG-induced MUC1-C translation is substantially blocked by silvestrol, a natural product isolated from the plant Aglaia silvestris (42). Silvestrol inhibits eIF4A by inducing dimerization of eIF4A and RNA (8) and preferentially blocks the translation of mRNAs with highly structured 5′UTRs that require efficient unwinding by the eIF4F complex (43). In that sense, translation of specific mRNAs varies substantially for different transcripts and is dependent in part on the presence of discrete hairpin structures in the 5′UTR (2). Notably, the MUC1 5′UTR includes such discrete hairpin structures, consistent with a potential requirement for unwinding by the eIF4A RNA helicase for translation initiation (Supplementary Fig. S2). In concert with this model, silvestrol blocked growth factor-induced MUC1-C translation, but had little effect on the abundance of β-actin, which is encoded by a mRNA with a relatively unstructured 5′UTR (2). To confirm these results, we showed that CR-1-31-B, a novel inhibitor of the eIF4A RNA helicase (36), similarly blocks MUC1-C translation in response to growth factor stimulation. By contrast and as a control for specificity, an inactive enantiomer of CR-1-31-B, designated CR-1-30-B, had no apparent effect on the induction of MUC1-C translation. These findings indicate that, like certain other oncoproteins (2), the translation of MUC1-C is preferentially induced by growth factor stimulation and activation of the eIF4A RNA helicase (Fig. 8).
Targeting cap-dependent translation to block overexpression of MUC1-C in human cancers
Dysregulation of protein synthesis has been linked to the development and progression of cancers as a result of aberrant cell signaling pathways that converge on translation initiation (7). For that reason, drugs have been developed to inhibit mRNA translation by blocking eIF4E, eIF4A and other targets that are components of the translational machinery (7). MUC1-C is aberrantly overexpressed in breast and other human cancers, and thereby contributes to growth and survival pathways (11). Thus, MUC1-C has become an attractive target for the treatment of cancers that overexpress this oncogenic subunit (11). The present results indicate that targeting MUC1-C translation represents a potential approach to inhibit the effects of MUC1-C overexpression in cancer cells. In the breast cancer cells studied in the present work, eIF4A and eIF4E levels were similar to those found in non-malignant MCF-10A cells. Notably, however, PDCD4 expression was decreased compared to that in MCF-10A cells, suggesting that the eIF4A RNA helicase could be of importance to the increased levels of MUC1-C in breast cancer cells. In concert with the findings in growth factor-stimulated MCF-10A cells, treatment of breast cancer cells with silvestrol was associated with decreases in MUC1-C abundance. Treatment with CR-1-31-B, but not CR-1-30-B, similarly resulted in downregulation of MUC1-C levels, indicating that the eIF4A RNA helicase activity is responsible, at least in part, for overexpression of MUC1-C in these cells (Fig. 8). CR-1-31-B was also effective in inhibiting breast cancer cell growth, consistent with the downregulation of diverse oncoproteins that are dependent on the eIF4A RNA helicase for translation (Supplemental Fig. S3). Cell-penetrating peptide and small molecule inhibitors have been developed that directly block the MUC1-C oncogenic function and induce death of breast cancer cells (25; 27). Targeting MUC1-C translation to decrease MUC1-C abundance could thus conceivably increase the effectiveness of these direct inhibitors. Finally, agents such as silvestrol and CR-1-31-B, are likely to be highly effective as anti-cancer agents given that, in addition to MUC1-C, multiple oncoproteins, including cyclin D1, MYC and MCL1, are downregulated by inhibiting the eIF4A RNA helicase. Indeed, preclinical studies with silvestrol in animal models have demonstrated promising anti-tumor activity with little toxicity, supporting the selectivity of blocking eIF4A function for cancer treatment (8–10).
Materials and Methods
Cell culture
Human MCF-10A mammary epithelial cells were grown in mammary epithelial growth medium (MEGM, Lonza). Human MCF-7 and MDA-MB-468 breast cancer cells were cultured in Dulbecco’s modified Eagle’s medium with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine. Human BT-549 cells were maintained in RPMI-1640 medium (ATCC) with 10% FBS and 0.023 IU/ml insulin. Cells were treated with EGF (Sigma), heregulin beta 1 (HRG; Neomarkers), cycloheximide (CHX, Calbiochem), U0126 (Calbiochem), LY294002 (Caymam Chemical Company), BEZ-235 (Selleckchem) and rapamycin (Cell Signaling Technology). In certain experiments, MCF-10A cells were serum-starved overnight, and inhibitors were added 2.5 h prior to EGF or HRG treatment.
eIF4A inhibitors
Silvestrol (44), CR-1-31-B (36), and its enantiomer CR-1-30-B (36) were synthesized according to literature procedures.
Immunoblot analysis
Lysates from subconfluent cells were immunoblotted with anti-MUC1-C (Ab5; Neomarkers), anti-β-actin (Sigma), anti-p-AKT, anti-AKT, anti-p-ERK1/2, anti-ERK1/2, anti-p-S6K1, anti-S6K1, anti-PCDC4, anti-eIF4E, eIF4A (Cell Signaling Technology) and anti-EGFR (Santa Cruz Technology). Immune complexes were detected with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (Amersham Biosciences). Intensity of certain signals was determined by densitometric scanning. Statistical significance was determined by the Student’s t-test.
Luciferase assays
Control pGL3 or pGL3-MUC1-promoter constructs were transfected with the Renilla plasmid into cells in the presence of Lipofectamine. At 24 h after transfection, cells were serum-starved overnight, and then treated with EGF (100 ng/ml) for 5 h. Luciferase reporter activity was measured using the Promega Dual Glo kit.
Real-Time PCR
Total RNA was isolated from cells using an RNeasy Mini kit (Qiagen). cDNAs were synthesized with 0.3–1 μg RNA using the first-strand cDNA synthesis kit (Invitrogen). The SYBR green qPCR assay kit (Applied Biosystems) was used with 5 μl of 20-fold diluted cDNA from each sample, and the samples were amplified with the ABI Prism 7300 machine (Applied Biosystems).
Coimmunoprecipitation experiments
MCF-10A cells were treated with EGF and lysed for 30 min on ice. Cell lysates were incubated with control IgG or anti-EGFR overnight at 4°C with agitation. Protein G-Sepharose beads (GE Health Care Life Sciences) were added and the cell lysates were incubated for another 2 h. Immune complexes were collected, washed in lysis buffer and subjected to immunoblotting with anti-MUC1-C and anti-EGFR.
Confocal microscopy
MCF-10A cells were seeded onto a 6-well plate with sterile cover slides. After overnight serum starvation, cells were treated with EGF for 24 h. Cells were fixed in 100% acetone, blocked with 5% milk in PBS and stained with an anti-EGFR monoclonal antibody (Santa Cruz Biotechnology), anti-MUC1-C (Neomarkers) and Hoechst 33258 (Invitrogen). The cover slides were mounted onto microscope slides using Prolong Gold antifade reagent (Invitrogen) and imaged by confocal microscopy.
Assessment of cell cycle distribution
Cells were fixed in ice-cold 100% ethanol overnight, washed with PBS, incubated with 100 μg/ml RNase for 30 min at 37°C, stained with 10 μg/ml propidium iodide at room temperature for 30 min, and analyzed by flow cytometry.
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Institute Grants CA97098, CA42802 and CA100707; the National Institutes of Health Grant GM-073855 (J.A.P., Jr.), and a postdoctoral fellowship from the American Cancer Society PF-11-077-01-CDD (C.M.R.).
Abbreviations
- MUC1
mucin 1
- MUC1-C
MUC1 C-terminal subunit
- MUC1-CD
MUC1 cytoplasmic domain
- EGF
epidermal growth factor
- HRG
heregulin
- mTORC1
mammalian target of rapamycin complex 1
- S6K1
ribosomal protein S6 kinase 1
- CHX
cycloheximide
- PDCD4
tumor suppressor programmed cell death protein 4
Footnotes
D.K. holds equity in Genus Oncology and is a consultant to the company. The other authors disclosed no potential conflicts of interest.
References
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 3.Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 5.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 6.Rogers GW, Jr, Komar AA, Merrick WC. eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 7.Blagden SP, Willis AE. The biological and therapeutic relevance of mRNA translation in cancer. Nat Rev Clin Oncol. 2011;8:280–291. doi: 10.1038/nrclinonc.2011.16. [DOI] [PubMed] [Google Scholar]
- 8.Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest. 2008;118:2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas DM, Edwards RB, Lozanski G, West DA, Shin JD, Vargo MA, et al. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood. 2009;113:4656–4666. doi: 10.1182/blood-2008-09-175430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med. 2011;208:1799–1807. doi: 10.1084/jem.20110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kufe D. Mucins in cancer: function, prognosis and therapy. Nature Reviews Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramasamy S, Duraisamy S, Barbashov S, Kawano T, Kharbanda S, Kufe D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell. 2007;27:992–1004. doi: 10.1016/j.molcel.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroeder J, Thompson M, Gardner M, Gendler S. Transgenic MUC1 interacts with epidermal growth factor receptor and correlates with mitogen-activated protein kinase activation in the mouse mammary gland. J Biol Chem. 2001;276:13057–13064. doi: 10.1074/jbc.M011248200. [DOI] [PubMed] [Google Scholar]
- 14.Schroeder JA, Masri AA, Adriance MC, Tessier JC, Kotlarczyk KL, Thompson MC, et al. MUC1 overexpression results in mammary gland tumorigenesis and prolonged alveolar differentiation. Oncogene. 2004;23:5739–47. doi: 10.1038/sj.onc.1207713. [DOI] [PubMed] [Google Scholar]
- 15.Pochampalli MR, el Bejjani RM, Schroeder JA. MUC1 is a novel regulator of ErbB1 receptor trafficking. Oncogene. 2007;26:1693–1701. doi: 10.1038/sj.onc.1209976. [DOI] [PubMed] [Google Scholar]
- 16.Raina D, Kosugi M, Ahmad R, Panchamoorthy G, Rajabi H, Alam M, et al. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol Cancer Therapeutics. 2011;10:806–816. doi: 10.1158/1535-7163.MCT-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raina D, Kharbanda S, Kufe D. The MUC1 oncoprotein activates the anti-apoptotic PI3K/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J Biol Chem. 2004;279:20607–20612. doi: 10.1074/jbc.M310538200. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks GSK3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 20.Yin L, Kufe D. Human MUC1 carcinoma antigen regulates intracellular oxidant levels and the apoptotic response to oxidative stress. J Biol Chem. 2003;278:35458–35464. doi: 10.1074/jbc.M301987200. [DOI] [PubMed] [Google Scholar]
- 21.Ren J, Agata N, Chen D, Li Y, Yu W-H, Huang L, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anti-cancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin L, Kharbanda S, Kufe D. Mucin 1 oncoprotein blocks hypoxia-inducible factor 1 alpha activation in a survival response to hypoxia. J Biol Chem. 2007;282:257–266. doi: 10.1074/jbc.M610156200. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad R, Raina D, Joshi MD, Kawano T, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the NF-κB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad R, Rajabi H, Kosugi M, Joshi M, Alam M, Vasir B, et al. MUC1-C oncoprotein promotes STAT3 activation in an auto-inductive regulatory loop. Science Signaling. 2011;4:ra9. doi: 10.1126/scisignal.2001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raina D, Ahmad R, Joshi M, Yin L, Wu Z, Kawano T, et al. Direct targeting of the MUC1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res. 2009;69:5133–5141. doi: 10.1158/0008-5472.CAN-09-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi MD, Ahmad R, Raina D, Rajabi H, Bubley G, Kharbanda S, et al. MUC1 oncoprotein is a druggable target in human prostate cancer cells. Mol Cancer Ther. 2009;8:3056–3065. doi: 10.1158/1535-7163.MCT-09-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Rajabi H, Kufe D. MUC1-C oncoprotein is a target for small molecule inhibitors. Mol Pharm. 2011;79:886–893. doi: 10.1124/mol.110.070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton R, Cambridge L, Hart LA, Stevens DA, Lindsay MA, Barnes PJ. The MAP kinase inhibitors, PD098059, UO126 and SB203580, inhibit IL-1beta-dependent PGE(2) release via mechanistically distinct processes. Br J Pharmacol. 2000;130:1353–1361. doi: 10.1038/sj.bjp.0703431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Workman P, Clarke PA, Raynaud FI, van Montfort RL. Drugging the PI3 kinome: from chemical tools to drugs in the clinic. Cancer Res. 2010;70:2146–2157. doi: 10.1158/0008-5472.CAN-09-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong D, Yamori T. Advances in development of phosphatidylinositol 3-kinase inhibitors. Curr Med Chem. 2009;16:2839–2854. doi: 10.2174/092986709788803222. [DOI] [PubMed] [Google Scholar]
- 31.Palamarchuk A, Efanov A, Maximov V, Aqeilan RI, Croce CM, Pekarsky Y. Akt phosphorylates and regulates Pdcd4 tumor suppressor protein. Cancer Res. 2005;65:11282–11286. doi: 10.1158/0008-5472.CAN-05-3469. [DOI] [PubMed] [Google Scholar]
- 32.Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005;65:6034–6041. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 33.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, et al. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterman H, Yarden Y. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 2001;490:142–152. doi: 10.1016/s0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]
- 35.Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci. 2004;24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodrigo C, Cencic R, Roche S, Pelletier J, Porco J., Jr Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: synthetic and biological studies. J Med Chem. 2012;55:558–562. doi: 10.1021/jm201263k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vermeer PD, Einwalter LA, Moninger TO, Rokhlina T, Kern JA, Zabner J, et al. Segregation of receptor and ligand regulates activation of epithelial growth factor receptor. Nature. 2003;422:322–326. doi: 10.1038/nature01440. [DOI] [PubMed] [Google Scholar]
- 38.Hsieh AC, Truitt ML, Ruggero D. Oncogenic AKTivation of translation as a therapeutic target. Br J Cancer. 2011;105:329–336. doi: 10.1038/bjc.2011.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Ren J, Yu W, Li G, Kuwahara H, Yin L, et al. The EGF receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and β-catenin. J Biol Chem. 2001;276:35239–35242. doi: 10.1074/jbc.C100359200. [DOI] [PubMed] [Google Scholar]
- 40.Wedeken L, Singh P, Klempnauer KH. Tumor suppressor protein Pdcd4 inhibits translation of p53 mRNA. J Biol Chem. 2011;286:42855–42862. doi: 10.1074/jbc.M111.269456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, et al. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 42.Hwang BY, Su BN, Chai H, Mi Q, Kardono LB, Afriastini JJ, et al. Silvestrol and episilvestrol, potential anticancer rocaglate derivatives from Aglaia silvestris. J Org Chem. 2004;69:3350–3358. doi: 10.1021/jo040120f. [DOI] [PubMed] [Google Scholar]
- 43.Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One. 2009;4:e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerard B, Cencic R, Pelletier J, Porco JA., Jr Enantioselective synthesis of the complex rocaglate (−)-silvestrol. Angew Chem Int Ed Engl. 2007;46:7831–7834. doi: 10.1002/anie.200702707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.