Abstract
Microvesicles (exosomes) are important mediators of intercellular communication, playing a role in immune regulation, cancer progression and the spread of infectious agents. The biological functions of these small vesicles are dependent upon their composition, which is regulated by mechanisms that are not well understood. Although numerous proteomic studies of these particles exist, little is known about their glycosylation. Carbohydrates are involved in protein trafficking and cellular recognition. Glycomic analysis may thus provide valuable insights into microvesicle biology. In this study, we analyzed glycosylation patterns of microvesicles derived from a variety of biological sources using lectin microarray technology. Comparison of the microvesicle glycomes with their parent cell membranes revealed both enrichment and depletion of specific glycan epitopes in these particles. These include enrichment in high mannose, polylactosamine, α-2,6 sialic acid, and complex N-linked glycans and exclusion of terminal blood group A and B antigens. The polylactosamine signature derives from distinct glycoprotein cohorts in microvesicles of different origins. Taken together our data point to the emergence of microvesicles from a specific membrane microdomain, implying a role for glycosylation in microvesicle protein sorting.
Keywords: Microvesicles, lectin microarray, exosomes, glycomics, carbohydrates, glycan
Introduction
The human body is made of a complex collection of cells, the interactions of which must be carefully orchestrated to ensure proper function and health. Our understanding of the mechanisms of cellular communication recently expanded with the discovery that secreted membranous particles such as microvesicles (exosomes) can act as a conduit for the exchange of information between cells. The terms microvesicle and exosome are often used interchangeably to describe particles between 40-100 nm in size, which contain mRNA, miRNA, proteins and lipids.1 They have been isolated from physiological fluids including urine, blood, saliva, tumor effusions, breast milk and amniotic fluid and from the culture media of a broad variety of cell types.2 These nanoparticles take part in a plethora of normal and pathogenic events including the spread of prion proteins,3 transformation of local cells with metastatic signaling receptors from tumors,4 induction of immune tolerance in the gut and in pregnancy,5, 6 and immune surveillance for intracellular pathogens.7 Contradicting biological functions have been observed for microvesicles. For example, several groups have reported that exosomes secreted from dendritic cells have the ability to stimulate T-cells8, 9 inducing the rejection of tumors when dendritic cells are pulsed with tumor antigens.10 In contrast, tumor-derived exosomes can activate T-cell apoptosis through presentation of the Fas ligand.11 These differences in biological function most likely arise from differences in the cargo present either on the surface of the vesicle or internally, which is dependent upon their cellular origin.2, 12
Numerous proteomic studies have defined a common set of membrane and cytosolic proteins in these particles. These include the transmembrane tetraspannins CD63 and CD81 and the cytosolic heat shock protein HSP70.13 In contrast, little attention has been paid to the glycosylation of microvesicles. Carbohydrates play a crucial role in the communication of cells with their extracellular environment, serving as points of cell-specific recognition for bacteria, virus particles and other cells.14-16 Recent work from our laboratory by Krishnamoorthy et al provided the first assessment of the glycomics of microvesicles.17 In this work, which focused on the comparison of HIV-1 and T-cell derived microvesicles, we utilized lectin microarray technology to demonstrate that microvesicles and virus from T-cell lines have a common glycome characterized by both the enrichment and exclusion of carbohydrate markers. This argues that these particles come from a specific portion of the T-cell membrane. Carbohydrate epitopes, including sialosides, high mannose residues and complex N-linked glycans, can direct protein sorting to membrane microdomains within cells.18-20 Identification of enriched glycomic structures across microvesicle populations would imply that similar sorting mechanisms may exist for the sorting of membrane proteins into microvesicles.
Herein we report the first direct glycomic comparison of microvesicles derived from a diverse panel of human cell lines (T-cells, melanoma, and colon cancer) and the physiological fluid breast milk using lectin microarray technology. Lectin microarrays consist of a panel of carbohydrate binding proteins (lectins) immobilized onto a solid substrate at a high spatial density.21, 22 Incubation of the arrays with a fluorescently labeled glycosylated sample enables elucidation of the glycome based on the distinct carbohydrate-binding specificities of the lectins. This method has been used to analyze the glycomic fingerprint of viruses,17 bacteria,23 mammalian cells24 and tissues25 in a high throughput manner. A more quantitative approach, pioneered by our laboratory for the direct comparison of glycosylated samples, is the ratiometric dual-color method. In this method fluorescently-labeled samples are analyzed against an orthogonally labeled reference that competitively binds to the lectins on the array.24 Using both single- and dual-color lectin microarray methods, we observe a conserved glycomic profile for microvesicles derived from a variety of biological sources. Comparison of the microvesicle glycomes with their parent cell membranes revealed both enrichment and depletion of specific glycan epitopes in these particles, pointing to their emergence from a specific microdomain. Cell-type dependent differences, reflective of the more diverse parent membrane glycosylation, were also observed. Our data suggest a role for glycosylation in protein sorting to microvesicles.
Methods
Cell culture
The SkMel-5, HT29 and HCT-15 cell lines were purchased from the National Cancer Institute (Frederick, MD). H9 and SupT1 were obtained from J. Bess (AIDS Vaccine Program, NCI, Frederick, MD). Jurkat-Tat-CCR5 cells were obtained from Q. Sattentau (University of Oxford). Cells were cultured in RPMI 1640 (Lonza) with 2 mM L-glutamine and 10% (v/v) fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA) at 37°C and 5% CO2. Penicillin-streptomycin (Mediatech, Manassas, VA) was added to non-adherent cells.
Breastmilk Collection
Human milk samples were obtained from a healthy 34 year old Caucasian mother who delivered a preterm infant at 30 weeks gestational age. This was the mother’s third pregnancy and delivery. The mother was rubella immune and negative for Hepatitis, HIV and Chlamydia infections with a history of infertility due to uterine adhesions. The sample used in this analysis was obtained by mechanical expression at 27 days post-partum, immediately frozen and stored at −20°C until analysis. This research was performed with NYU School of Medicine Institutional Review Board approval and the mothers’ written informed consent.
Microvesicle Isolation
Cells were rinsed twice with Hank’s buffered salt solution (HBSS) (Mediatech, Manassas, VA) to remove traces of FBS and growth medium was replaced with serum-free medium (RPMI 1640 with 2 mM L-glutamine). After 24 h, conditioned media was collected and microvesicles were isolated by differential centrifugation as described previously.26 Briefly, cell debris and larger vesicles were pelleted out of the media by sequential centrifugation (300 x g, 10 min, 2,000 x g, 20 min and 10,000 x g, 30 min; Beckman Coulter). The cleared supernatant was subjected to ultracentrifugation at 100,000 x g for 1 h to obtain a microvesicle pellet. Microvesicles were resuspended in either PBS (0.1 M phosphate buffer, 0.15 M NaCl, pH 7.4) or Cy-labeling buffer (0.1 M NaHCO3, pH 9.3). A slightly altered protocol was utilized for the isolation of microvesicles from breast milk. Differential centrifugation (as above) of 2 ml of breast milk was followed by sequential filtration through 0.45 μm and 0.22 μm filters. The filtrate was overlayed onto a 30% sucrose cushion and centrifuged at 100,000 x g for 1 h. The pellet was resuspended in PBS and centrifuged at 100,000 x g for 1 h. The washed pellet was then diluted in Cy-labeling buffer. The protein levels of all microvesicle preparations were quantified using the micro-BCA assay (Thermo Scientific).
Cell membrane preparation
Labeled cell membranes were prepared as described previously.24 After collecting conditioned media, cells were washed twice in cold PBS and either scraped (adherent cells) or resuspended (non-adherent cells) in cold PBS containing protease inhibitor cocktail. Cells were then sonicated on ice (3 × 5 s, 70% power, Branson sonicator) to disrupt cell membranes. Membranes were pelleted by ultracentrifugation at 100,000 x g for 1 h. The pellet was resuspended in Cy-labeling buffer and homogenized by sequential passing through 18- and 24- gauge needles. We determined the protein concentration using the DC protein assay (Bio-Rad).
Cy3- and Cy5- labeling
Samples were fluorescently labeled with 60 mg of Cyanine 3 (Cy3) or Cyanine 5 (Cy5) mono reactive-NHS (GE Life Sciences) per mg of protein in Cy-labeling buffer for 30 min at room temperature with gentle rocking. The labeling reaction was quenched by the addition of a Tris-buffered saline stock solution (2 M Tris-HCL, 1.2 M NaCl, pH 6.8) to a final concentration of 250 mM Tris-HCl and 150 mM NaCl (final pH ~7.6-7.8) for 30 min. Microvesicle samples were used without further purification from the quenched excess dye. For cell membrane samples, excess dye was removed by dialyzing into PBS overnight at 4°C. This prevented the high background observed with these samples when used after quenching alone, which is most likely due to the exposure of hydrophobic lipids in unsealed membrane bilayers following the cell disruption. Protein concentrations for membrane samples were obtained after dialysis.
Lectin Microarray Print
All lectins were purchased from either EY Laboratories (San Mateo, CA) or Vector Labs (Burlingame, CA) with the following exceptions: cyanovirin (CVN), scytovirin (SVN) and griffithsin (GRFT) were gifts from Dr. B. O’Keefe (NCI-Frederick); galectin-9 was a gift from Dr. L. Baum (UCLA Medical School); and Gaf-D, PA-IL, PA-IIL, PapGII, PapGIII and RS-IIL were made recombinantly as previously described.27, 28 Three spots per lectin were printed using a Nanoplotter 2.1 piezoelectric printer (GeSIM, Germany) at 14°C and 45% humidity. See Supplementary Table 1 for lectin list, print concentrations and buffers. Lectin microarrays were tested for activity using Cy5-labeled glycoprotein standards (5 μg, ovalbumin, asialofetuin, fetuin and transferrin).
Microarray Hybridization and Analysis
Microarray slides were submerged in blocking buffer (50 mM ethanolamine, mM sodium borate ph 8.5) for 1 h followed by 3 washes with PBST (PBS with 0.005% Tween) and a final wash with PBS. The microarray slides were then fitted to a 24-well frame (ArrayIt, Sunnyvale, CA). For single color experiments, we added 1.5 μg (by protein concentration) of labeled sample in a final volume of 100 μl in PBST to each subarray. For dual color experiments, 1.5 μg of Cy3-labeled microvesicles were incubated with equal amounts of either Cy5-labeled H9 reference or Cy5-labeled parent membrane in 100 μl total volume PBST. The order of the samples was randomized on a total of two slides for all dual color assays. Labeled glycoprotein standards were hybridized on all slides as controls and no statistical difference in lectin activity was observed between slides (data not shown). Samples were hybridized to the lectin microarrays for 2 h at room temperature with gentle rocking. The individual subarrays were then washed with PBST for 5 × 5 min with a final 10 min wash in PBS. Slides were scanned and analyzed using a GenePix 4300B fluorescent slide scanner (Molecular Devices, Sunnyvale, CA) with GenePix Pro 7 software. The background subtracted median fluorescence of the three replicate spots per lectin was tested for outliers using the Grubbs outlier test with alpha = 0.05. The log2 values of the average median fluorescence of the 3 replicate spots were median centered over the array in each channel to account for differences in labeling efficiency.29 The resultant data sets were analyzed by hierarchical clustering using the Pearson correlation coefficient with average linkage analysis using Cluster 3.030 and visualized with Java Treeview.31 Lectins with signal to noise ratios (SNR) of less than 5, as determined by the GenePix Pro 7 software, in 90% or more of samples were excluded from the cluster. If the Pearson correlation values were statistically significant (P < 0.0001 for a two tailed t test, DF = N-2, calculated using http://faculty.vassar.edu/lowry/tabs.html#r ) then arrays were considered to be statistically similar.
Western and Lectin Blots
Microvesicle and cell membrane proteins were probed with antibodies to CD63 (1:500 dilution) (RFAC4, Millipore, Billerica, MA), CD81 (1: 500 dilution) (H-121, Santa Cruz Biotechnology, Santa Cruz, CA), galectin-3 (1:1,000 dilution) (ZG001, Invitrogen, Carlsbad, California), and galectin-4 (1:500 dilution) (1E8, Novus Biologicals, Littleton, CO) followed by incubation with the appropriate HRP-conjugated secondary antibody (1:10,000 dilution) (Bio-Rad, Hercules, CA). Biotin conjugated DSA lectin (EY Laboratories, San Mateo, CA) was blotted at a concentration of 1 μg/ml followed by streptavidin-HRP (1:500 dilution) (Jackson ImmunoResearch, West Grove, PA). Blots were visualized using Supersignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Glycosidase Treatment
All enzymes were purchased from New England Biolabs. Cy3-labeled microvesicles (3 μg) were treated with either 1.5 kU EndoHf cloned from Streptomyces plicatus, 100 U Neuraminidase cloned from Clostridium perfringens or 1 kU PNGaseF purified from Flavobacterium meningosepticum per μg of microvesicle protein in the manufacturer provided buffer without the addition of detergent at 37°C for 18 h. Negative controls were mock treated with enzyme buffer and incubated at 37°C for 18 h.
Electron Microscopy
Images were obtained from the New York University Langone Medical Center Office of Collaborative Sciences Microscopy Core. Samples were negatively stained with 1% Uranyl Acetate in ddH2O on 400 mesh Cu grids coated with a Carbon membrane. Grids were viewed on a Philips CM12 tungsten emission TEM at 120kV and imaged with a Gatan 4k × 2.7k digital camera.
Results and Discussion
Purification of Microvesicles from Multiple Sources
To study the glycan composition of microvesicles from a variety of sources, we isolated them from the cell culture media of six different human cancer cell lines derived from three distinct cancer types and from the human physiological fluid, breast milk. The cell lines included three T-cell lines (Jurkat, SupT1 and H9), two colon cancer lines (HCT-15 and HT-29) and a skin cancer line (SkMel-5). Microvesicles were isolated for glycomic analysis by the strategy outlined in Figure 1A. Our initial isolation protocol for microvesicles used media containing fetal bovine serum (FBS) that was pre-cleared by ultracentrifugation.26 This pre-clearing step should eliminate contaminating bovine microvesicles that may pellet with the desired cell-derived vesicles. However, we found that even with overnight ultracentrifugation at 100,000 x g, our pre-cleared media still contained trace contaminants that could be observed on our lectin microarrays (data not shown). This is problematic as, unlike proteins where species-specific isoforms exist, it is not possible to clearly distinguish the glycans of the bovine contaminant from our human samples. Due to this complication, we cultured our cells in serum-free media, which has no protein content. To avoid viability issues, cells were cultured in serum-free media for no longer than 24 hours and cell viability was tested by the trypan blue exclusion assay. No culture was used for microvesicle isolation if cell death was greater than 10% due to concerns of contamination from cell debris.
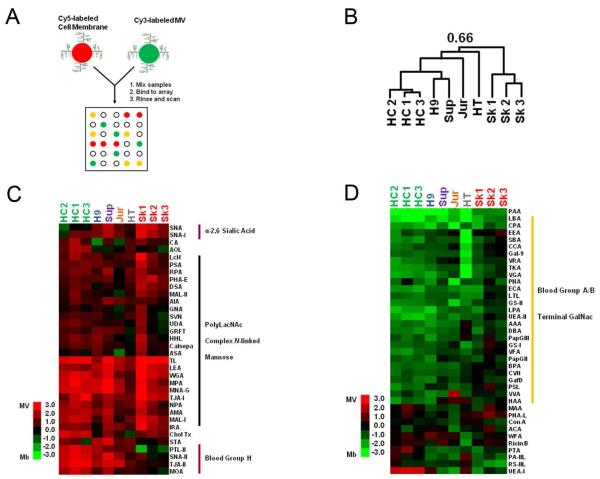
Figure 1.
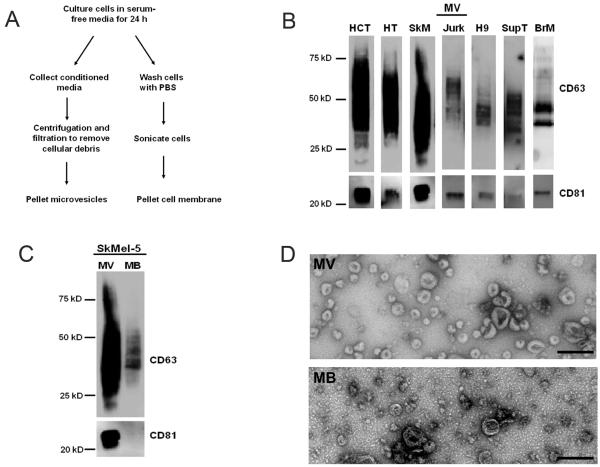
Isolation of exosomes and cell membrane for glycomic comparison. (A) Schematic of microvesicles and membrane isolation. (B) Western blots of the microvesicle preparations (MV) from HCT-15 (HCT), HT29 (HT), SkMel-5 (SkM), Jurkat-TAT-CCR5 (Jurk), H9, Sup-T1 (SupT), and breast milk (BrM). Markers CD63 and CD81 are present in all samples. (C) Equal amounts (3 ug protein) of SkMel-5 MV and cell membrane (MB) were probed for CD63 and CD81. (D) Transmission electron microscopy images of SkMel-5 MV and MB. Bar is 200 nm.
To ensure the uniformity of our preparations, we tested all microvesicle isolations for the markers CD63 and CD81 by western blot analysis. Only microvesicle isolations containing both markers were used on our microarrays. We observed heterogeneity in both the levels of the markers and in their observed molecular weights between samples (Figure 1B). The latter may be due to different glycoforms of the proteins from different sources. As expected, microvesicle markers were highly enriched in the microvesicles when compared to the parent cell membranes (Figure 1C, Supplementary Figure 1) and the prototypical cup shape morphology for these particles was observed (Figure 1D).
Microvesicles and membranes display discrete glycomes
We hybridized 1.5 μg (based on protein concentration) of either Cy3-labeled microvesicles or cell membrane preparations to our lectin microarrays. Electron microscopy images of general cell membrane preparations from Sk-Mel-5 revealed a size distribution of vesicle-like structures (20-200 nm) with a few larger membranous structures (Figure 1D). The majority of particles observed were under 100 nm in size, comparable to our microvesicle samples. Both enrichment and exclusion of glycan epitopes were observed in microvesicles when compared to their parent membrane preparations as measured by fluorescence intensities (Figure 2). In general, lectin analysis revealed that microvesicles were enriched in high mannose (HHL, PSA, NPA, SVN, UDA), complex N-linked glycan (Calsepa,32 LcH, PHA-E, PHA-L, TL), α-2,6 sialic acid (SNA, TJA-I) and poly N-acetyllactosamine (LEA, DSA) epitopes but showed little to no binding to lectins that recognize terminal N-acetyl-D-galactosamine (CAA, DBA, SBA, SJA, BPA) and Blood Group A/B (EEA, LTL). An example of the raw data is shown in Figure 2B and Supplementary Figure 2 for the SkMel-5 cell line.
Figure 2.
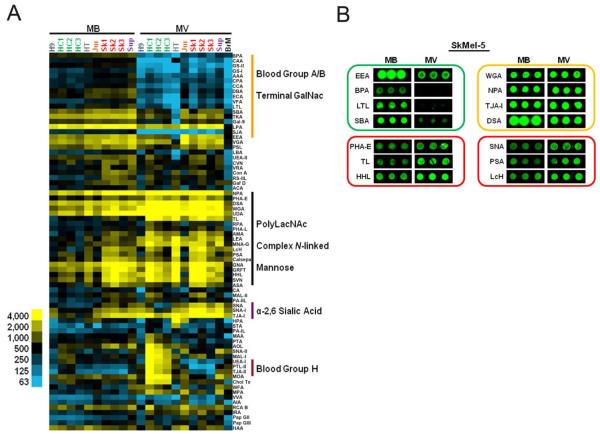
SkMel-5 microvesicles and cell-membranes have discrete glycomic profiles. (A) Equal amounts (1.5 μg by protein) of Cy3-labeled samples were added to the lectin microarray. Lectins were hierarchically clustered by using the Pearson correlation coefficient as the distance metric and average linkage analysis (n = 74 lectins). The tree is not shown. The arrays are manually organized. Heat map of the background corrected median fluorescence (average of three spots) is shown. Yellow indicates median fluorescence units > 500 and blue indicates median fluorescence < 500. Abbreviations: GalNAc (N-acetylgalactosamine), PolyLacNAc (poly-N-acetyllactosamine) (B) Comparison of SkMel-5 MV and MB demonstrates exclusion (green block) and enrichment (red blocks) of glycan epitopes in MV as well as lectins that bind equally to both (yellow block) as determined by the average median fluorescence signal.
Single color analysis can only distinguish differences in carbohydrate levels between samples if lectin binding is in the linear range.17, 24 In SkMel-5, WGA, TJA-I (sialic acid binding lectins), NPA (a high mannose lectin), and DSA (a polyLacNAc binder) appear to bind microvesicles and the corresponding cell membrane preparations at similar levels as measured by the median fluorescence intensity, which is irrespective of spot size and morphology (Figure 2B). However, lectins with similar specificity profiles (α-2,6 sialic acid: SNA ; high mannose: HHL, PSA) show clear enrichment in binding to the microvesicles, suggesting that some lectins are not in the linear range of detection. To address this issue and validate our single color results, we utilized the ratiometric dual-color approach developed in our laboratory.17, 24 In this approach, competitive binding between the sample and an orthogonally labeled common reference enables comparison of the relative binding levels thereby increasing sensitivity to differences between samples. We used Cy5-labeled H9 cell membrane preparations as a biological reference for our comparative analysis. Glycoprofiles of our panel of microvesicles and parent cell membranes were obtained by incubation of 1.5 μg of Cy3-labeled sample against an equal amount of reference (Figure 3A). Using this method, we observed the expected enrichment of DSA, WGA, NPA and TJA-I epitopes in SkMel-5 microvesicles (Figure 3B).
Figure 3.
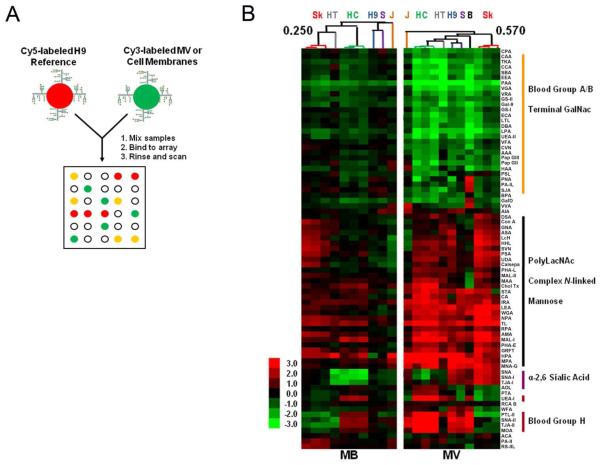
Ratiometric lectin microarray comparison using H9 membrane as a reference. (A) Experimental schematic. Equal amounts of Cy3-labeled MV and parent MB were incubated against Cy5-labeled H9 MB. (B) Median-normalized log2 ratios (Sample/Reference) of the background subtracted median fluorescence signal were hierarchically clustered using the Pearson correlation coefficient as the distance metric and average linkage analysis (n=75 lectins). Heat map is shown. Red indicates log2 ratio of sample over reference is greater than the log2 ratio of the median sample signal divided by the median reference signal within an array (log2(Sample/Reference) > log2(Samplemedian/Referencemedian); green indicates log2 ratio lower than the ratio of medians. Select Pearson correlation values are shown on the tree. Data for the corresponding uncentered clusters are shown in Supplementary Figures 3 and 4. SkMel-5 (Sk), HT29 (HT), HCT-15 (HC), SupT1 (S), Jurkat (J), breast milk (B).
Hierarchical clustering of the arrays using the Pearson correlation coefficient as the distance metric and average linkage analysis revealed a conserved lectin binding pattern for our panel of microvesicles, including those derived from the physiological fluid, breast milk (Figure 3B, R=0.57, n=75, P < 0.0001, two-tailed). In contrast, clustering of the parent cell membranes did not show a conserved glycomic signature (R = 0.25, n = 75, P = 0.03). The data shown is median-centered to aid in the comparison between differentially labeled samples.29 Uncentered data gives similar correlation coefficients (Supplementary Figures 3 and 4). In this work we consider a P value less than or equal to 0.0001 as our threshold for significance to indicate a conserved glycome. Our data is consistent with our previous work on T-cell microvesicles (Jurkat, H9 and SupT1) and suggests that the glycomic pattern observed for microvesicles is not a phenomenon of the culture conditions.17
Our analysis revealed two sets of GalNAc-specific lectins with divergent behaviors. The first set (DBA, BPA, SBA, VVA), identified in our single color analysis, bind to terminal α- and β-GalNAc epitopes, such as Blood Group A. They display lower binding to the microvesicles, arguing that the epitopes they bind to are excluded. Heirarchichal clustering of the lectins in our dual-color analysis identified a second cohort of GalNAc binders (MNA-G, MPA, SNA-II, IRA, HPA, AIA) specific for terminal α-GalNAc structures such as the Tn antigen, with highly enriched binding to the microvesicles. Overall, the microvesicle signature observed was consistent with our single color data, revealing a glycomic pattern of enrichment in high mannose, complex N-linked glycans and polyLacNAc epitopes and a reduction in Blood Group A/B antigens. The enriched epitopes have been associated with apical trafficking of glycoproteins by endogenous lectins including galectins, suggesting that glycans may be involved in trafficking to these particles.
Microvesicles displayed some cell specific differences in glycosylation. For example, neither SkMel-5 microvesicles nor the parent cell membranes show binding to Blood Group H binding lectins (UEA-I, PTL-II, TJA-II). However this epitope, which is present in HCT-15 membranes, is clearly enriched in HCT-15 microvesicles. In addition, HCT-15 and HT29 cells, which have diminished levels of α-2,6 sialic acid in their membranes when compared to the entire panel of cell lines, consequently displayed lower levels overall of this epitope in their cognate microvesicles (SNA, SNA-I and TJA-I, Figure 3B), although enrichment is still observed in comparison to the parent membranes. Thus, the gross glycomic composition of the parent membrane is reflected in the microvesicle glycome.
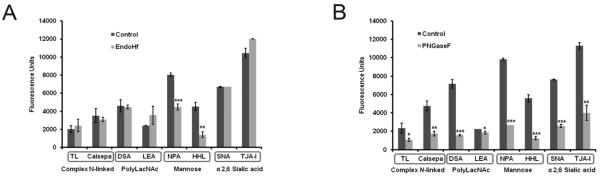
We performed multiple control experiments to confirm the specificity of lectin binding to our microvesicle samples. First, we inhibited binding on our arrays by pre-incubation of the lectins with the appropriate mono- and disaccharides and observed the expected results overall (see Supplementary Figure 5). It should be noted that some lectins, especially complex epitope binders such as TL, are not inhibited by simple sugars. Next we subjected our microvesicles to enzymatic treatments to gain further insight into the specificity of lectin binding. Treatment of SkMel-5 microvesicles with Endo Hf, an enzyme that cleaves within the core of high mannose and hybrid N-linked glycans, preferentially inhibited binding to mannose lectins (NPA, HHL, Figure 4A). In contrast, the enzyme PNGase F, which cleaves all mammalian N-linked glycans, significantly reduced the binding of lectins to both mannose and complex epitopes as expected (Figure 4B). A strong reduction was also observed in α-2,6 sialic acid binding (SNA, TJA-I) upon PNGaseF treatment, arguing that this epitope is mainly in N-linked glycans on the microvesicles. The specificity of these lectins (SNA, TJA-I) for sialic acid was confirmed by the loss of microvesicle binding to these lectins upon treatment with neuraminidase (sialidase), which cleaves this glycan (Supplementary Figure 6). Disparate results were seen upon treatment with PNGaseF for the polyLacNAc binders DSA and LEA. While a large reduction in binding upon N-glycan cleavage was observed for DSA (78%), LEA displayed only a modest reduction (17%). Although both bind to polyLacNAc, the two lectins have discrete specificities, with LEA preferring long polyLacNAc chains.33, 34 Our data indicates that polyLacNAc may be present in multiple contexts within SkMel-5 microvesicles, including possible O-linked and glycolipid epitopes.
Figure 4.
Enzymatic treatment of microvesicles confirms specificity of lectins. Cy3-labeled SkMel-5 MV were treated with (A) EndoHf and (B) PNGaseF for 18 h prior to hybridization on the array. Fluorescence values are expressed as a mean ± SD of replicate spots. Statistically significant differences are indicated by their p-values: * p < 0.05; **p < 0.001; *** p < 0.0001. These values were calculated using the unpaired t-test.
Microvesicle enrichment in conserved glycans suggests carbohydrate-mediated sorting
To directly examine whether our observed microvesicle signature could result from glycan-mediated sorting to microvesicles, we compared microvesicles and their cognate membranes using our dual-color assay. Equal amounts of Cy3-labeled microvesicles and their corresponding Cy5-labeled parent cell membranes were combined and incubated on our lectin microarrays. We again observed the previous pattern of enrichment and exclusion of specific glycans as a dominant feature of the microvesicle glycosylation profiles when compared to their parent membranes (Figure 5). This implies that the glycan signature is due to sorting of glycoproteins and glycolipids to the microvesicles, though whether this is due to the glycans themselves is not known. Although there was no common biological reference, heirarchical clustering using the Pearson correlation and average linkage analysis again showed a clear glycopattern (Figure 5B, R = 0.66, n = 72, P < 0.0001 and Supplementary Figure 7). We validated the enrichment and exclusion of epitopes by lectin blot analysis (Supplementary Figures 8 and 9). This data strongly suggests that microvesicles are emerging from a specific portion of the membrane with a conserved glycome.
Figure 5.
Direct comparison of microvesicles with their parent cell-membranes. (A) Experimental schematic. Equal amounts of Cy3-labeled MV were analyzed against Cy5-labeled parent MB on the lectin microarray. (B) Median-centered log2 ratios were hierarchically clustered using the Pearson correlation coefficient as the distance metric and average linkage analysis (n=72 lectins). The tree is shown with the overall Pearson correlation value. Heat map shows (C) enriched and (D) lower lectin binding to glycan epitopes in MV. The entire heat map is shown in Supplementary Figure 7.
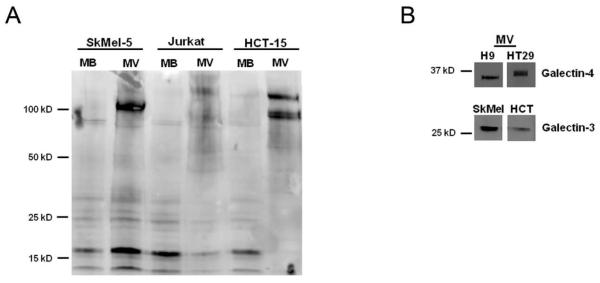
The microvesicle glycome could arise from the presence of predominant conserved glycoproteins or lipids containing the same glycoforms. These could be concentrated in microvesicles by mechanisms other than glycan-mediated sorting. To examine this possibility, we performed a lectin blot assay with DSA, a lectin whose binding epitope, polyLacNAc, is enriched in microvesicles (Figure 5C). Equal amounts (by protein) of microvesicles and membranes from 3 cell lines (SkMel-5, HT-29 and Jurkat) were probed. Enrichment in DSA binding was observed in the microvesicles in concordance with the microarray data (Figure 5C, 6A and Supplementary Figures 9 and 10). However, the microvesicles from each cell line showed a distinct pattern of DSA-positive bands. This provides preliminary evidence that different glycoproteins are responsible for DSA-reactivity within the microvesicles from distinct cell lines.
Figure 6.
A possible role for glycans in MV protein sorting. (A) DSA lectin blot of equal protein amounts of SkMel-5, Jurkat and HCT-15 MV and MB shows enrichment of DSA epitopes on multiple proteins in MV. (B) The cell derived MV were probed for galectin-3 and galectin-4.
Multiple lectins have been implicated in carbohydrate-based protein sorting to specific membranes. Among these lectins, galectins -3 and -4, which bind LacNAc epitopes, have been identified in microvesicles.35, 36 These galectins are known to mediate glycoprotein sorting to apical domains in polarized epithelia cells.37, 38 We tested for the presence of galectins -3 and -4 by Western blot analysis (Figure 6). Galectin-3 was observed in microvesicles derived from two of our cell lines, SkMel-5 and HCT-15. We found galectin-4 in the microvesicles of the T-cell line H9 and the colon cancer cell line HT29. Microvesicles derived from the other two T-cell lines, Jurkat and SupT1, did not display either of these galectins, although other galectin isoforms may be present. We do not yet know whether these lectins are playing a role in the origination of our observed microvesicle glycan signature. However, their presence in four of our six microvesicle samples, coupled with our observation that polyLacNAc epitopes are enriched in these particles suggests that these lectins may be involved, a subject of future research.
Conclusions
Microvesicles (exosomes) are important agents of cellular communication, ferrying RNA, proteins and lipids between cells. The function of microvesicles is intimately tied to their composition. Protein content is important in defining the role of the particle, distinguishing immunosuppressive particles from immunogenic ones, for example. We are only starting to understand the molecular mechanisms underlying the biogenesis of these particles. Previous observations from our laboratory suggested that in T-cells, microvesicles derive from a conserved portion of cellular membrane with a unique carbohydrate composition.17 In this work we demonstrate that the glycosylation profiles of microvesicles obtained from multiple sources including melanoma cells, colon cancer cells and breast milk are strikingly similar to T-cell microvesicle profiles, although they retain characteristics of their parent membranes. High mannose, complex N-linked glycans, α-2,6 sialic acid and polyLacNAc epitopes are enriched on the microvesicles while Blood Group A and B antigens are excluded from these particles. In concordance with our work, enrichment in α-2,6 sialic acid has been observed for bronchial epithelia- and ovarian cancer-derived microvesicles.39, 40 In addition, the enrichment of complex N-linked glycans has been seen in the exosomal glycosylation pattern of the protease ADAM-10.41 These studies further validate our findings.
Our data strongly suggest that glycans play a role in the sorting of glycoproteins to microvesicles. Currently, the process by which proteins are targeted to these particles is not fully understood. Multiple groups have demonstrated that oligomerization of membrane proteins is one potential sorting signal, as multimerization by artificial means targets them to secreted vesicles.42, 43 In polarized epithelial cells, oligomerization of glycoproteins via lectins including VIP36 and galectins, mediate their carbohydrate-directed recruitment to the apical membrane.18, 44, 45 Oligomerization of glycoproteins by lectins may also sort proteins into microvesicles, a potential explanation for the conserved glycan signature of these particles observed on our arrays. Two of the major components of the microvesicle glycan signature, polyLacNAc and high mannose are associated with glycoprotein sorting by galectins and VIP36, respectively. These lectins have previously been observed in microvesicles.35 In this study, galectin-3 and -4 were found. Whether these lectins play any role in our observed glycopattern has yet to be determined and is the subject of future work.
Overall, our work has defined a common glycome for microvesicles/exosomes. In providing tantalizing hints of the potential importance of glycosylation to microvesicle protein recruitment, this work generates the hypothesis that carbohydrate-mediated protein sorting is a major mechanism for protein incorporation. Much work needs to be done to test this hypothesis but if correct, this would extend carbohydrate-mediated sorting to membrane domains beyond apical and basolateral trafficking, provide a new means to control microvesicle content and have implications for the sorting of proteins to viruses that use this machinery.2, 17
Supplementary Material
ACKNOWLEDGMENT
The authors acknowledge support from the National Institutes of Health (L.K.M. , 7 DP2 OD004711-02; K.H.-M., 1R21HD059047-01A1). Electron microscopy was obtained at the Microscopy Core at New York University Lagone Medical Center.
Footnotes
Supporting Information Available: Six Supplementary Figures: TEM images of MB preparations, single color lectin microarray image of SkMel-5, saccharide inhibition of lectins, neuraminidase treatment of MV, hierachical cluster tree for direct comparison of MV and MB, DSA blot quantitation. Supplementary Table of lectins printed on microarray. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Johnstone RM. Exosomes biological significance: A concise review. Blood Cells Mol Dis. 2006;36(2):315–21. doi: 10.1016/j.bcmd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6(3):267–83. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 3.Porto-Carreiro I, Fevrier B, Paquet S, Vilette D, Raposo G. Prions and exosomes: from PrPc trafficking to PrPsc propagation. Blood Cells Mol Dis. 2005;35(2):143–8. doi: 10.1016/j.bcmd.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–24. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 5.Karlsson M, Lundin S, Dahlgren U, Kahu H, Pettersson I, Telemo E. “Tolerosomes” are produced by intestinal epithelial cells. Eur J Immunol. 2001;31(10):2892–900. doi: 10.1002/1521-4141(2001010)31:10<2892::aid-immu2892>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 6.Taylor DD, Akyol S, Gercel-Taylor C. Pregnancy-associated exosomes and their modulation of T cell signaling. J Immunol. 2006;176(3):1534–42. doi: 10.4049/jimmunol.176.3.1534. [DOI] [PubMed] [Google Scholar]
- 7.Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110(9):3234–44. doi: 10.1182/blood-2007-03-079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Admyre C, Johansson SM, Paulie S, Gabrielsson S. Direct exosome stimulation of peripheral human T cells detected by ELISPOT. Eur J Immunol. 2006;36(7):1772–81. doi: 10.1002/eji.200535615. [DOI] [PubMed] [Google Scholar]
- 9.Andre F, Chaput N, Schartz NE, Flament C, Aubert N, Bernard J, Lemonnier F, Raposo G, Escudier B, Hsu DH, Tursz T, Amigorena S, Angevin E, Zitvogel L. Exosomes as potent cell-free peptide-based vaccine. I. Dendritic cell-derived exosomes transfer functional MHC class I/peptide complexes to dendritic cells. J Immunol. 2004;172(4):2126–36. doi: 10.4049/jimmunol.172.4.2126. [DOI] [PubMed] [Google Scholar]
- 10.Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4(5):594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 11.Abusamra AJ, Zhong Z, Zheng X, Li M, Ichim TE, Chin JL, Min WP. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells Mol Dis. 2005;35(2):169–73. doi: 10.1016/j.bcmd.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 13.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8(19):4083–99. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 14.Ilver D, Johansson P, Miller-Podraza H, Nyholm PG, Teneberg S, Karlsson KA. Bacterium-host protein-carbohydrate interactions. Methods Enzymol. 2003;363:134–57. doi: 10.1016/S0076-6879(03)01049-8. [DOI] [PubMed] [Google Scholar]
- 15.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126(5):855–67. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–9. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnamoorthy L, Bess JW, Jr., Preston AB, Nagashima K, Mahal LK. HIV-1 and microvesicles from T cells share a common glycome, arguing for a common origin. Nat Chem Biol. 2009;5(4):244–50. doi: 10.1038/nchembio.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara-Kuge S, Ohkura T, Ideo H, Shimada O, Atsumi S, Yamashita K. Involvement of VIP36 in intracellular transport and secretion of glycoproteins in polarized Madin-Darby canine kidney (MDCK) cells. J Biol Chem. 2002;277(18):16332–9. doi: 10.1074/jbc.M112188200. [DOI] [PubMed] [Google Scholar]
- 19.Morelle W, Stechly L, Andre S, Van Seuningen I, Porchet N, Gabius HJ, Michalski JC, Huet G. Glycosylation pattern of brush border-associated glycoproteins in enterocyte-like cells: involvement of complex-type N-glycans in apical trafficking. Biol Chem. 2009;390(7):529–44. doi: 10.1515/BC.2009.075. [DOI] [PubMed] [Google Scholar]
- 20.Potter BA, Weixel KM, Bruns JR, Ihrke G, Weisz OA. N-glycans mediate apical recycling of the sialomucin endolyn in polarized MDCK cells. Traffic. 2006;7(2):146–54. doi: 10.1111/j.1600-0854.2005.00371.x. [DOI] [PubMed] [Google Scholar]
- 21.Gupta G, Surolia A, Sampathkumar SG. Lectin microarrays for glycomic analysis. OMICS. 14(4):419–36. doi: 10.1089/omi.2009.0150. [DOI] [PubMed] [Google Scholar]
- 22.Krishnamoorthy L, Mahal LK. Glycomic analysis: an array of technologies. ACS Chem Biol. 2009;4(9):715–32. doi: 10.1021/cb900103n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu KL, Pilobello KT, Mahal LK. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat Chem Biol. 2006;2(3):153–7. doi: 10.1038/nchembio767. [DOI] [PubMed] [Google Scholar]
- 24.Pilobello KT, Slawek DE, Mahal LK. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proc Natl Acad Sci U S A. 2007;104(28):11534–9. doi: 10.1073/pnas.0704954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuno A, Matsuda A, Ikehara Y, Narimatsu H, Hirabayashi J. Differential glycan profiling by lectin microarray targeting tissue specimens. Methods Enzymol. 478:165–79. doi: 10.1016/S0076-6879(10)78007-1. [DOI] [PubMed] [Google Scholar]
- 26.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3, Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 27.Hsu KL, Gildersleeve JC, Mahal LK. A simple strategy for the creation of a recombinant lectin microarray. Mol Biosyst. 2008;4(6):654–62. doi: 10.1039/b800725j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pace KE, Hahn HP, Baum LG. Preparation of recombinant human galectin-1 and use in T-cell death assays. Methods Enzymol. 2003;363:499–518. doi: 10.1016/S0076-6879(03)01075-9. [DOI] [PubMed] [Google Scholar]
- 29.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30(4):e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura-Tsuruta S, Uchiyama N, Peumans WJ, Van Damme EJ, Totani K, Ito Y, Hirabayashi J. Analysis of the sugar-binding specificity of mannose-binding-type Jacalin-related lectins by frontal affinity chromatography--an approach to functional classification. FEBS J. 2008;275(6):1227–39. doi: 10.1111/j.1742-4658.2008.06282.x. [DOI] [PubMed] [Google Scholar]
- 33.Merkle RK, Cummings RD. Relationship of the terminal sequences to the length of poly-N-acetyllactosamine chains in asparagine-linked oligosaccharides from the mouse lymphoma cell line BW5147. Immobilized tomato lectin interacts with high affinity with glycopeptides containing long poly-N-acetyllactosamine chains. J Biol Chem. 1987;262(17):8179–89. [PubMed] [Google Scholar]
- 34.Ito N, Yokota M, Kawahara S, Nagaike C, Morimura Y, Hirota T, Matsunaga T. Histochemical demonstration of different types of poly-N-acetyllactosamine structures in human thyroid neoplasms using lectins and endo-beta-galactosidase digestion. Histochem J. 1995;27(8):620–9. [PubMed] [Google Scholar]
- 35.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9(21):4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 36.Rapoport EM, Kurmyshkina OV, Bovin NV. Mammalian galectins: structure, carbohydrate specificity, and functions. Biochemistry (Mosc) 2008;73(4):393–405. doi: 10.1134/s0006297908040032. [DOI] [PubMed] [Google Scholar]
- 37.Delacour D, Gouyer V, Zanetta JP, Drobecq H, Leteurtre E, Grard G, Moreau-Hannedouche O, Maes E, Pons A, Andre S, Le Bivic A, Gabius HJ, Manninen A, Simons K, Huet G. Galectin-4 and sulfatides in apical membrane trafficking in enterocyte-like cells. J Cell Biol. 2005;169(3):491–501. doi: 10.1083/jcb.200407073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delacour D, Greb C, Koch A, Salomonsson E, Leffler H, Le Bivic A, Jacob R. Apical sorting by galectin-3-dependent glycoprotein clustering. Traffic. 2007;8(4):379–88. doi: 10.1111/j.1600-0854.2007.00539.x. [DOI] [PubMed] [Google Scholar]
- 39.Kesimer M, Scull M, Brighton B, DeMaria G, Burns K, O’Neal W, Pickles RJ, Sheehan JK. Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense. FASEB J. 2009;23(6):1858–68. doi: 10.1096/fj.08-119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 11:108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Escrevente C, Morais VA, Keller S, Soares CM, Altevogt P, Costa J. Functional role of N-glycosylation from ADAM10 in processing, localization and activity of the enzyme. Biochim Biophys Acta. 2008;1780(6):905–13. doi: 10.1016/j.bbagen.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Vidal M, Mangeat P, Hoekstra D. Aggregation reroutes molecules from a recycling to a vesicle-mediated secretion pathway during reticulocyte maturation. J Cell Sci. 1997;110(Pt 16):1867–77. doi: 10.1242/jcs.110.16.1867. [DOI] [PubMed] [Google Scholar]
- 43.Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007;5(6):e158. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delacour D, Cramm-Behrens CI, Drobecq H, Le Bivic A, Naim HY, Jacob R. Requirement for galectin-3 in apical protein sorting. Curr Biol. 2006;16(4):408–14. doi: 10.1016/j.cub.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 45.Mishra R, Grzybek M, Niki T, Hirashima M, Simons K. Galectin-9 trafficking regulates apical-basal polarity in Madin-Darby canine kidney epithelial cells. Proc Natl Acad Sci U S A. 107(41):17633–8. doi: 10.1073/pnas.1012424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.