Abstract
Metagenomic shotgun sequencing data can identify microbes populating a microbial community and their proportions, but existing taxonomic profiling methods are inefficient for increasingly large datasets. We present an approach that uses clade-specific marker genes to unambiguously assign reads to microbial clades more accurately and >50× faster than current approaches. We validated MetaPhlAn on terabases of short reads and provide the largest metagenomic profiling to date of the human gut
Microbial communities are responsible for a broad spectrum of biological activities carried out in virtually all natural environments including oceans1, soil2 and human-associated habitats3–5. Profiling the taxonomic and phylogenetic compositions of such communities is critical for understanding their biology and characterizing complex disorders like inflammatory bowel diseases4, 6 and obesity7 that do not appear to be associated with any individual microbes.
Metagenomic shotgun sequencing provides a uniquely rich profile of microbial communities, each dataset yielding billions of short reads sampled from the DNA in the community. A community's taxonomic composition can be estimated from such data by assigning each read to the most plausible microbial lineage, often with a taxonomic resolution not achievable by profiling the universal 16S rRNA marker gene alone. Both alignment- and composition-based approaches have been developed for this task, and the two approaches have also been integrated in hybrid methods (see Methods). However, none have simultaneously achieved both the efficiency and the species-level accuracy required by current highly-complexity datasets due to computational limitations, untenable accuracy for short (<400 nt) reads, and the need to normalize read counts into clade-specific relative abundances (Supplementary Note 1).
We thus present MetaPhlAn (Metagenomic Phylogenetic Analysis), a tool that accurately profiles microbial communities and requires only minutes to process millions of metagenomic reads. MetaPhlAn estimates the relative abundance of microbial cells by mapping reads against a reduced set of clade-specific marker sequences that are computationally pre-selected from coding sequences that unequivocally identify specific microbial clades at the species or higher taxonomic levels and cover all main functional categories (Supplementary Fig. 1). Starting from the 2,887 genomes available from the Integrated Microbial Genomes (IMG) system (July 2011)8, we identified more than 2 million potential markers from which we selected a subset of 400,141 genes most representative of each taxonomic unit (Online Methods). The resulting catalog spans 1,221 species with 231 (standard deviation 107) markers per species and >115,000 markers at higher taxonomic levels (available at http://huttenhower.sph.harvard.edu/metaphlan).
The MetaPhlAn classifier compares each metagenomic read from a sample to this marker catalog to identify high-confidence matches. This can be done very efficiently, as the catalog contains only ~4% of sequenced microbial genes, and each read of interest has at most one match due to the markers' uniqueness. Since spurious reads are very unlikely to have significant matches with a marker sequence, no pre-processing of metagenomic DNA (for example error detection, assembly, or gene annotation) is required. The classifier normalizes the total number of reads in each clade by the nucleotide length of its markers and provides the relative abundance of each taxonomic unit, taking into account any markers specific to subclades. Microbial reads belonging to clades with no sequenced genomes available are reported as an “unclassified” subclade of the closest ancestor with available sequence data.
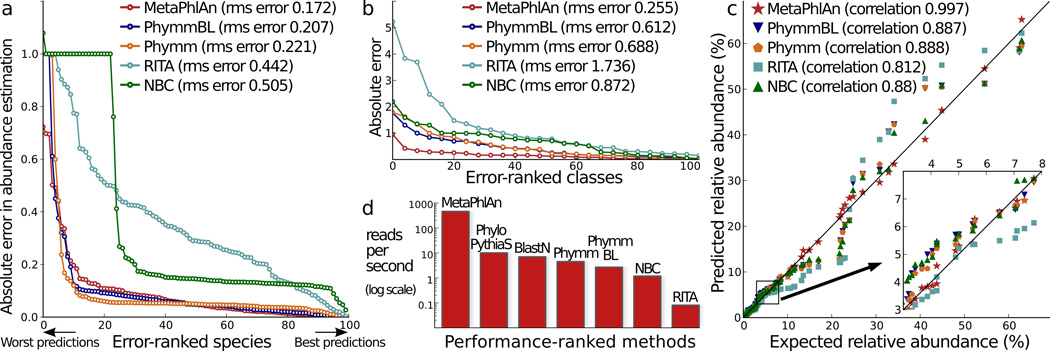
We first evaluated MetaPhlAn's performance in estimating microbial community composition using synthetic data. We constructed ten datasets comprising 4 million noisy reads from 300 organisms. MetaPhlAn mapped a number of reads consistent with the fraction of lineage-specific genomic regions (7.7% and 8.4%), correctly identified all 200 organisms in the two high-complexity datasets, and accurately estimated their species relative abundances (RMSE of 0.17 and 0.14), with 75% of species within 10% deviation from expected value (Fig. 1a and Supplementary Fig. 2). Similar performance was observed for higher taxonomic ranks (Fig. 1b and Supplementary Figs. 2 and 3) and for the eight non-evenly distributed datasets that better mimic the abundance distributions of real communities (Pearson r >0.991, species-level Pearson P <2×10−21) (Fig. 1c and Supplementary Fig. 4).
Fig. 1. Comparison of MetaPhlAn to existing methods.
We used 10 total synthetic metagenomes to compare MetaPhlAn to PhymmBL12, BLAST13, theRITA pipeline14, and NBC15. a–b) Absolute and root mean squared (rms) errors with respect to 100 total organisms in one synthetic metagenome at the species (a) and class (b) level c) Correlations of inferred and true species abundances on 8 non-evenly distributed synthetic metagenomes;. d) Reads-per-second rates for the tested methods on single CPUs.
In contrast to all existing methods, which are optimized for read-based statistics rather than microbial cell relative abundance estimation, we considered the latter more biologically informative. Microbial clade abundances were thus estimated by normalizing read-based counts by the average genome size of each clade. MetaPhlAn compares favorably to existing methods on all tested synthetic metagenomes (Fig. 1 and Supplementary Figs. 2–4), with PhymmBL being the closest (but substantially slower) alternative. This also held true for the more challenging scenario in which metagenomes contained microbes without reference genomes (Supplementary Tables 1 and 2).
Notably, MetaPhlAn achieved a classification rate of about 450 reads-per-second on standard single-processor systems, thus greatly outperforming all existing methods (Fig. 2d; note that PhyloPythiaS provides only genus-level predictions). This allowed us to provide the first practical high-throughput assessments of several real-world metagenomes at the species-level as detailed below.
Fig. 2. Composition of healthy vaginal microbiota.
MetaPhlAn species and genus abundances and 16S phylotype abundances for 51 healthy vaginal microbiomes from the Human Microbiome Project. Samples were naively grouped by assigning each based on its dominant (>50%) Lactobacillus species or by the absence (<2%) of any Lactobacillus. For each cluster (named from I to V5) we report averages across samples for all genera and species as inferred by MetaPhlAn and, for genera, as estimated by the combination of mothur and the RDP classifier (see Methods) applied to 16S rRNA gene sequences from the same specimen.
We first characterized the vaginal microbiota of asymptomatic pre-menopausal adult women enrolled in the Human Microbiome Project (HMP)3, analyzing 51 metagenomes sampled from the posterior fornix. MetaPhlAn detected 98 clades with abundances >0.5% in at least one sample (32 species from 17 genera). Lactobacillus was consistently the most abundant genus, representing >50% of the bacterial community in 49 of the 51 samples and >98% in half of the samples, confirming its well-established role in healthy vaginal microbiomes5, 9. 16S rRNA gene sequencing in an independent cohort5 previously identified five distinct community types, each characterized by a specific Lactobacillus species or by the absence of any of them; in MetaPhlAn's results, these five groups are easily identifiable (Supplementary Fig. 5) and cluster naturally by species (Fig. 2). Although in 16S pyrosequencing data the characterization of Lactobacillus species-level operational taxonomic units is sensitive to the region being sequenced, we performed a direct comparison with 16S data from the same HMP specimens. Despite extensive technical differences between 16S pyrosequencing and shotgun sequencing, the estimated relative abundances were remarkably similar in all clusters. Moreover, MetaPhlAn's native coverage of all species with sequenced members further details the structure of these clusters. For example, Lactobacillus is not the only genus with species-specific differences among these microbiome types; Bifidobacterium species are present in cluster II as B. breve and B. dentium, whereas in cluster IV we identified an unclassified member distinct from all sequenced species. Similarly, Prevotella is represented by P. multiformis in cluster II in contrast to P. amnii and P. timonensis in cluster IV.
MetaPhlAn's methodology is restricted neither to bacteria alone nor to human-associated microbiomes, and it allowed us to investigate the microbial flora collected from oxygen minimum zones at intermediate depths in the Eastern Tropical South Pacific10. This marine ecosystem proved to include a substantial fraction of archaea, but Proteobacteria (mainly Alphaproteobacteria) was the most abundant phylum, representing approximately half of the community. Depth-associated shifts were observable for Bacteroidia, Chlamydiae, and Gammaproteobacteria, whereas the Cenarchaea dropped off specifically within the deepest sample. MetaPhlAn’s relative abundances (Supplementary Fig. 6) were consistent with BLAST-based approaches and confirmed its applicability for communities with limited coverage of reference genomes and its suitability for environmental metagenomic samples. Moreover, as the number of sequenced organisms continues to increase, MetaPhlAn's species-specificity in such environments will automatically improve without computational drawbacks.
MetaPhlAn’s read-based estimation of relative abundances enabled straightforward integration of multiple cohorts sequenced with different technologies and depths; specifically, to comprehensively characterize the asymptomatic human gut microbiota, we combined 224 fecal samples (>17 million reads) from the HMP3 and MetaHIT project4, the two largest gut metagenomic collections available. MetaPhlAn detected 102 species present at least once at >0.5% abundance (Fig. 3a) with strong consistency among different markers for the same clade (Supplementary Fig. 7). The MetaHIT project has previously characterized gut microbiomes as arising from three distinct and stable microbiome types (enterotypes11), and we investigated this hypothesis by hierarchically clustering the 224 samples separately at the genus and species levels (Fig. 3b,c). In some cases, enterotype-like discrete prevalence patterns were readily apparent, the genus Prevotella being the most striking example with Butyrivibrio showing similar behavior but for fewer samples. Conversely, many samples were characterized by high fractions of Bacteroides resembling Enterotype 111, but this genus’ relative abundance overall formed a continuum across samples, as did those of several other genera including Eubacterium and Alistipes.
Fig. 3. The gut microbiota in asymptomatic Western populations as inferred by MetaPhlAn on 224 samples combining the HMP and MetaHIT cohorts.
a) A taxonomic cladogram reporting all clades present in one or both cohorts (≥0.5% abundance in ≥1 sample). Circle size is proportional to the log of average abundance; color represents relative enrichment of the most abundant taxa (≥1% average in ≥1 cohort) between the HMP (139) and MetaHIT (85, healthy only) populations. b–c) Genus- (b) and species-level (c) taxonomic profiles of the most abundant clades hierarchically clustered (average linkage) with the Bray-Curtis similarity reveal sets of samples with similar microbial community compositions. With the exception of the cluster dominated by genus Bacteroides (B. vulgatus and B. ovatus in particular), samples from both studies are present in all groups, confirming substantial consistency of the gut microbiota characterized by independent and geographically distant western-diet asymptomatic cohorts. Only species and genera with at least 7.5% abundances at the 95th percentile of their distribution are reported.
MetaPhlAn's estimates of species-level abundance allowed us to refine this investigation (Fig. 4c). While Enterotype 2 remained clearly identifiable, the Bacteroides were diversified in a manner quite similar to lactobacilli in the vaginal microbiota, although with more species and less exclusive dominance. This suggests the existence of more complex community patterns than those captured by the proposed genus-level enterotypes (Supplementary Note 2). The integrated dataset also furnished the opportunity to investigate differences between independent and geographically unrelated healthy Western-diet populations (Fig. 4a and Supplementary Note 3). Overall, this analysis showed that MetaPhlAn is effective in processing very large metagenomic datasets with different short read lengths at high taxonomic resolution, enabling meta-analyses difficult to achieve using other technologies.
Shotgun metagenomic data are rapidly decreasing in cost to a per-sample level comparable to that of 16S gene surveys. Community-wide sequence reads already provide unique insights into gene function, metabolism, and polymorphisms that are unavailable from individual marker genes. By enabling efficient, high-resolution taxonomic profiling in such data, MetaPhlAn provides a further advantage with respect to 16S rRNA-based investigations, which can be difficult to extend past a genus level of resolution. Metagenomic sequencing further provides better statistical support (~108 reads/sample) than 16S pyrosequencing approaches (typically <104 reads/sample), and the sequencing protocols do not require potentially biased amplification steps. Finally, the MetaPhlAn database of clade-specific markers is constructed by a fully automated computational pipeline, which will allow improved accuracy as additional microbial genomes become available and improve support for gene markers' intra-clade universality and inter-clade uniqueness.
Online Methods
Implementation and availability
An open-source implementation of MetaPhlAn, the corresponding core gene catalog, and species-level results for the analyzed gut and vaginal metagenomes are available online at http://huttenhower.sph.harvard.edu/metaphlan. The taxonomic profiles of all the 691 HMP shotgun samples from 18 different body habitats can be accessed at http://hmpdacc.org/HMSMCP.
MetaPhlAn pipeline, database, and classifier overview
MetaPhlAn estimates microbial relative abundances by mapping metagenomic reads against a catalog of clade-specific marker sequences currently spanning the bacterial and archaeal phylogenies. Clades are groups of genomes (organisms) that can be as specific as species or as broad as phyla. Clade-specific markers are coding sequences (CDS) that satisfy the conditions of (i) being strongly conserved within the clade's genomes and (ii) not possessing substantial local similarity with any sequence outside the clade. The definition of such markers is to some extent sensitive to the availability of sequenced genomes, especially point (i), because a gene can be present in all available sequenced genomes in a clade but missing from some yet-to-be-sequenced strains. However, we did demonstrate in several experiments that the definition is effective even with only partial coverage of reference genomes (Supplementary Tables 1 and 2).
Thus, starting from the 2,887 genomes currently available from IMG (July 2011)8, we identified more than 2 million potential markers meeting this level of stringency and allowing for sequencing and annotation errors. We then selected the subset of 400,141 genes most representative of each taxonomic unit. This employed a two-step computational pipeline that performed an intra-clade CDS clustering followed by an extra-clade sequence uniqueness assessment, based loosely on our previous system for detecting core genes16. The resulting marker catalog spans 1,221 species with an average of 231 s.d. 107 markers per species and only 12 species (<1% of the total) with fewer than 15 markers. 9 of these are species in the Brucella genus, which is known to have high genotypic homogeneity and unclear taxonomy17. Even these rare cases in which species are not completely characterized by specific markers, however, are themselves well-covered at higher taxonomic levels. For example, the Brucella genus (as opposed to its constituent species) has 346 markers covering almost 300 k-nt; in total, 375 of 652 genera had >250 markers, as did 80 of 278 families and 22 of 130 orders. These markers were in addition to those for corresponding subclades, allowing MetaPhlAn to recover relative abundances within broader clades even in the absence of sequenced genomes for all organisms in a community. The generation of this catalog of marker genes is an offline procedure that we perform regularly as a relevant set of newly sequenced microbial genomes are available, and it is downloaded automatically with the associated classifier.
The MetaPhlAn classifier finally compares metagenomic reads against this precomputed marker catalog using nucleotide BLAST searches13 in order to provide clade abundances for one or more sequenced metagenomes. This achieves an approximately two order of magnitude speedup compared to applying BLAST to the full catalog of microbial genomes, due primarily to the reduced size of our reference catalog. In order to estimate each clade's relative abundance in terms of cell counts (i.e. whole-genome counts, rather than single-read counts), the MetaPhlAn classifier normalizes the total number of reads in each clade by the nucleotide length of its markers.
Genomic data sources and preprocessing
The input genomic data for training MetaPhlAn consists in a catalog of finished and draft genomes and the corresponding functionally unannotated coding sequences (CDS) calls. Specifically, the system automatically retrieves the nucleotide sequences of bacterial and archaeal genomes from the Integrated Microbial Genomes (IMG) system8, (2,887 total for this publication, 1,449 finished and 1,438 draft). These are screened for minimum length (>50,000 nt), minimum number of CDSs (>50), minimum percentage of coding genome (>75%), and taxonomic label not higher than the genus level. A total of 2,834 genomes (2,727 bacteria and 107 archaea) pass this quality control screening, and after a minimal manual curation of the corresponding taxonomy they span 2 domains, 33 phyla, 66 classes, 130 orders, 278 families, 652 genera, and 1,222 species for a total of 2,383 taxonomic clades. The raw nucleotide sequences, the CDS calls (list of gene or hypothetical gene start/end nucleotide positions), and the taxonomic classification of the genomes are the three only inputs for our system and can be retrieved from any available genomic database. The current version of MetaPhlAn is based on the genomic data contained in IMG version 3.4 (July 2011); different and future versions of the IMG database or other genomic databases can be used with minimal configuration changes in the pipeline.
Identification of clade-specific core genes
The offline identification of clade-specific gene markers was developed from a previous method for identifying core genes at multiple taxonomic levels16. As a first step, each genome is independently processed and all its CDSs are clustered for obtaining a bag-of-genes representation of the genome; only one representative (called the seed) for clusters with more than one CDS is retained. The second step of the procedure considers the seeds of all the genomes in a given species and identifies the seeds with sequence homology in all genomes using UCLUST18 with 75% of nucleotide identity threshold. Several refinements are introduced in order to make the core gene identification procedure robust to the missing genomic region in the draft genomes and to errors and noise in the original CDS call and in the taxonomic assignment of the genomes. First, the clade-specific gene families are blasted against the raw genomes and when new high-confidence matches are found (75% identity), CDS calls are modified accordingly and the gene family expanded. Second, we relaxed the definition of core genes: given an empirically estimated probability that a CDS is missing from a genome for experimental rather than biological reasons (called the misannotation rate and set to 0.1), we computed the posterior probability density function (using the beta distribution) to model the presence/absence of a gene family in the genomes and we used it to estimate (minimum confidence at 5%) whether the observed CDS homology pattern is consistent with the null hypothesis that the CDS is present in all genomes of the clade.
The procedure is recursively applied to higher taxonomic levels (from genera to phyla) considering all seeds of CDS families in direct descendant clades that possess a sufficiently large core confidence score to potentially satisfy the 5% confidence score of the parent clade. Notice that some genomes are taxonomically assigned by IMG directly to genus level and they are thus not inserted in the species-level core gene identification process. Differently from the standard core-gene definition16 we are considering core gene families that are conserved within clade but not within its parent clades as the purpose here is to identify genes that characterize each microbial clade. These alignment parameters are tuned here to identify nucleotide representatives along all CDS lengths and thus do not necessary capture broader functional categories.
Screening core genes for unique taxonomic marker genes
Each clade-specific core gene family is blasted against all the raw genomes in order to identify those core genes that are not uniquely present in the clade and exclude them from the candidate marker list. In this way the gene catalog eliminates the genes that cannot be unequivocally associated to a clade; this is the case, for example, for the 16S rRNA gene and all other genes ubiquitous within the microbial taxonomy19, 20 and for genes likely to have been horizontally transferred. For each core gene family, a uniqueness index is estimated which is inversely proportional to the number of genomes containing a local homology with the seed of the core gene family. Multiple sets of blasting parameters have been applied (e-values from 1e-5 to 1e-50, minimum alignment length from 20 to 50 nt) to this procedure in order to rank the uniqueness of a gene with the stringency of the homology match and consequently drive the preference of inclusion in the final reduced marker catalog.
Multi-copy genes (including paralogs detected above) were excluded at this point for all clades with at least 300 markers, because single-copy genes provide a more direct quantitative estimate of genome abundance when available. Interestingly, the widely used 16S rRNA marker gene is itself a multi-copy gene with a strain-specific number of copies and, to the best of our knowledge, is typically used for community profiling without adjusting for the number of copies. Finally, a heuristic considering the uniqueness index, the confidence of the core genes, the different stringency parameters, and the length of the genes is applied to obtain a consistent number of core genes (with a maximum number of 350 core genes per clade) for each medium- to low-level taxonomic clade. This offline automatic pipeline produced the unique taxonomic markers which populate the database of 400,141 CDS currently used by the taxonomic classifier.
Mapping metagenomes to the marker gene catalog and estimation of organismal relative abundances
The selection of the marker genes described above is relatively computationally intensive (typically requiring several CPU-days), but needs to be performed only once when the set of reference genomes is modified, typically because of the addition of newly sequenced genomes. MetaPhlAn users do not need to perform this task, as we provide the most updated reference marker set. The algorithm for estimating the taxonomic composition of the sample using the marker catalog is available for download (see "Implementation and availability") and described below.
MetaPhlAn first needs to compare each metagenomic read from a sample with the catalog of marker genes for finding sequence matches. Although the software performs this operation internally using NCBI blastn (default threshold on the evalue of 1e-6), any other sequence matching method (including accelerated methods like MapX (Real Time Genomics, San Francisco, CA), MBLASTX (Multicoreware, St. Louis, MO), and USEARCH18 can be used by providing to MetaPhlAn the generated blasting results in a tabular format. MetaPhlAn also provides an option to perform the blasting operation with multiple processors, internally splitting the input reads in multiple independent sets that can be blasted using different CPUs. The classifier then assigns read counts to the microbial clades accordingly to the blasting prediction; in the rare case of multiple matches with markers from different clades only the best hit is considered. For each clade, read counts can be assigned directly using the clade-specific markers, or indirectly considering the reads assigned to all direct descendants. Total read counts are estimated with the direct approach if the clade has a sufficient total size of the marker set (default 10,000 nt), otherwise the indirect approach is preferred. Moreover, for all clades except the leaves of the taxonomic tree (species), the two estimations are compared and an “unclassified” subclade is added when the clade-specific read count is larger than the sum of descendant counts, and the difference between the two estimations is assigned to the new descendant. Relative abundances are estimated weighting read counts assigned using the direct method with the total nucleotide size of all the markers in the clade, and normalizing by the sum of all directly-estimated weighted read counts.
Synthetic metagenomes and comparison with existing approaches
The generation of the ten synthetic communities used in our evaluation was inspired by Mavromatis et al.21 and consisted of two high-complexity evenly-distributed metagenomes (HC1-2) and eight low-complexity lognormally-distributed metagenomes (LC1-8). The 100 genomes for each HC community and the 25 genomes for each LC community were randomly chosen from the KEGG v5422 genome catalog. 100nt long reads were sampled from the selected genomes using a MAQ23 error model constructed using real Illumina reads. The number of reads sampled from each genome was proportional to the size of the genome, and the distribution of the genome abundances was even for HC1-2 and log-normal for LC1-8. The resulting synthetic metagenomes are available at the MetaPhlAn web site.
We additionally assessed the accuracy with which MetaPhlAn detects novel organisms (those without closely related sequenced genomes) as compared to existing approaches. We selected 20 genomes from RefSeq24 versions 46–51 that belong to species not represented in the IMG repository. Although these 20 genomes may be included in IMG in the near future, they were not included in MetaPhlAn's current core gene inputs due to incomplete sequence validation, unverified gene calls and annotations, or missing database depositions. Fortunately, this made them appropriate candidates for held-out test data. 25,000 short reads for each of these 20 genomes were thus generated using the same strategy adopted for creating synthetic metagenomes in order to assess MetaPhlAn's classification of novel taxa.
We compared MetaPhlAn's performance on these data (Supplemental Tables 1 and 2) and on the synthetic data above (Fig. 1 and Supplemental Figs 2–4) relative to several existing classes of methods for taxonomic classification of metagenomic reads, based on alignment- and composition-based approaches. Alignment-based methods rely on comparing reads to a set of reference genomes and include NCBI BLAST13 and other BLAST-based approaches like MEGAN25, 26, MTR27, PaPaRa28, and CARMA329. Composition-based methods exploit features like k-mer frequency and include PhyloPythia/PhyloPythiaS30, 31, Phymm12, 32, NBC15, 33, RAIphy34, and MetaCluster 3.035. These two approaches have also been integrated in hybrid methods like PhymmBL12, 32 and RITA14, generally resulting in improved accuracy relative to either method alone. We thus compared MetaPhlAn to the six most popular methods representative of the current state-of-the-art: Phymm, PhymmBL, and the blast-based component of PhymmBL have been installed locally on the same machine on which MetaPhlAn ran, whereas RITA and NBC provides a convenient webserver as well as PhyloPythiaS.
Description of novel Human Microbiome Project and MetaHIT metagenome analyses
The human metagenomic data we analyzed with MetaPhlAn are all available in public repositories. The 51 vaginal and 139 fecal samples from the HMP3 can be accessed at http://hmpdacc.org/HMASM, from which we used the "WGS" reads (not the "PGA" assemblies). The 16S rRNA profiling of HMP vaginal microbiomes used for comparison (Fig. 2) was performed from the mothur pipeline36 data followed by phylotype profiling with the RDP classifier37 (http://hmpdacc.org/HMMCP). The 85 fecal samples from MetaHIT4 were downloaded from the European Nucleotide Archive (http://www.ebi.ac.uk/ena/, study accession number ERP000108), and the marine minimum oxygen zone samples10 are available in the NCBI Sequence Read Archive under accession number SRA023632.
The full tables of results for the vaginal and gut metagenomes are available at http://huttenhower.sph.harvard.edu/metaphlan.
Supplementary Material
Acknowledgments
We would like to thank F. Stewart and E. DeLong for their helpful input during this study, as well as D. Gevers and K. Huang for feedback on the methodology. This work was supported by NIH 1R01HG005969 and NSF DBI-1053486 to CH.
Footnotes
Author Contributions
N.S., A.B., O.J. and C.H. conceived the method, N.S. implemented the software, N.S. and C.H. performed the experiments, N.S., L.W., V.N. and C.H. analyzed the data, N.S. and C.H. wrote the manuscript.
References
- 1.DeLong EF. Nat Rev Microbiol. 2005;3:459–469. doi: 10.1038/nrmicro1158. [DOI] [PubMed] [Google Scholar]
- 2.Daniel R. Nat Rev Microbiol. 2005;3:470–478. doi: 10.1038/nrmicro1160. [DOI] [PubMed] [Google Scholar]
- 3.Methé B, et al. Nature. 2012 in press. [Google Scholar]
- 4.Qin J, et al. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravel J, et al. P Natl Acad Sci USA. 2011;108:4680–4687. [Google Scholar]
- 6.Veiga P, et al. P Natl Acad Sci USA. 2010;107:18132. [Google Scholar]
- 7.Turnbaugh PJ, et al. Nature. 2008;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markowitz VM, et al. Nucleic Acids Res. 2010;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fredricks DN, Fiedler TL, Marrazzo JM. New Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 10.Stewart FJ, Ulloa O, DeLong EF. Environ Microbiol. 2012;14:23–40. doi: 10.1111/j.1462-2920.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- 11.Arumugam M, et al. Nature. 2011 [Google Scholar]
- 12.Brady A, Salzberg S. Nat Methods. 2011;8:367–367. doi: 10.1038/nmeth0511-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Parks D, MacDonald N, Beiko R. BMC Bioinformatics. 2011;12:328. doi: 10.1186/1471-2105-12-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosen GL, Reichenberger ER, Rosenfeld AM. Bioinformatics. 2011;27:127. doi: 10.1093/bioinformatics/btq619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segata N, Huttenhower C. PloS one. 2011;6:e24704. doi: 10.1371/journal.pone.0024704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohlin J, et al. BMC evolutionary biology. 2010;10:249. doi: 10.1186/1471-2148-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. Bioinformatics. 2010;26:2460. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 19.Wu M, Eisen JA. Genome biology. 2008;9:1–11. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciccarelli FD, et al. Science. 2006;311:1283. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 21.Mavromatis K, et al. Nat Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Ruan J, Durbin R. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruitt KD, Tatusova T, Klimke W, Maglott DR. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huson DH, Auch AF, Qi J, Schuster SC. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Genome Res. 2011 doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gori F, Folino G, Jetten MSM, Marchiori E. Bioinformatics. 2011;27:196. doi: 10.1093/bioinformatics/btq649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger SA, Stamatakis A. Bioinformatics. 2011;27:2068–2075. doi: 10.1093/bioinformatics/btr320. [DOI] [PubMed] [Google Scholar]
- 29.Gerlach W, Stoye J. Nucleic Acids Res. 2011;39:e91. doi: 10.1093/nar/gkr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McHardy AC, Rigoutsos I, Hugenholtz P, Tsirigos A, Martin HG. Nat Methods. 2007;4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 31.Patil KR, et al. Nat Methods. 2011;8:191–192. doi: 10.1038/nmeth0311-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brady A, Salzberg SL. Nat Methods. 2009;6:673–676. doi: 10.1038/nmeth.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen G, Garbarine E, Caseiro D, Polikar R, Sokhansanj B. Advances in Bioinformatics. 2008;2008:1–12. doi: 10.1155/2008/205969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozkan N, Samuel W, Khalid S. BMC Bioinformatics. 2011;12 [Google Scholar]
- 35.Leung H, et al. Bioinformatics. 2011;27:1489–1495. doi: 10.1093/bioinformatics/btr186. [DOI] [PubMed] [Google Scholar]
- 36.Schloss PD, et al. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole JR, et al. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.