Summary
The advent of methods for mapping the location of specific proteins across genomes is substantially enlightening our understanding of gene regulation. One recent discovery is that Pol II is concentrated at the 5’ end of thousands of genes in mammalian and Drosophila cells. Prior to this, much research had focused on understanding how sequence-specific, DNA-binding proteins orchestrate the actions of regulators of chromatin structure and the general transcriptional machinery to control transcription initiation. The concentration of Pol II at the 5’ ends of genes indicates that key steps regulating transcription occur after Pol II has associated with a gene’s promoter.
Introduction
Nuclear run-on assays and in vivo crosslinking analyses first revealed that Pol II was concentrated at the 5’ end of a handful of genes including hsp70 in Drosophila, and c-myc and c-fos in mammals [1,2]. Notably, crosslinking detected the Pol II but the nuclear run-on analysis established that the Pol II was transcriptionally engaged [3,4]. High-resolution analyses of the nuclear run-on products of the hsp70 and c-fos genes revealed that Pol II pauses in the region 20 to 40 nucleotides downstream from the transcription start site. Permanganate genomic footprinting, which maps the location of transcription bubbles associated with elongation complexes in living cells, corroborated the results of the run-on assays.
Biochemical experiments primarily in the laboratories of Handa and Price have provided a framework for understanding the behavior of Pol II in the promoter proximal region [5]. Both groups were investigating the mechanism by which a nucleoside analog called DRB inhibits transcription elongation. By fractionating nuclear extracts and assaying for DRB sensitive transcription, Handa’s laboratory discovered two proteins called DSIF and NELF that inhibit elongation [6,7] and Price’s laboratory discovered a protein called P-TEFb that promotes elongation [8] (Fig. 1). In nuclear extracts, DSIF and NELF collaborate to inhibit transcription elongation after Pol II transcribes approximately 30 nucleotides [7]. Recent results show that the distance between the start site and the paused Pol II could be dictated by length of transcript needed to cause the association of DSIF with the elongation complex [9,10]. P-TEFb is a kinase that alleviates inhibition by NELF and DSIF. In a defined transcription system, optimal reversal of inhibition by NELF and DSIF is achieved by phosphorylating an intact elongation complex containing Pol II, DSIF, and NELF [11,12].
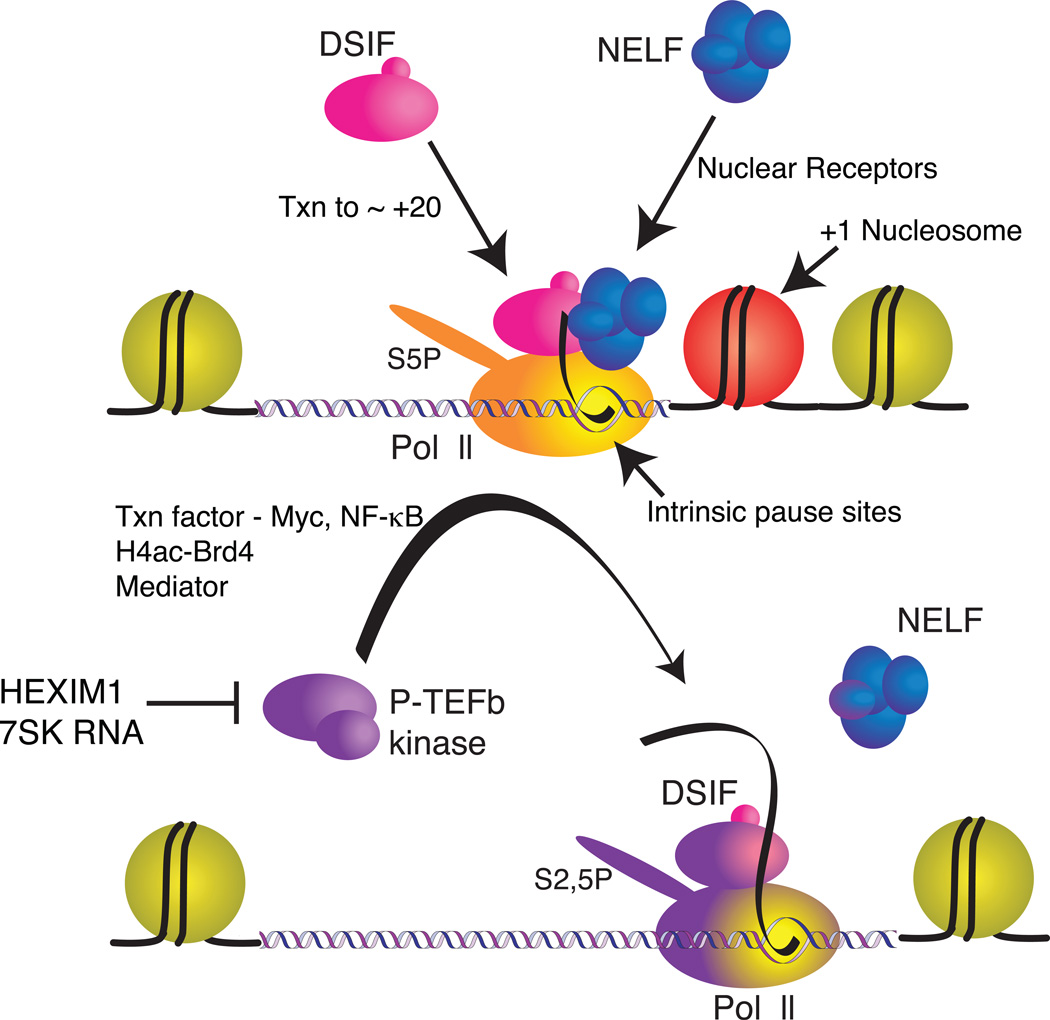
Figure 1.
Interactions impacting promoter proximal pausing. DSIF and NELF provide the basis for promoter proximal pausing [45]. NELF’s association with the elongation complex is dependent on DSIF, and DSIF’s association requires the Pol II transcribe approximately 20 nucleotides [9,10]. NELF’s recruitment to particular promoters can also be facilitated by sequence-specific, DNA-binding proteins such as nuclear hormone receptors [17,18]. Nucleic acid sequences affecting the stability of the RNA-DNA heteroduplex in the transcription bubble may also influence where Pol II pauses [38]. The +1 nucleosome may also contribute to the pause [24]. P-TEFb is a kinase that phosphorylates DSIF, NELF, and the CTD of Pol II, and appears to reactivate the paused Pol II [46]. P-TEFb activity is repressed by the 7SK RNA/HEXIM1 complex in mammals[t4] [47]. P-TEFb’s recruitment to a gene may occur through association with DNA binding proteins such as c-Myc [13] and NF-κB [14], association with Brd4 that is in turn associated with acetylated tails of histone H4 [15], or mediator [16].
Much attention has been directed at understanding how P-TEFb’s activity is regulated at genes as this is thought to influence the reactivation of the paused Pol II as well as contribute to the processivity of the Pol II and to transcription-coupled RNA processing. Multiple mechanisms appear to exist for recruitment of P-TEFb to genes including direct association with DNA binding proteins such as Myc and NF-κB [13,14], indirect association with acetylated histones via Brd4 [15], and association with the mediator [16](Fig. 1). While P-TEFb appears to trigger the dissociation of NELF from the elongation complex, it is unclear to what extent factors control the initial association of NELF or DSIF with the elongation complex. The finding that NELF associates with the estrogen receptor and other nuclear receptors, and that this association functions to attenuate the level of induction of target genes indicates that NELF may be actively recruited by DNA binding proteins to the elongation complex [17,18].
Functions of promoter associated Pol II
Given that NELF and DSIF inhibit transcription elongation in vitro, one might have anticipated that promoter proximal pausing represses gene expression and, indeed, this appears to be true for many genes. Several estrogen-regulated genes were found to be upregulated upon RNAi-mediated depletion of NELF [19] and junB and c-fos behaved similarly [20]. However, genome-wide expression analysis revealed that more genes decrease than increase expression in NELF-depleted cells suggesting that pausing plays a positive role in gene expression [t1][21,22]. In accordance with this conclusion, almost half of the genes expressed above the 90th percentile level in Drosophila are associated with NELF [23].
One way in which pausing appears to positively influence gene expression is by maintaining the accessibility of the promoter region. Genome-wide mapping of nucleosomes in Drosophila revealed that the first nucleosome downstream from the transcription start is often located adjacent to paused Pol II and that the core promoter is nucleosome-free [24]. Depletion of NELF from Drosophila cells resulted in the appearance of a nucleosome over the promoters of the TepII and Tl genes, which also exhibited reduced expression upon depletion of NELF [21]. In the case of a human breast cancer cell line, the depletion of NELF resulted in the loss of histone modifications typically associated with active transcription [22]. Thus, the presence of paused Pol II could function to prevent formation of repressive chromatin both by excluding nucleosomes from the promoter region and by maintaining patterns of histone modification that are conducive to activation.
It has been proposed that pausing of Pol II in the promoter proximal region serves as a checkpoint for coupling transcription and mRNA processing, and for delaying the elongation complex until modifications occur that increase the processivity of Pol II [25]. These hypotheses clearly require more rigorous testing in vivo. DSIF and serine 5 phosphorylation of the CTD of Pol II stimulate mRNA capping but there is no in vivo data demonstrating that disruption of pausing results in aberrant capping [20]. NELF has been shown to be involved in 3’ end processing of the nonpolyadenylated histone RNAs [26] and to influence splicing [18], but it is unclear if these activities are connected to promoter proximal pausing. It is also unclear whether promoter proximal pausing is necessary for establishing the transcriptional competency of Pol II in vivo. To address these issues, one needs to disrupt pausing and monitor the behavior of Pol II during activation.
Promoter proximal pausing allows the process of recruiting Pol II to a promoter to be uncoupled from transcription of a gene, thus allowing genes to be potentiated for rapid induction in response to external stimuli. The heat shock genes are excellent examples since these genes are fully induced within minutes of a heat shock [27]. Paused Pol II is also detected on genes that respond rapidly to LPS and estrogen [15,19,28]. However, the connection between pausing and rapid induction remains correlative and has not been directly tested by demonstrating that disruption of pausing slows the induction rate of a gene. For example, the depletion of NELF in the case of estrogen responsive genes results in precocious expression in the absence of hormone [19], and the kinetics of junB induction in response to IL6 remained unchanged following depletion of NELF and loss of paused Pol II [20].
Several recent studies have linked pausing to developmental processes. Knockout of a NELF subunit, which would presumably impair promoter proximal pausing, results in embryonic lethality in mice and Drosophila [29,30]. Many developmental control genes in Drosophila have paused Pol II at their promoters [31,32]. An analysis of the spatial and temporal pattern of transcription of specific genes in the Drosophila embryo revealed that genes with paused Pol II are synchronously induced in fields of nuclei [33]. In contrast, those genes without paused Pol II tend to be induced with significantly less synchrony. The Drosophila embryo is a special case because the time in development at which these analyses were done was at the late syncitial blastoderm stage, just as cellularization was beginning. This situation does not exist in developing mammals. In addition, it remains to be demonstrated that perturbing pausing affects the synchrony of expression.
An intriguing activity to be linked to the paused Pol II is enhancer blocking activity [34,35]. Levine and colleagues noted that the genes flanking the bithorax and antennapedia complexes of Drosophila have paused Pol II while the genes within the loci do not. They hypothesized that the paused Pol II might insulate the genes within the developmental loci from the effects of enhancers in flanking regions of the chromosome. When the paused Pol II was positioned between an enhancer and a target promoter, it blocked enhancer activity. Although pausing was not directly measured on the transgenes, it was inferred from the finding that embryos heterozygous for NELF or DSIF mutations impaired insulator activity. Intriguingly, the NELF mutation also impaired the ability of the Fab7 and Fab8 insulator elements to block enhancer action. Since paused Pol II is not known to be associated with the Fab7 and Fab8 insulators, it was proposed that these insulator elements might associate with paused Pol II to regulate the targeting of enhancers. Consistent with such a model is the recent finding that an insulator-associated protein called BEAF is commonly located near paused Pol II [36]. Perhaps paused Pol II can function in establishing higher order structure of the chromatin fiber. It will be interesting to see if perturbing pausing alters patterns of chromosome looping and the targeting of enhancer function for endogenous loci.
The state of Pol II at the 5’ ends of genes in Drosophila and mammalian cells
Chromatin immunoprecipitation (ChIP) analyses of individual genes and genome-wide clearly establish that Pol II tends to concentrate at the 5’ ends of genes. However, ChIP is unable to assess the transcriptional status of the Pol II, so one cannot know definitively from these analyses alone whether the Pol II is in a preinitiated or transcriptionally engaged state. Currently, nuclear run-on and permanganate genomic footprinting are the only techniques that directly monitor transcriptionally engaged Pol II. Lis and colleagues recently developed a nuclear run-on technique that queries the distribution of transcriptionally engaged Pol II throughout the genome [37]. This technique, called GRO-seq, incorporates bromouridine into nuclear run-on products. The bromouridinated RNA products are isolated and analyzed by massively parallel sequencing. Not only does the method identify transcriptionally engaged Pol II, which is a prerequisite for run-on, but it also reveals the direction of transcription. Transcriptionally engaged Pol II is detected in the promoter proximal region of thousands of genes in human cells. Unexpectedly, many of these genes also have Pol II transcribing in the opposite direction, upstream from the gene’s start site. The significance of this antisense transcription is not known.
Other methods for assessing the behavior of Pol II also provide evidence that the Pol II concentrated in the promoter proximal region is transcriptionally engaged. Sequencing of 5’ capped RNAs in the size range from 25 to 120 nucleotides, and sequencing of short uncapped RNAs indicate that Pol II is at some point transcriptionally engaged in the promoter proximal region [38,39]. Importantly, neither of these analyses distinguishes whether these RNAs are nascent transcripts associated with engaged Pol II or products of premature termination or RNA degradation. For example, for a collection of promoters that have similar levels of cross-linked Pol II in a ChIP-seq analysis, the corresponding amounts of 5’ capped RNAs can range at least 10-fold, raising the possibility that transcripts and Pol II are dissociating from the DNA (38). The small uncapped RNAs known as tiny RNAs or tiRNAs are intriguing and could be hinting at a possible fate of some of the Pol II. It was proposed that tiRNAs might be the products of TFIIS-induced cleavage of backtracked Pol II [40]. However, these tiRNAs are typically 18 nucleotides in size, and there is no data indicating that TFIIS-induced cleavage occurs at 18 nucleotide intervals. Instead, 18 nucleotides is the length of RNA associated with an elongation complex that is protected from ribonucleolytic attack [41]. Hence, it is possible that these tiRNAs are the products of nucleolytic attack of the nascent transcripts in vivo. This would suggest that some paused Pol II might abort from the template rather than resuming transcription. [t2]
The phosphorylation state of the carboxyl-terminal domain of the largest subunit of Pol II has been used to infer the transcriptional status of Pol II. Serine 5 phosphorylation of the CTD is an indication that TFIIH has acted on the Pol II and that Pol II has initiated transcription [25]. Hence, the absence of serine 5 phosphorylation has been taken as an indication that Pol II is in a preinitiated state. By this criterion, several investigators have identified cases where Pol II is deemed to be in a preinitiation complex [19,42,43]. Byun et al. [43] have proposed that Pol II in a preinitiation complex bookmarks genes like c-fos for rapid induction. In contrast, others have shown by permanganate footprinting and nuclear run-on that the Pol II associated with c-fos is in a transcriptionally engaged, but paused state [20]. The recent finding that serine 5 phosphorylation is not required for Pol II to initiate transcription in yeast serves as a precaution against relying solely on this marker to assess the state of Pol II [44[t3]]. The inconsistencies over the status of Pol II at the c-fos promoter serve to highlight a weakness in our current methods for evaluating the status of the Pol II. No one method allows quantification of the proportions of Pol II in the preinitiated and the paused states.
Conclusion
The discovery that Pol II is concentrated at the 5’ ends of many genes in metazoans has directed much interest towards understanding the control of Pol II after it initiates transcription. Promoter proximal pausing mediated by NELF and DSIF represents as early stage in the control of transcription elongation, and cells appear to use this pausing in different ways to either positively or negatively regulate gene expression. The wide-spread application of chromatin immunoprecipitation has made it relative simple to determine if Pol II is concentrated at the promoter of a particular gene but nuclear run-on and permanganate footprinting assays are required to assess whether these Pol II are pausing in the promoter proximal regions of genes. In lieu of this information, it remains possible that regulation is targeting the behavior of the Pol II in a preinitiation complex, or the rate of elongation and processivity of the Pol II as it advances beyond the promoter proximal region and into the body of the gene.
Acknowledgements
This work was supported by National Institutes of Health Grant GM047477 to D.S.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- 2. Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. This review describes how the level of paused Pol II detected at the 5' end of the gene will depend on the rates of initiation and release of the paused Pol II. Recognizing this feature of the transcription process is key to realizing that promoter proximal pausing is not solely involved in gene repression.
- 3.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 4.Gilmour DS, Lis JT. RNA Polymerase II Interacts With the Promoter Region of the Noninduced hsp70 Gene in Drosophila melanogaster Cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 8.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 9.Cheng B, Price DH. Analysis of factor interactions with RNA polymerase II elongation complexes using a new electrophoretic mobility shift assay. Nucleic Acids Res. 2008;36:e135. doi: 10.1093/nar/gkn630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A. 2010;107:11301–11306. doi: 10.1073/pnas.1000681107. Using purified proteins, gel-shift assays were used to show that the association of NELF with the elongation complex depends on DSIF, and that the association of DSIF with the elongation complex requires Pol II to transcribe greater than 18 nucleotides.
- 11.Cheng B, Price DH. Properties of RNA Polymerase II Elongation Complexes Before and After the P-TEFb-mediated Transition into Productive Elongation. J Biol Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 12.Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. A highly purified RNA polymerase II elongation control system. J Biol Chem. 2001;276:42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- 13.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 15.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aiyar SE, Sun JL, Blair AL, Moskaluk CA, Lu YZ, Ye QN, Yamaguchi Y, Mukherjee A, Ren DM, Handa H, et al. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, Blair AL, Aiyar SE, Li R. Cofactor of BRCA1 modulates androgen-dependent transcription and alternative splicing. J Steroid Biochem Mol Biol. 2007;107:131–139. doi: 10.1016/j.jsbmb.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kininis M, Isaacs GD, Core LJ, Hah N, Kraus WL. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol Cell Biol. 2009;29:1123–1133. doi: 10.1128/MCB.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gilchrist DA, Nechaev S, Lee C, Ghosh SKB, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008:1921–1933. doi: 10.1101/gad.1643208. The researchers show that RNAi-mediated depletion of NELF from Drosophila cells results in the appearance of a nucleosome over the core promoter of genes. This provides an explanation for how a negative elongation factor positively regulates gene expression.
- 22.Sun J, Li R. Human negative elongation factor activates transcription and regulates alternative transcription initiation. J Biol Chem. 2010;285:6443–6452. doi: 10.1074/jbc.M109.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008 doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 26.Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF interacts with CBC and participates in 3' end processing of replication-dependent histone mRNAs. Mol Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci U S A. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amleh A, Nair SJ, Sun J, Sutherland A, Hasty P, Li R. Mouse cofactor of BRCA1 (Cobra1) is required for early embryogenesis. PLoS ONE. 2009;4:e5034. doi: 10.1371/journal.pone.0005034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Hang S, Prazak L, Gergen JP. NELF potentiates gene transcription in the Drosophila embryo. PLoS One. 2010;5:e11498. doi: 10.1371/journal.pone.0011498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Lee C, Gilmour DS, Gergen JP. Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev. 2007;21:1031–1036. doi: 10.1101/gad.1521207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. Using a sophisticated imaging analysis technique that monitors nascent transcripts in nuclei of Drosophila embryos, the researchers show that genes with paused Pol II tend to be synchronously expressed within a 5 minute period whereas genes lacking paused Pol II tend to be expressed asynchronously over a 30 minute period.
- 34. Chopra VS, Cande J, Hong JW, Levine M. Stalled Hox promoters as chromosomal boundaries. Genes Dev. 2009;23:1505–1509. doi: 10.1101/gad.1807309. This study provides evidence for an unanticipated possible role of paused Pol II in organizing higher order chromatin structure. Reference 35 provides an excellent discussion of the implications of this finding.
- 35.Core LJ, Lis JT. Paused Pol II captures enhancer activity and acts as a potent insulator. Genes Dev. 2009;23:1606–1612. doi: 10.1101/gad.1827709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang N, Emberly E, Cuvier O, Hart CM. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol Cell Biol. 2009;29:3556–3568. doi: 10.1128/MCB.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. The researchers have developed a method for analyzing the nuclear runon products genome wide and provide definitive evidence that Pol II pauses in the promoter proximal region of thousands of genes in human cells.
- 38. Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. By comparing short, capped transcripts in in control and TFIIS-depleted Drosophila cells, the authors propose that the position where Pol II pauses is strongly influenced by the intrinsic stability of the RNA heteroduplex associated with the active site of the elongation complex.
- 39.Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, Holst J, Ritchie W, Wong JJ, Rasko JE, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1841. [DOI] [PubMed] [Google Scholar]
- 40.Taft RJ, Kaplan CD, Simons C, Mattick JS. Evolution, biogenesis and function of promoter-associated RNAs. Cell Cycle. 2009;8:2332–2338. doi: 10.4161/cc.8.15.9154. [DOI] [PubMed] [Google Scholar]
- 41.Gu W, Wind M, Reines D. Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. Proc Natl Acad Sci U S A. 1996;93:6935–6940. doi: 10.1073/pnas.93.14.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, Wu SY, Chiang CM, Karn J. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. Embo J. 2006;25:3596–3604. doi: 10.1038/sj.emboj.7601248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Byun JS, Wong MM, Cui W, Idelman G, Li Q, De Siervi A, Bilke S, Haggerty CM, Player A, Wang YH, et al. Dynamic bookmarking of primary response genes by p300 and RNA polymerase II complexes. Proc Natl Acad Sci U S A. 2009;106:19286–19291. doi: 10.1073/pnas.0905469106. The researchers provide evidence the Pol II concentrated at the promoter of c-fos is in a preinitiated state under certain cell culture conditions.
- 44.Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, et al. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol. 2010;17:1154–1161. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiba K, Yamamoto J, Yamaguchi Y, Handa H. Promoter-proximal pausing and its release: Molecular mechanisms and physiological functions. Exp Cell Res. 2010 doi: 10.1016/j.yexcr.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 46.Price DH. Poised polymerases: on your mark…get set…go! Mol Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]