Abstract
Carbohydrates encode biological information necessary for cellular function. The structural diversity and complexity of these sugar residues have necessitated the creation of novel methodologies for their study. This review highlights recent technological advancements that are starting to unravel the intricate web of carbohydrate biology. New methods for the analysis of both glycoconjugates and glycan structures are discussed. With the use of these innovative tools, the field of glycobiology is poised to take center-stage in the post-genomic era of modern biology and medicine.
Glycomics or the systems-level study of carbohydrates (glycans) has emerged as an important area of research in the post-genomic era. Although proteins are tagged with a variety of post-translational modifications including phosphorylation, acetylation, and methylation, glycosylation is the most prevalent modification, occurring on at least 50% of all proteins (1). The vast majority of glycoproteins and glycolipids are found at the cell membrane creating a carbohydrate-coated surface which communicates with the extracellular world. Interactions with cell surface oligosaccharides play a critical role in numerous biological events including differentiation, cellular adhesion, immune responses, and host-pathogen interactions (2–7). For example, the expression of an α-2,6-sialyltransferase (ST6GALNAC5) has been shown to mediate metastasis of breast cancer cells to the brain by enhancing adhesion of tumor cells to brain tissue (8). Although there is increasing evidence for critical biological roles of carbohydrates, the challenges posed by the diversity of glycans have slowed progress in understanding this class of biopolymers. This review delineates the challenge of glycomics and how it is being met by the advent of new technologies to analyze both structural and functional aspects of carbohydrates.
Analytical challenge of glycomics
Structural features of the glycome
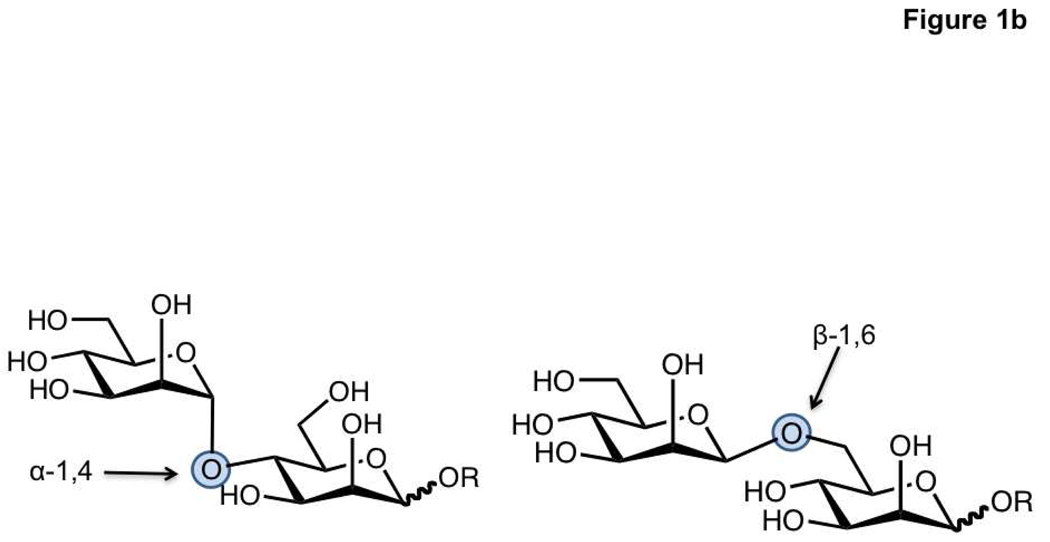
Glycoconjugates, defined as both glycolipids and glycoproteins, are known to occur in organisms ranging from bacteria to mammals. Most work in glycomics has focused on mammalian glycans which consist of 10 monosaccharide units that form the building blocks for complex oligosaccharide polymers (Figure 1, panel a) (9). In comparison to nucleic acids and proteins, carbohydrates possess certain unique properties that render their analysis difficult. First, monosaccharide monomers can be connected via one of many hydroxyl groups, resulting in a diverse set of linkages that form either linear or branched structures. In addition, stereochemistry at the linkage site can be either α or β, resulting in a high number of possible glycan epitopes (10, 11). For example, two mannose residues can theoretically be connected by α-1,2, α-1,3, α-1,4, α-1,6, β-1,2, β-1,3, β-1,4, or β-1,6 linkages, although only some of these connections have been observed (Figure 1, panel b). Carbohydrate residues can be further modified by sulfation, phosphorylation and acetylation (2, 12–14). This stereochemical and structural diversity provides a major analytical challenge to glycomics.
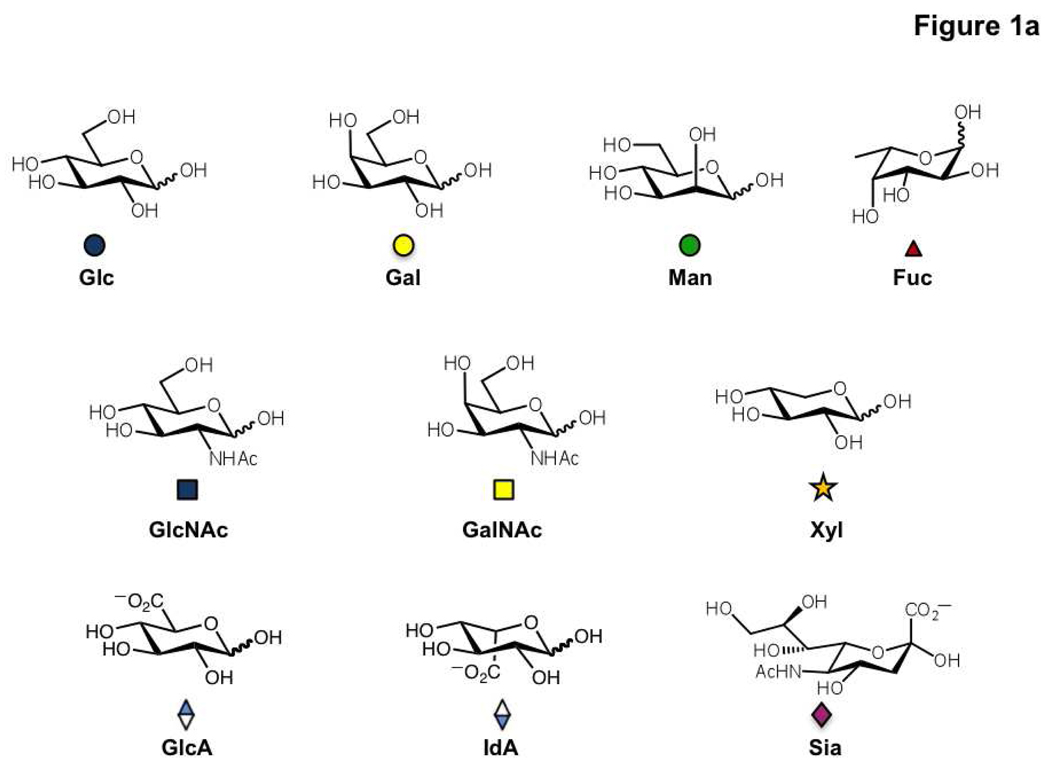
Figure 1. Building blocks of the mammalian glycome.
a) The structures of the ten carbohydrate residues found in mammalian cells. These are glucose (Glc), galactose (Gal), mannose (Man), fucose (Fuc), N-acetylglucosamine (GlcNAc), N-acetylgalactosamine (GalNAc), xylose (Xyl), glucuronic acid (GlcA), iduronic acid (IdA) and sialic acid (Sia). The more commonly used three letter codes and the symbolic notations used by the Consortium for functional glycomics are shown. b) The diversity of possible stereochemical and structural linkages in a mannose disaccharide is illustrated.
Molecular contexts of the glycome
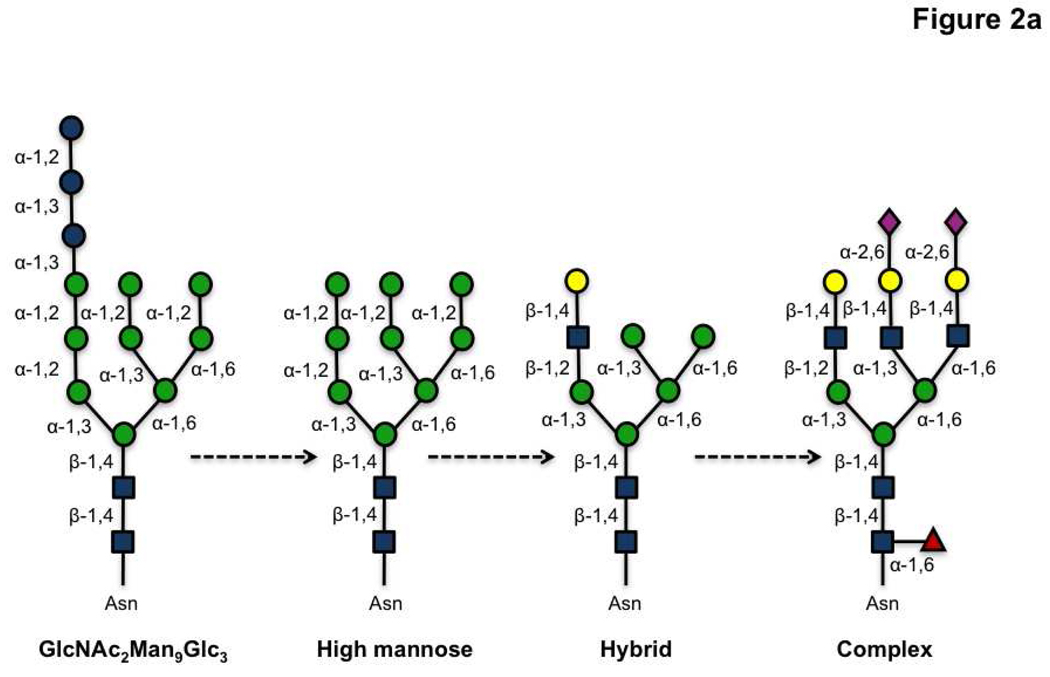
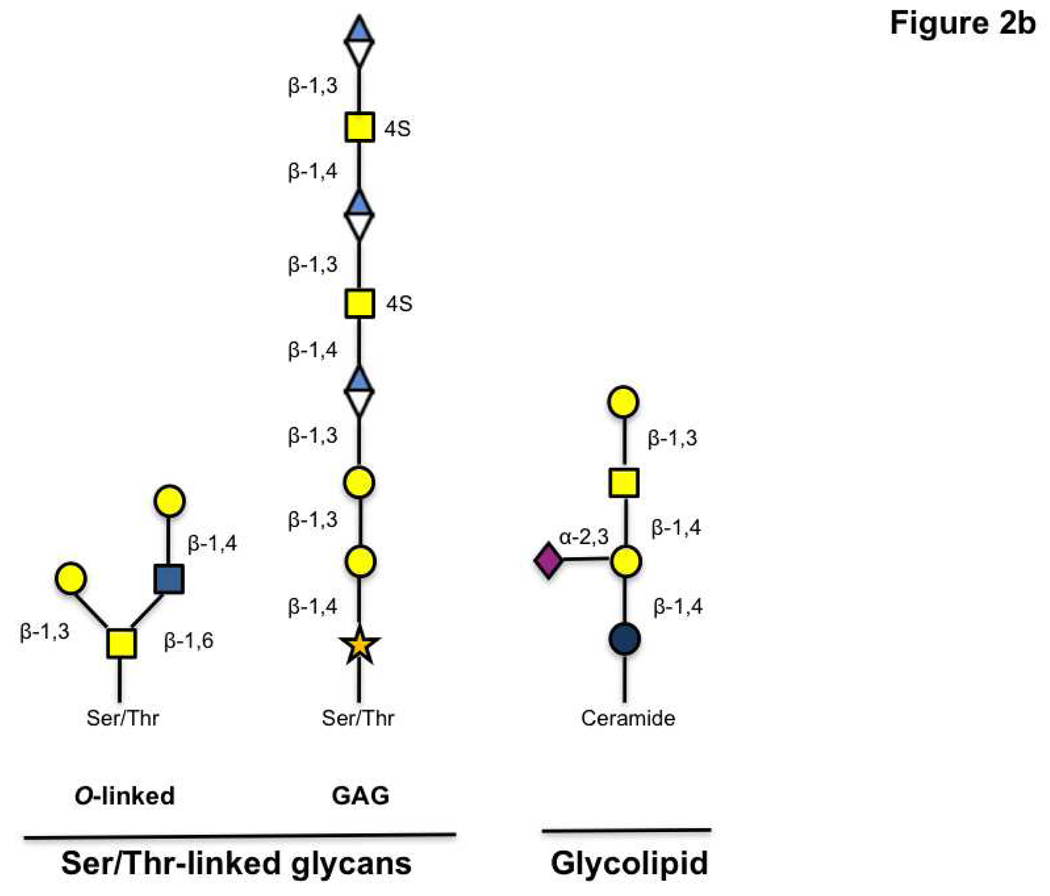
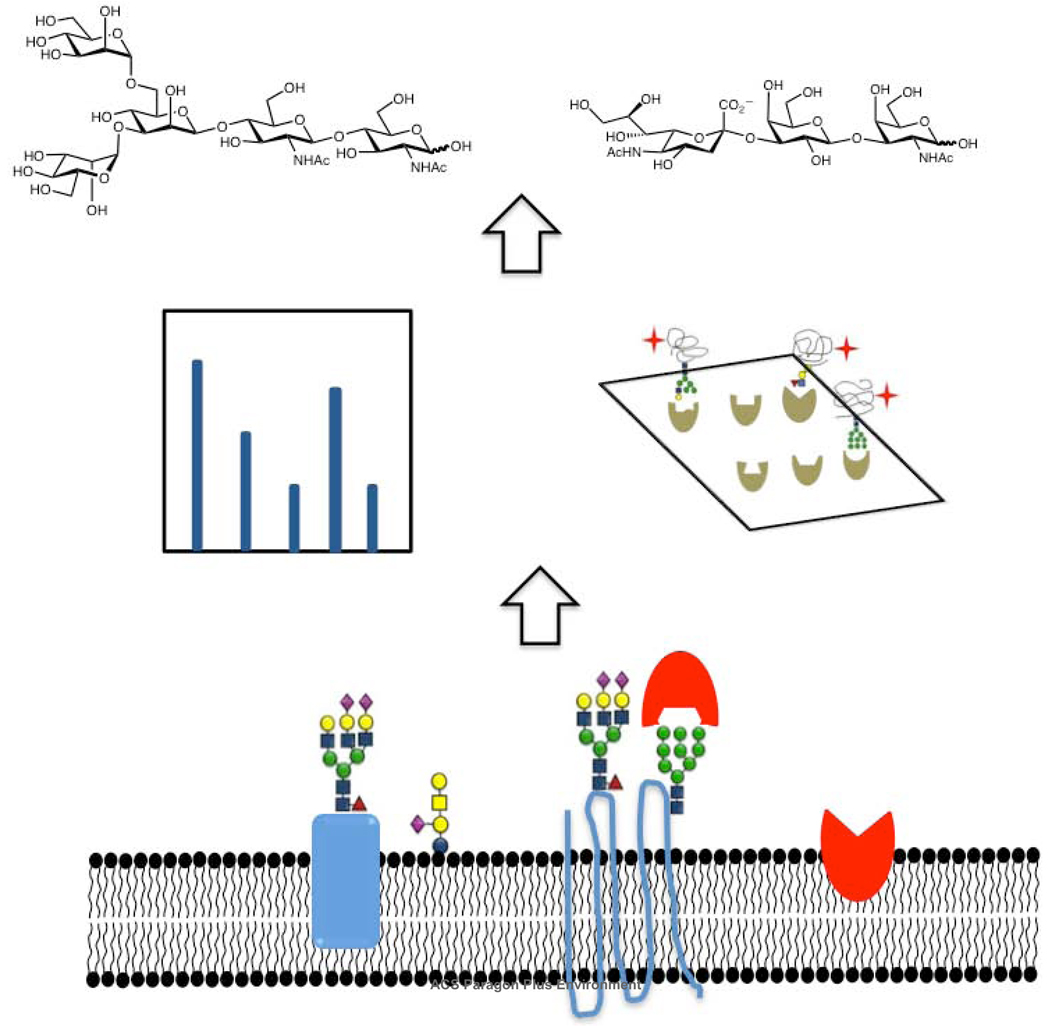
Glycan analysis is also complicated by the multitude of contexts in which this modification is found. In proteins, glycans can be attached at N-linked (Asn), O-linked (Ser/Thr) and C-linked (Trp) sites. Additionally they are found in glycolipids including globosides and glycosphingolipids (9, 13). N-linked glycosylation occurs at the consensus sequence Asn-X-Ser/Thr (X= any amino acid except proline), and starts with the transfer of the core glycan GlcNAc2Man9Glc3 from a dolichol phosphate precursor to the asparagine residue. Not all predicted sites are modified and these differences in glycan occupancy contribute to the heterogeneous composition of glycoproteins (microheterogeneity) (2, 12, 14–16). Subsequent trimming and modification of the common core N-glycan creates a series of discrete glycan structures (Figure 2, panel a). In contrast, O-linked glycans are typically elaborated by subsequent additions of monosaccharides to an α-linked N-acetylgalactosamine (GalNAc) moiety linked to Ser/Thr residues (Figure 2, panel b). Unlike N-linked glycosylation, no consensus sequence has yet been found for this modification. Glycosylaminoglycans (GAGs), another common form of Ser/Thr linked sugars, are elaborated off of a xylose core attached to the amino acid. In addition, fucose, mannose, and glucose have also been observed on Ser/Thr residues at the cell surface (2, 13, 17). In the nucleus and cytoplasm, the simple monosaccharide, β-O-N-acetylglucosamine (O-GlcNAc) is known to modify Ser/Thr residues and has been implicated in cellular signaling (18, 19). Glycolipids can also differ in their structures. For example, ceramide containing lipids can be elaborated from either a galactose or glucose core residue attached to the lipid portion (9). Taken together, this diversity of contexts presents a significant challenge for glycomics.
Figure 2. Representative structures of glycans found in the mammalian glycome.
a) A series of N-linked glycans are derived from trimming and addition of various residues on the GlcNAc2Man9Glc3 precursor. b) Ser/Thr-linked glycans and glycolipids are shown.
Biological challenges presented by the glycome
Our understanding of glycans is also complicated by the fact that synthesis of glycan motifs is not template driven i.e there is no direct genetic code for this complex modification. Several factors including levels of glycosyltransferases, glycosidases, nucleotide sugar transporters, and protein and lipid trafficking contribute significantly to the glycome of an organism or cell (12, 20). Redundancy in the glycosyltransferases further hampers our ability to dissect glycan synthesis as demonstrated by gene knockout experiments (21). Consequently, we are still striving to predict the glycome from genomic information, a current challenge in the field (22–24).
A final complicating factor in our understanding of glycosylation is the multivalent nature of glycan-protein interactions in biology. Monovalent interactions between carbohydrates and proteins such as lectins are typically in the millimolar to micromolar range (25). However, physiological interactions are often effected by increased avidity due to multivalent binding of carbohydrates to proteins. Local glycan concentration is determined by both the mutivalent presentation of glycans within a single protein and the clustering of glycoproteins and glycolipids on the cell surface membrane (2, 25–28). Thus, it is important to examine glycan-protein interactions using appropriate methodologies which take into account the effects of multivalency and presentation. The biology of the glycome is a reflection of all these different factors, and requires a multi-pronged approach to comprehend in its entirety.
Meeting the challenge of glycomics
Meeting the challenge of glycomics requires the integration of techniques from multiple disciplines including chemistry, biochemistry, molecular biology, bioinformatics and biotechnology. Herein we provide an overview of these new and exciting methods. The first section focuses on the application of several technologies towards expanding our knowledge of the glycoproteome. The second section covers technologies for determining the structure of the glycome including mass spectrometry, HPLC and lectin microarrays. The final section provides an overview of an emerging technology for studying glycan-protein interactions – carbohydrate microarrays. Taken together, these techniques are stimulating a new understanding of the glycome.
Tools for identifying the glycoproteome
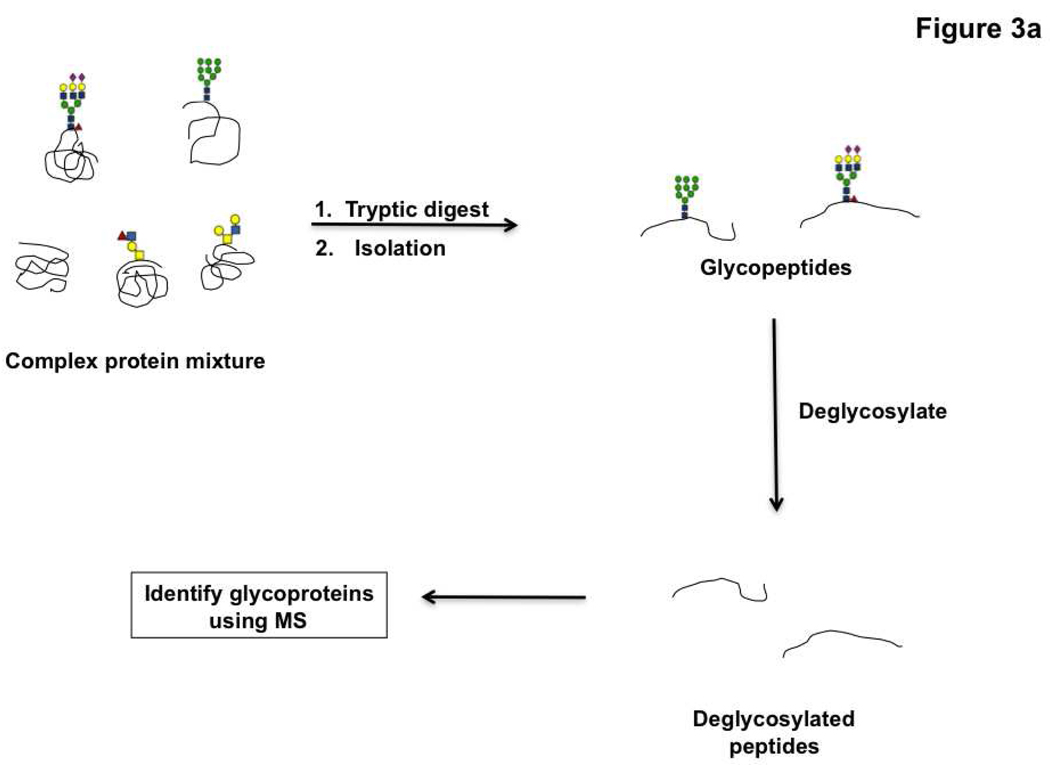
To understand the role of glycans in modulating protein and lipid function, it is necessary to identify the molecular context of these oligosaccharides. The majority of work in this area has focused on glycoproteomics, a sub-discipline of proteomics that deals with the identification of proteins bearing glycans within the complex proteomic milieu, rather than on the carbohydrates themselves. In a typical glycoproteomic analysis, proteins are enzymatically cleaved using proteases and the glycopeptides are isolated using a variety of techniques. These peptides are then deglycosylated and the glycoproteins identified using mass spectrometry (Figure 3, panel a).
Figure 3. Glycoproteomic strategies.
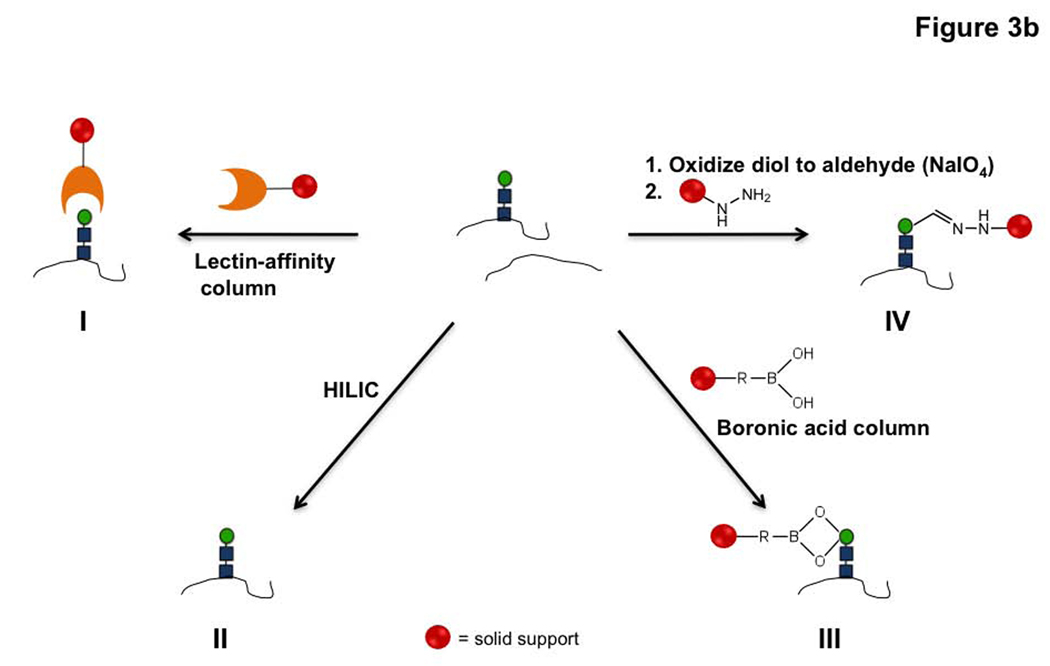
a) General schematic for glycoproteomic analysis by mass spectrometry. b) Different methods used for the isolation of glycopeptides include the following, I. Lectin affinity chromatography to pulldown proteins with specific glycans. II. HILIC based isolation. III. Boronic acid based enrichment methods. IV. Oxidation of carbohydrates followed by capture with hydrazide beads.
The isolation techniques employed are the main points of difference between different glycoproteomic strategies (Figure 3, panel b). The most widely applied isolation technique involves the use of lectin affinity chromatography for the separation of proteins modified with glycans (29–31). Lectins are carbohydrate-binding proteins that are not enzymes or antibodies, but which can bind to specific sugar residues (32, 33). Thus, these reagents can be used to differentially isolate subpopulations of the glycosylated pool that contain a particular sugar moiety. Glycopeptides that are selectively enriched on the lectin columns are often eluted by incubation with competing sugar molecules (29, 31). This general procedure has been used primarily for isolation of N-linked glycoproteins (34–36), although the O-linked subset of the proteome can also be characterized (37, 38). The choice of lectin affinity column determines whether glycosylated proteins containing a specific sugar residue (39, 40), for example fucosylated glycoproteins (36, 41), or a wider subset of the glycoproteome is targeted (42–44). Glycoproteins from a diverse array of samples including sera (42, 45), cancer cells (34, 35) and tissues (46, 47) have been identified using this method. For example, specific changes in the fucosylation of target biomarkers in the sera of patients with hepatocellular carcinoma using this technique (41). One limitation of this technology however is that the lectin column chosen biases which glycoproteins are identified based on glycan composition, an issue for identifying the whole glycoproteome rather than a specific subset.
In order to increase coverage of the glycoproteome, several chemical approaches have been developed for isolation (Figure 3, panel b). One of the simplest methods is the enrichment of glycopeptides through hydrogen bonding interactions with cellulose or sepharose matrices (hydrophilic affinity chromatography, HILIC) (48–51). Although this method can be used for identifying both N-and O-linked glycoproteins, the reproducibility of the results is not ideal (52). Boronic acid based enrichment techniques have also been employed for the separation of glycoproteins. Boronic acid conjugated to a solid support forms a covalent bond with molecules containing cis-diol groups such as mannose and galactose and thus can selectively isolate glycopeptides (53, 54). An alternate method for identifying the N-linked glycoproteome relies on a combination of mild oxidation and the N-linked specific enzyme, PNGase F. Sodium periodate treatment oxidizes cis-diol groups in carbohydrate residues to aldehydes. These modified proteins are then reacted with either biotin hydrazide, for isolation with avidin beads, or with hydrazide beads, thereby pulling down glycoproteins selectively. Treatment of the solid support with proteases followed by PNGase F releases the N-linked glycopeptides which are subsequently identified (55–59). Recently, this technique was selectively applied to the identification of cell surface glycoproteins and used to visualize dynamic glycoproteomic changes that occur during T-cell activation and stem cell differentiation (60, 61).
Combinations of these approaches have recently been used to maximize coverage of the glycoproteome. McDonald and co-workers demonstrated that isolation of N-linked glycoproteins by the hydrazide capture method yielded dramatic differences in the proteins identified when compared to lectin affinity chromatography. Thus, these technologies can be used in tandem to isolate a broader spectrum of the N-linked glycoproteome (62). Boronic acid based enrichment has also been used together with lectin affinity chromatography to enhance the range of glycoproteins identified (63). Using hydrophilic affinity chromatography and hydrazide capture methods in parallel, a broader dataset of ~300 identified glycoproteins was obtained for the secretome of cancer cells (64). The use of these methodologies, either in parallel or in tandem, is still only capable of identifying fractions of the glycoproteome, thus emphasizing the need for further improvements in this area.
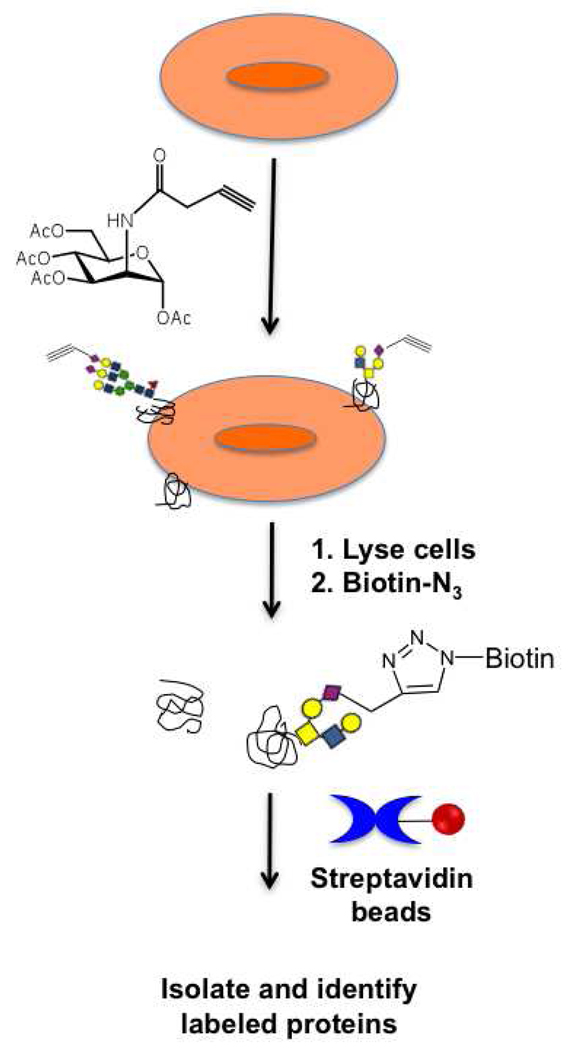
An alternative technique for selective isolation of a subset of the glycosylated pool takes advantage of chemical probes. These probes are unnatural analogs of simple sugars containing a bioorthogonal reactive handle. Typically, cells are grown in the presence of these probes, which are metabolized and incorporated into newly synthesized glycans via native biosynthetic machinery. A compatible reagent that reacts with the bioorthogonal handle is then used to either track these modified glycoproteins in living cells for in vivo imaging (65–67) or to isolate tagged proteins for glycoproteomic analysis (68) (Figure 4). For example, recent work has identified the sialylated portion of the glycoproteome using an alkynyl modified N-acetylmannosamine precursor to orthogonally label the sialoproteome. Selective biotinylation via a Huigsen [3+2] cycloaddition to a biotin-azide, followed by pulldown with streptavidin beads allowed for the identification of the tagged glycoproteins. Although this method cannot be used for clinical samples, it is still a valuable technique as unnatural analogs of different sugars can be used to identify specific subsets of the glycoproteome (68). This strategy has also been adapted for the metabolic labeling of proteins modified with O-GlcNAc using an azide derivative of GlcNAc (69, 70). An alternate scheme to identify O-GlcNAcylated proteins exploits the promiscuity of a mutant glycosyltransferase to label O-GlcNAc residues with a ketone modified sugar analog. The ketone can then be used for isolation and subsequent identification of the glycoproteins (71–73). The development and application of these novel chemical tools has added a new dimension to the field of glycoproteomic analysis.
Figure 4. Metabolic labeling strategies for glycoproteomics.
Cells grown in the presence of alkynyl sugars are lysed, and the lysates are reacted with biotin-azide. Glycoproteins are isolated via pulldown with streptavidin beads and subjected to identification by mass spectrometry.
One drawback common to these methods is that they cannot accurately identify the sites of glycan modification in proteins. Typically, the potential N-linked glycosylation sites in glycopeptides are assigned based on the presence of the consensus sequence (36, 49, 52). For a predicted N-linked glycan the site occupancy can vary (microheterogeneity), multiple consensus sequences may exist within a peptide, and nonglycosylated peptides can contaminate glycopeptide isolations, thus it is important to incorporate additional methods to accurately identify the sites of glycosylation in a glycoproteomic analysis (74, 75). One method relies on the endoglycosidases Endo D and H which cleave the bond connecting the two GlcNAc residues in N-linked sugars. The use of these enzymes, in tandem with other glycosidases, leaves the first GlcNAc residue at sites of glycosylation allowing site specific detection via mass spectrometry (50, 76). A more widely used method for identifying these sites is based on the use of PNGase F, which converts the asparagine into aspartic acid during deglycosylation. This change can be detected via either the subtle shift in mass (1 Da) (77) or the incorporation of an isotopic label. Deglycosylation in the presence of H2 18O incorporates the oxygen isotope at sites of N-glycosylation creating a more detectable mass signature (3 Da) (74, 75). For both methods the mass signature combined with the presence of the consensus sequence is used to identify specific sites of occupancy. Isotopic incorporation has been used to identify novel sites of glycosylation in human platelet proteins (78). Additionally, sites of O-GlcNAcylation have been identified using beta elimination followed by a Michael addition by dithiothreitol to tag the modified site (79). The use of both traditional and newly developed chemical tools has significantly enhanced the field of glycoproteomics.
Tools for identifying structure of the glycome
Mass spectrometry
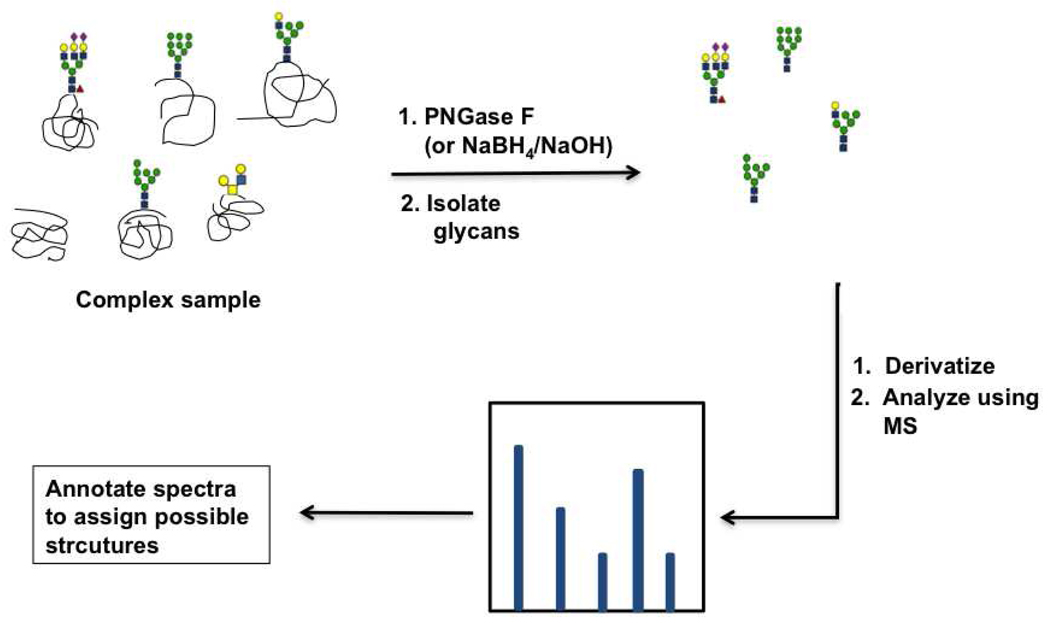
Glycomic analysis by mass spectrometry presents an inherently greater challenge than glycoproteomic analysis. The structural variations in glycan linkages coupled with the identical mass of epimeric monosaccharides makes identification of glycan structures difficult (14, 80). Mass spectrometric analysis of glycans typically consists of the following steps 1) glycan release by either PNGase F treatment (for N-linked glycans) or β-elimination (for O-linked glycans) 2) glycan isolation using solid phase separation techniques 3) glycan derivatization and ionization and 4) annotation of the resulting spectra to list possible glycan structures (Figure 5) (80–87). In the past few years, significant advances have been made in the isolation, derivatization, and ionization techniques used for glycomic analysis. These methods increased reproducibility and considerably reduced the time required for glycomics by mass spectrometry (for a comprehensive review on this topic, see (80)). However, the interpretation of spectral data is still a major bottleneck for high-throughput analysis. This is due both to the mass degeneracy of glycans and to the complicated fragmentation patterns which arise from the more complex MS strategies required to obtain precise structural information. Although, programs exist to automate some aspects of data interpretation such as Cartoonist, GlycoPep DB and SysBio Ware, software based interpretation of glycan spectral data is still in its infancy (88–90). The recent advances in mass spectrometry have made it a widely used technique for the detection of glycan epitopes from a wide range of samples including bacterial and mammalian cells, sera, and tissue samples (80, 82, 91–97). In recent work, mass spectrometry was used to identify glycan epitopes which were specifically elevated in hepatocellular carcinoma patients in a study involving ~200 participants, and this underscores the power of this technique for glycomic analysis (94).
Figure 5. Mass spectrometry based analysis of glycans.
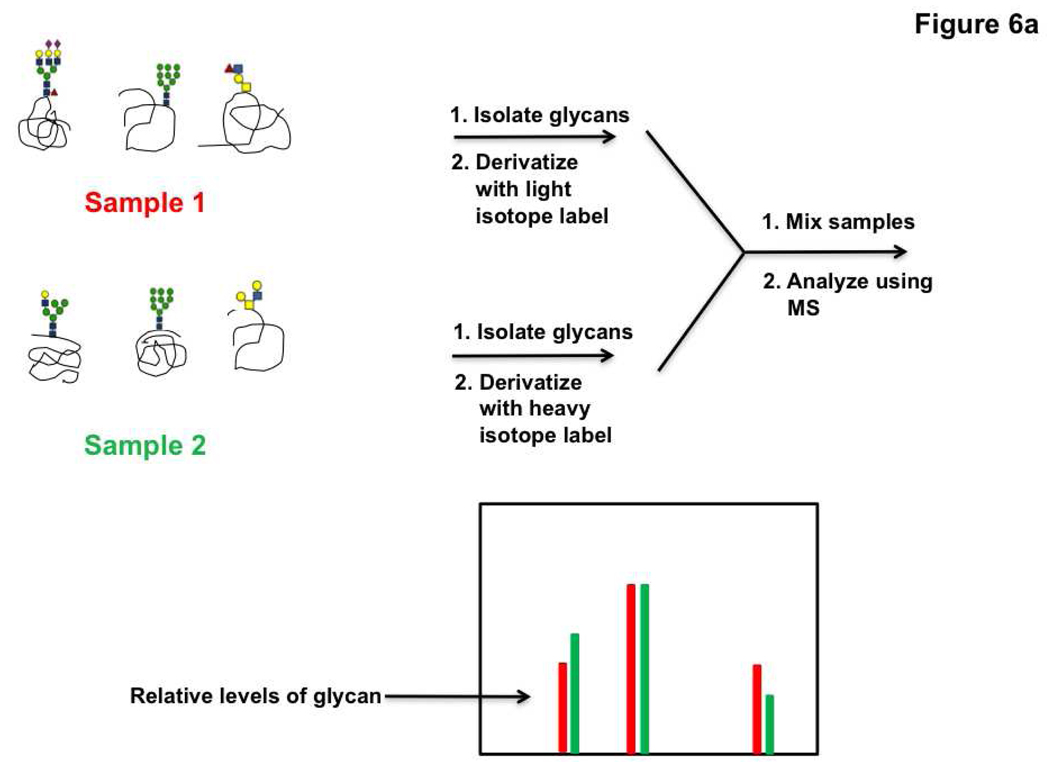
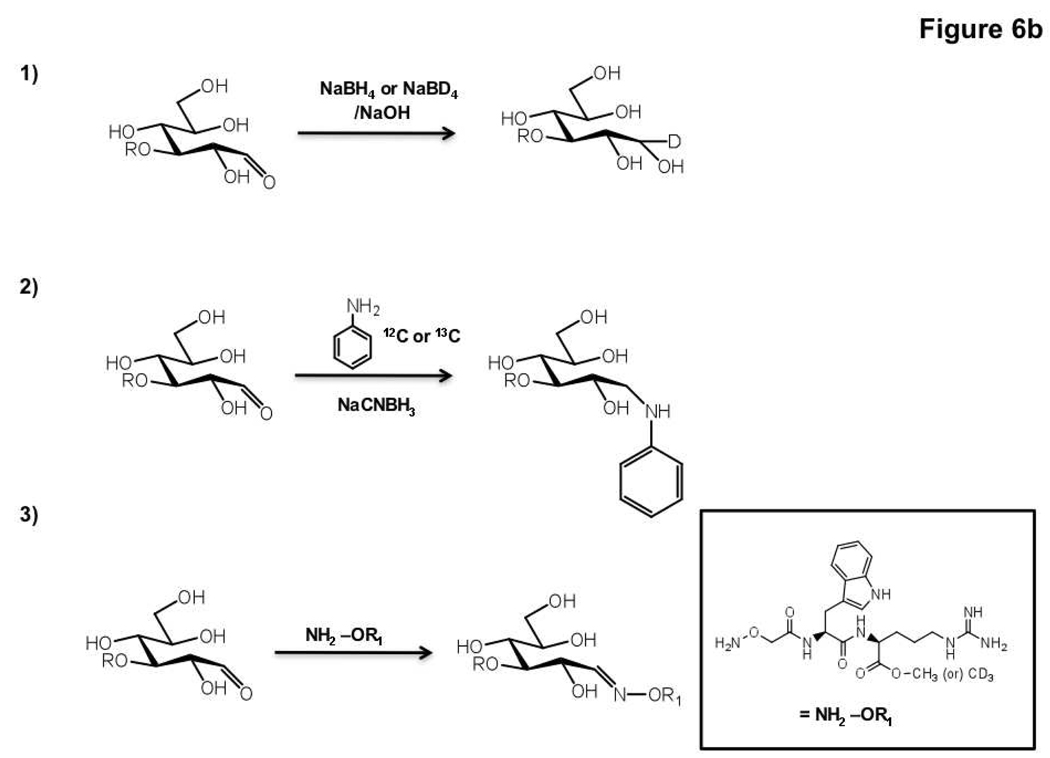
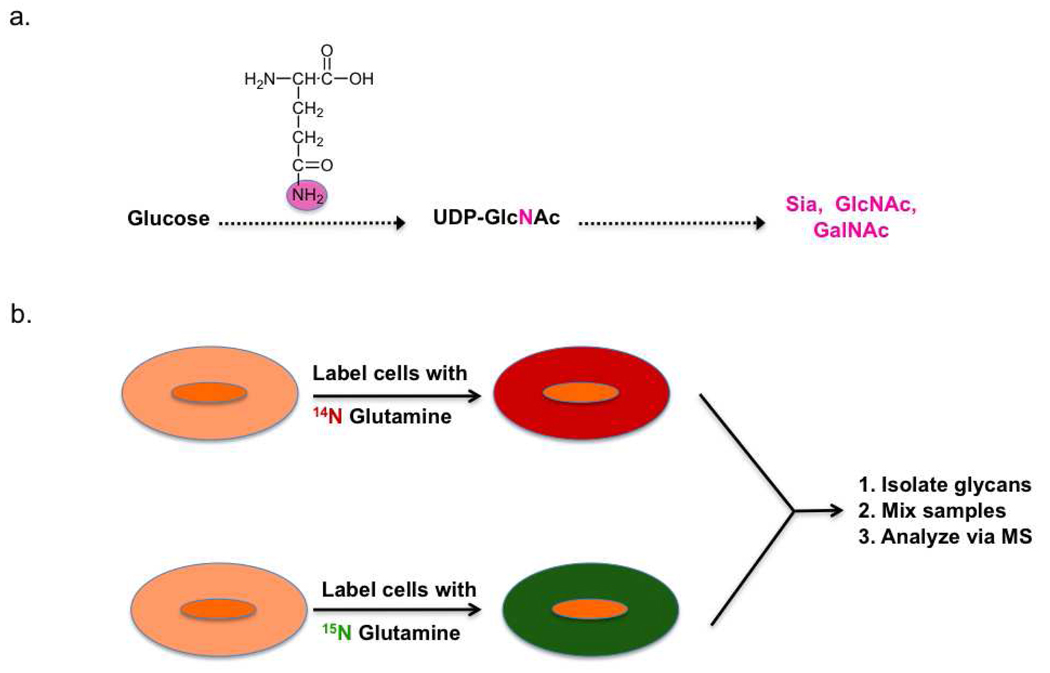
Following the example of work enabling comparative proteomic profiling, isotopic labeling strategies have recently been developed for comparative glycomics by mass spectrometry. In brief, glycans are differentially labeled with isotope tags, mixed and analyzed via mass spectrometry (Figure 6, panel a). Labeling of the glycans can be achieved via either chemical or metabolic means. Chemical derivatization methods take advantage of the unique chemistry at the reducing end of a cleaved glycan, namely the cyclic hemiacetal which is in equilibrium with the open-chain aldehyde (Figure 6, panel b). Several different labeling reagents have been developed for the incorporation of an isotopic tag. The simplest of these is the reduction of the open-chain aldehyde by a labeled reducing agent such as deuterated sodium borohydride (98). Reductive amination using 13C labeled aniline has also been used to selectively derivatize glycans from serum samples and obtain relative concentrations of sugars (99). Differential labeling of samples is also achieved with deuterated hydroxyl amines which can react with cleaved glycans to form the corresponding oximes (100). Relative quantitation of either O-linked or N-linked glycans can be obtained using these methods (98, 100–105). Recently metabolic labeling of glycans has been achieved by a clever new strategy called IDAWG (isotopic detection of aminosugars with glutamine). In this method, glutamine containing heavy or light isotopes of nitrogen in the amide of the side chain is incubated with cells. Since the amide nitrogen is the sole nitrogen source in the synthesis of UDP-GlcNAc, this isotope is subsequently incorporated to GlcNAc, GalNAc and sialic acid residues (Figure 7, panel a). Comparative glycomic profiles of cells grown in the light and heavy medium are then obtained (Figure 7, panel b). This approach has been used to differentially label and visualize glycan epitopes during differentiation of embryonic stem cells (106). Given that these sugars are found in both N-linked and O-linked glycans, this technique holds great promise for tracking dynamic changes in glycan synthesis and degradation.
Figure 6. Differential glycan analysis using isotope labeling and mass spectrometry.
a) Schematic of a typical isotope labeling experiment for quantitative mass spectrometric analysis of glycans. b) Examples of isotopic labeling reactions. The reactive open-chain aldehyde form of the glycan is shown.
Figure 7. Metabolic isotope incorporation for quantitative glycomics (IDAWG).
a) Labeled glutamine is incorporated into select glycans via native biosynthetic machinery. b) Cells are labeled with differential isotopes of glutamine and their glycan compositions are compared by mass spectrometry as in Figure 6, panel a.
Although mass spectrometry can be a powerful technique for glycomics, there are several issues that are yet to be resolved. Because of the need to simplify and purify the glycome prior to analysis mass spectrometric techniques often provide a snapshot of a select portion of the glycome. Most commonly only the N-linked glycome is analyzed as in the example given previously for hepatocellular carcinoma (94). Relatively few methods exist for the identification of glycans from GAGs (107, 108), and glycolipids (109, 110). Thus, these portions of the glycome are currently underrepresented in the glycomic data set. A second major issue for mass spectrometry as a high-throughput glycomic technique is the fact that further analysis is typically required to obtain linkage information from a mass spectrometric glycan fingerprint (85). Annotation of the N-linked glycome from a simple spectra typically relies upon assumptions based on glycan biosynthetic pathways, limiting these analyses to well characterized biological systems.
High performance liquid chromatography (HPLC)
An alternate method for glycomic analysis is based on the separation of oligosaccharides using HPLC. This technique relies on the unique chemistry of carbohydrates to label and visualize their separation. Typically, N-linked glycans are released using PNGase F, tagged at the reducing end with a fluorescent label such as 2-aminobenzamide or anthranilic acid, and analyzed using HPLC. The differential HPLC traces generated can be used to compare the general N-linked glycomic profiles of multiple samples. This methodology has been used for examining the N-linked glycan pools in samples such as serum (111, 112). A recent study utilized this method to observe high variability in the human N-linked glycome, providing initial evidence for the role of genetics and environmental factors in the composition of human glycome (112). A caveat to this method is that the HPLC profiles can contain multiple glycans in each peak and thus changes in the HPLC profiles are difficult to interpret at the level of individual glycan structures. Further analysis via enzyme degradation and sophisticated data interpretation is needed to obtain more specific structural information from this technique. The development of automated tools for annotation of HPLC-derived glycomic profiles and new glycan HPLC standards may help to offset some of these disadvantages (113).
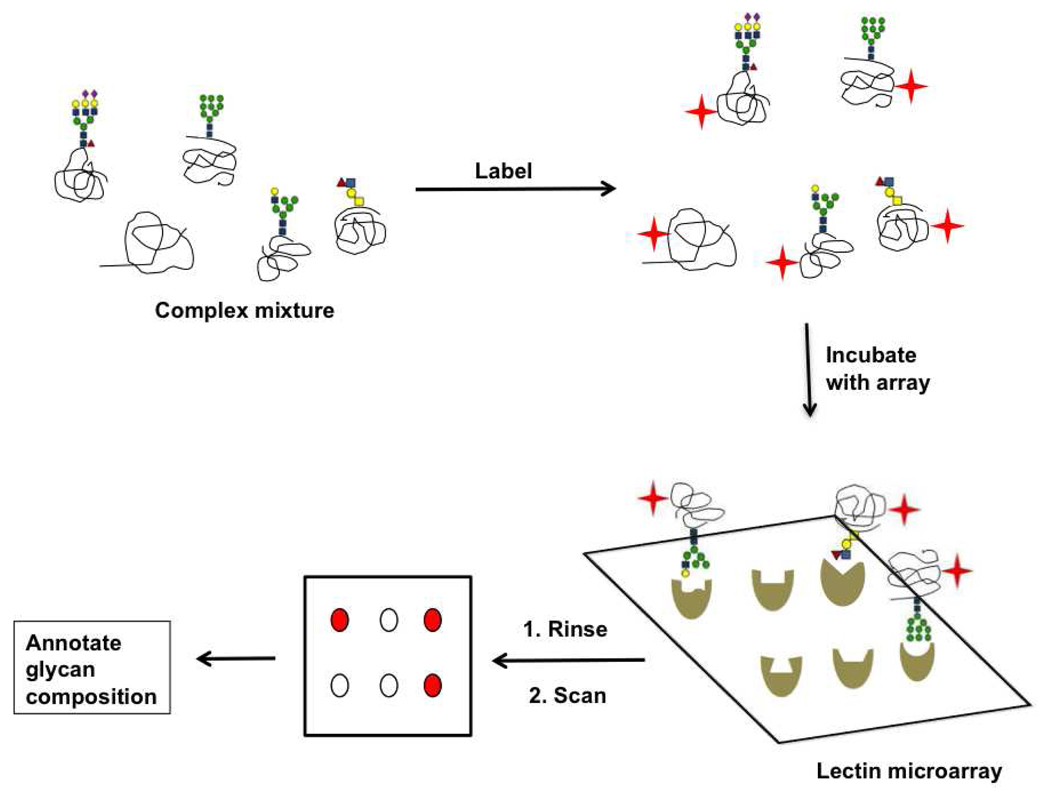
Lectin microarray
A simple yet powerful new technology for comprehensive glycan analysis, lectin microarrays, consist of a panel of lectins with distinct binding specificities that are spotted onto a solid support. The microarray is then interrogated with fluorescently labeled analytes and the known binding specificities of lectins are used to interpret the resultant pattern and annotate glycan epitopes present in the sample of interest (Figure 8) (114–116). Glycan epitopes from a variety of samples including glycoproteins (116–119), cellular membranes (120), whole mammalian cells (121, 122), pathogenic bacteria (115, 123), and viruses (124) have been profiled using this technique.
Figure 8. Lectin microarray technology for glycomic analysis.
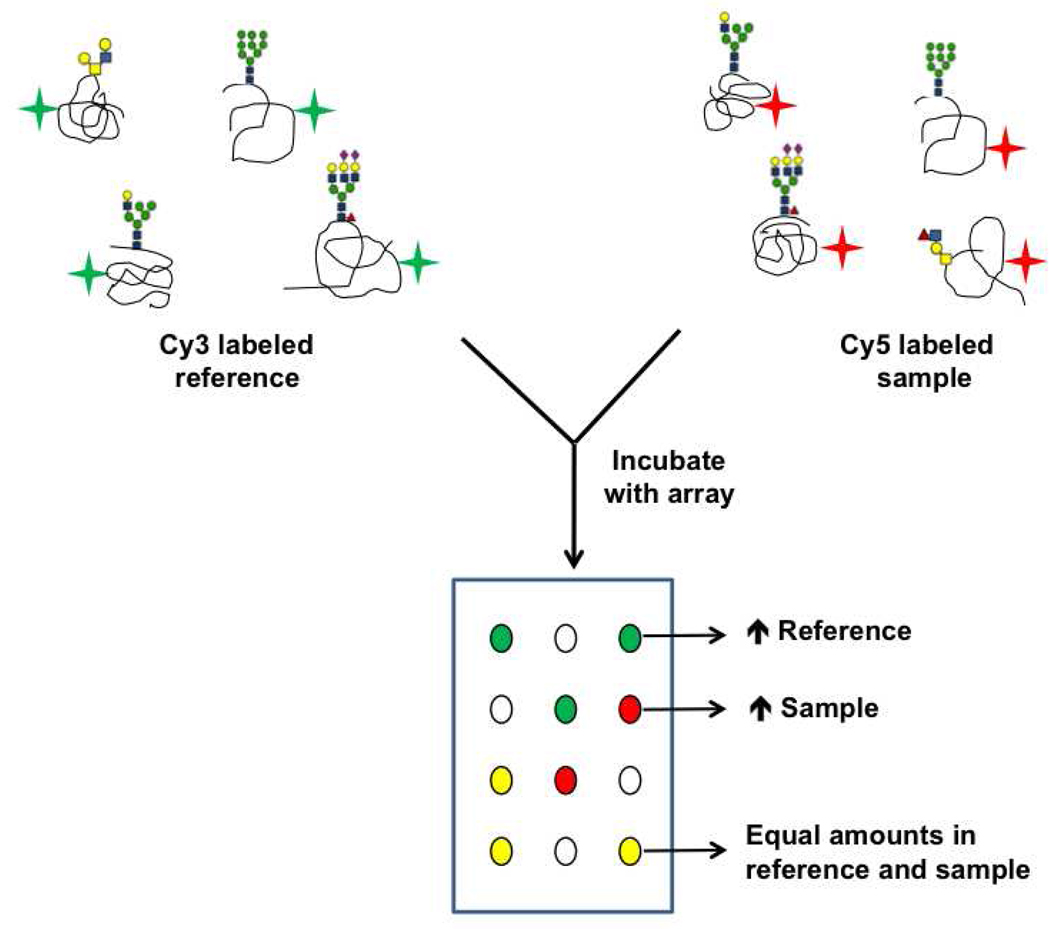
Our laboratory has advanced this technology further by developing a sensitive and robust two-color approach for the semi-quantitative analysis of glycans from complex mixtures (Figure 9). In brief, the sample and a common biological reference are differentially labeled with Cy3- and Cy5- dyes respectively, mixed in equal amounts, and used to probe a lectin microarray. Competitive binding between the two analytes gives ratiometric data. The use of a common reference addresses issues arising from differences in sample labeling and inherent differences in lectin activity that can arise from minor alterations in the print conditions and variations observed with different lectin preparations (120). Moreover, the direct comparison of multiple samples to a single internal standard allows subtle changes in levels of glycans to be unraveled using this approach, in contrast to the more commonly used single color approach (120, 125). Recently, we utilized our two-color method for the comparative glycomics of whole HIV-1 virions and immunomodulatory nanoparticles called microvesicles. This analysis revealed that the glycomes of HIV-1 and microvesicles derived from T-cells are nearly identical, lending strong support to the theory that HIV-1 co-opts the microvesicular exocytic mechanism to exit T-cells (124).
Figure 9. Ratiometric method for lectin microarray analysis.
Two-color approach enables comparative glycomic profiling of samples.
Lectin microarray technology has several advantages over other glycan analysis techniques. Unlike analysis by mass spectrometry, it is possible to simultaneously observe glycolipids and N- and O-linked oligosaccharides using the array (120). Additionally the lectin microarray format is multivalent, mimicking the physiologically relevant context of glycan-protein interactions. Because the binding specificities of many of these lectins have been extensively characterized using carbohydrate arrays (see below), it is possible to obtain linkage-specific information on the glycome, which is another advantage of the arrays. Lectin microarrays can be easily fabricated and analyzed using widely available equipment (114, 115). In addition, there are ~100 commercially available lectins, the majority of which are naturally purified from plant sources and are currently used in the array format (9, 14, 126, 127). These factors combine to make lectin arrays a powerful technique for deconvoluting the glycome.
It should be noted, however, that this technology still have several drawbacks. First, glycan structural elucidation is restricted to the motifs that are recognized by the lectins on the array. Thus, the availability of carbohydrate binding proteins is a limiting factor. For example, there is a dearth of carbohydrate-binding proteins that are known to be specific for unique bacterial glycans; therefore these epitopes are invisible to current lectin microarrays. Additionally, some of the plant lectins are glycosylated which poses a potential problem when the array is probed with samples that may contain endogenous lectins (128). Currently, we and others are using a wide variety of approaches including recombinant lectins (128), the creation of synthetic receptors (129) and the evolution of new lectin epitopes in efforts to increase the number of unglycosylated carbohydrate binders for the detection of diverse glycan epitopes (128, 130, 131). A final drawback is that unlike the more detailed analysis that can be obtained by higher order mass spectrometry, this technique allows the identification of structural epitopes but not unique molecular structures. However, at present it is the specific epitopes, rather than individual structures, that are thought to encode biological activity and this methodology provides a rapid method of assessing those epitopes for a large number of samples.
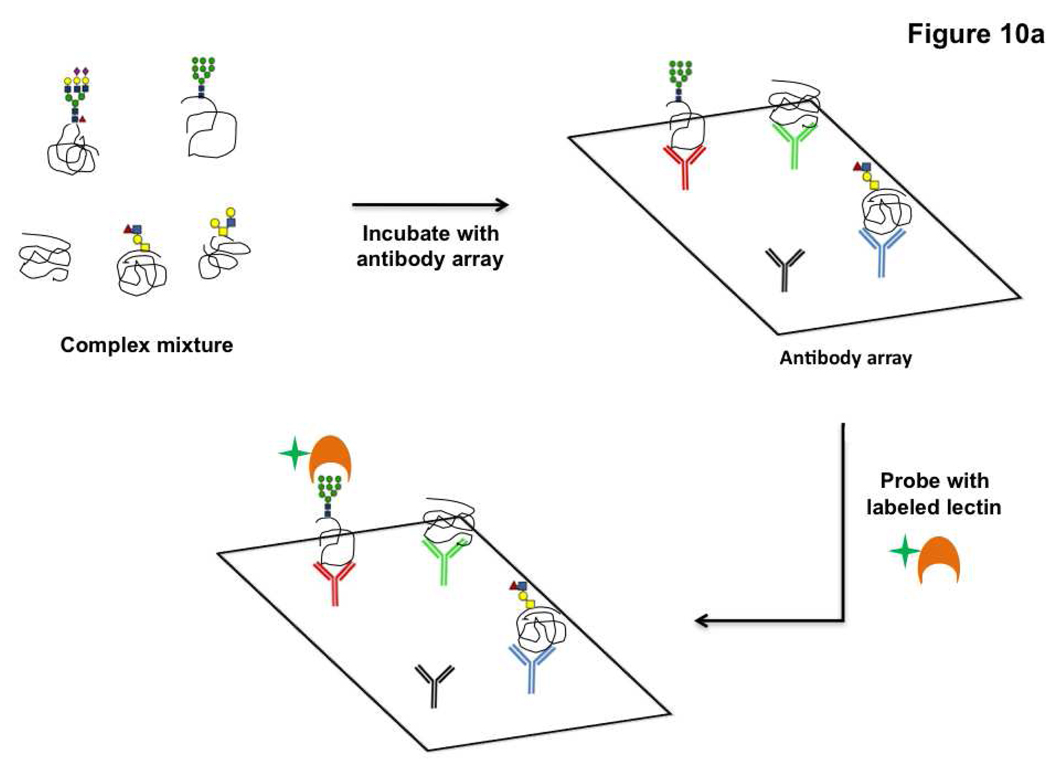
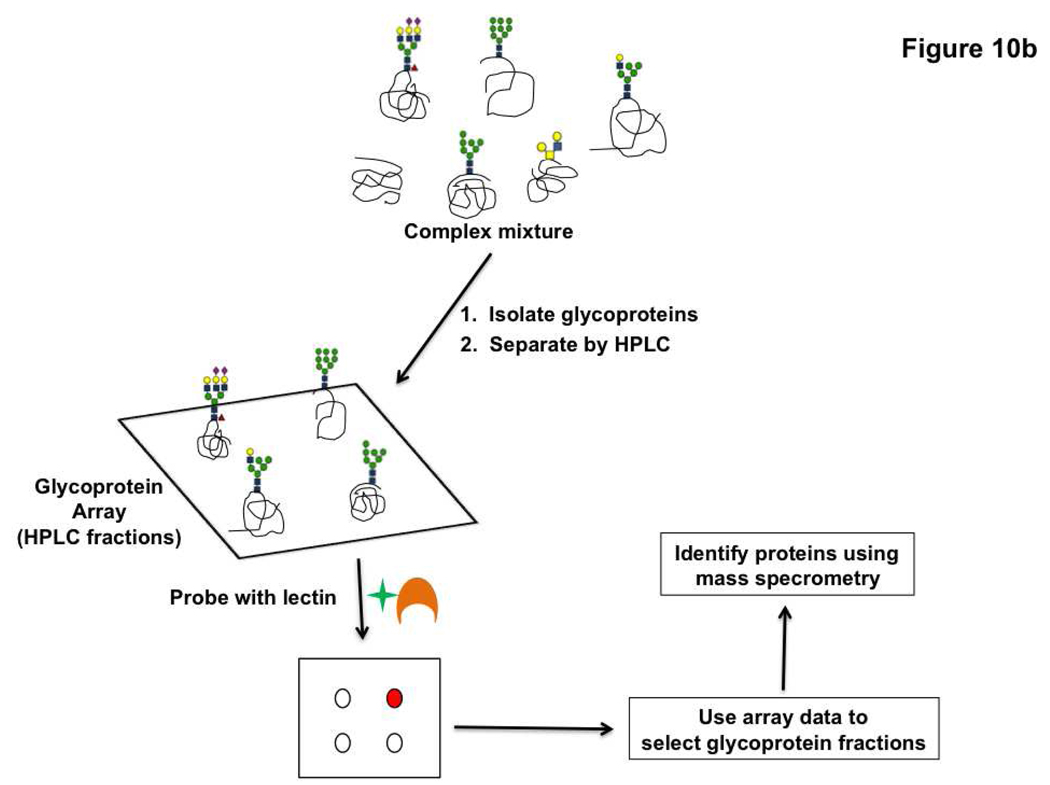
Lectins have also been used as a probe for detecting changes in glycosylation on target proteins displayed in an array format. There are two variations of this technology. The first uses antibody arrays to pull down specific glycoproteins and then assays these proteins for glycan changes using lectins and glycan-specific antibodies (Figure 10, panel a) (132). In recent work, this technique was used to detect alterations in the glycosylation of MUC1 and CEA (carcinoembryonic antigen), both potential cancer biomarkers, in sera from pancreatic cancer patients (132) and to investigate cancer-related changes in glycosylation of alpha1-β-glycoprotein (133). In the second microarray variation, glycoproteins are printed directly onto the array and assayed. In brief, glycoproteins from samples (typically serum) are isolated by lectin affinity chromatography and then fractionated using HPLC (Figure 10, panel b). These fractions are spotted in array format and probed with select lectins to identify potential glycan changes. Glycoprotein fractions that display differences in the cancer state are resolved further and the individual proteins responsible for the differential glycosylation are identified using mass spectrometry (134–136). Candidate glycoprotein biomarkers displaying increased sialylation and fucosylation in colorectal cancer samples were identified using this technique (136). These methods, while useful for specific biomarker discovery, are not meant as a general method to survey the glycoproteome. They focus on identifying specific portions of the glycoproteome, restricted by either the anti-glycoprotein antibodies arrayed or the limited number of lectins with which the glycoproteome is surveyed for changes. Thus, they are targeted for a select purpose, namely biomarker discovery and validation. In general, the use of microarray technologies is not only advancing translational biomarker research but is also transforming the field of glycan structural characterization via the lectin microarrays, and provides new technology for dissecting the intimate relationship between the structural and functional aspects of glycans.
Figure 10. Microarray based methods for glycoprotein biomarker discovery.
a) Glycoproteins captured on antibody arrays are assayed for changes in glycosylation using lectins. b) Glycoprotein arrays derived from HPLC fractionation of the glycoproteome can be interrogated with lectins to screen for glycoprotein markers.
Tools for the study of glycan-protein interactions
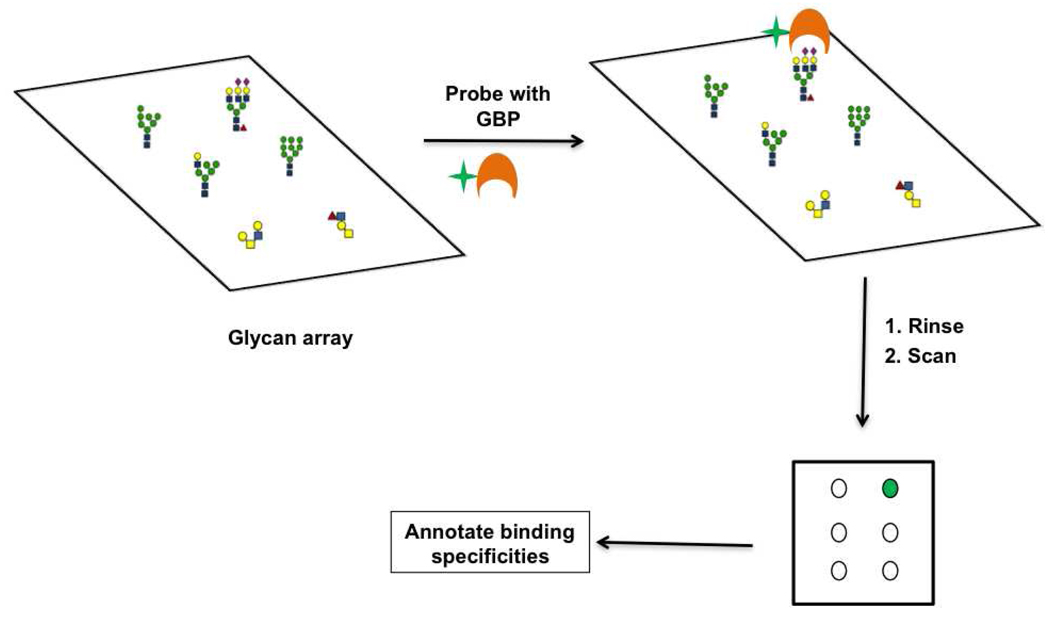
To develop a more comprehensive understanding of the glycome we must explore the fundamental interactions by which the glycome is decoded i.e. the binding of proteins to glycans. Techniques for the study of glycan-protein interactions include surface plasmon resonance (SPR), frontal affinity chromatography (FAC) and the more traditional ELISA methods (29, 137–139). A new technology, the glycan microarray, has rapidly advanced our knowledge of glycan-binding proteins by enabling the simultaneous evaluation of multiple glycan-protein binding interactions. These arrays consist of oligosaccharides from chemical, enzymatic and natural sources spotted in a high density format. Immobilization of the glycans onto a solid support is most commonly through a covalent linkage. Chemistries used for this purpose include the covalent coupling of thiols and maleimides, NHS-esters and amines, and amines and epoxides, among others (for comprehensive reviews regarding this topic, see (14, 140–146)). Glycan display is multivalent, enabling detection of glycan-protein interactions that are often weak at the monovalent level. Interactions of the glycan-binding protein (GBP) with the array are typically detected via fluorescence, where the fluorophore is either directly attached to the GBP, or is on a secondary reagent used to detect the GBP (Figure 11). Carbohydrate microarrays have been used to characterize a diverse range of GBPs (138, 147–154) including those involved in viral pathogenesis and immune recognition (155–157). For example, recent work using carbohydrate microarrays to characterize the binding of mutant influenza viruses showed that mutations previously found in pandemic strains, when introduced into current avian influenza strains of H5N1, cause a shift in glycan binding towards human sialic acid epitopes, thus demonstrating the increased propensity of these new H5N1 viruses towards the generation of a pandemic strain (158). For comprehensive reviews on the use of glycan arrays in biomedical applications see (159–161).
Figure 11. Glycan microarray technology.
The multivalent presentation of carbohydrates is one of the attractive features of the array format. It has long been known that subtle differences in carbohydrate presentation and density can alter binding specificities of carbohydrate binding proteins (162–164). Recent work has shown that varying glycan density within an array format can reveal higher order binding patterns in seemingly identical GBPs, confirming the strong influence of density on glycan recognition (165). This study underscores the need for incorporating multiple presentations of the same glycan epitope in the array format to obtain biologically relevant specificities.
At present carbohydrate microarrays are limited by the variety of epitopes presented and the mode of glycan presentation. In spite of the tremendous improvements in carbohydrate synthesis, synthesis of glycans is still a major limiting factor in expanding this array technology as it both time consuming and difficult (145, 166, 167). In addition well defined molecular architectures for carbohydrate display on a solid support are still needed to better represent the variety of glycan presentations found in nature in the context of the array.
Future work and directions
The challenge of glycomics is being met with an arsenal of novel analytical tools which are revealing the structure and functions of these crucial biopolymers. These novel technologies have generated an immense amount of glycomic information in a short period of time. This is necessitating the development of bioinformatic tools for data integration. Publicly available databases include those from the Consortium for Functional Glycomics (CFG) (168), KEGG (Kyoto Encyclopedia of Gene and Genomics) (169) and Glycosciences.de (170). Although these provide an initial framework for including glycomic information in databases, there is still a long way to go in the development of advanced databases and algorithms which will allow scientists rapid access information generated via different glycomic tools across multiple labs. Generation of parallel glycomic, genomic and glycoproteomic datasets are becoming possible, bringing the opportunity to integrate these datasets to gain an understanding of the fundamental control mechanisms underpinning the glycome and ultimately to model the glycome from genomic information (22–24). Given the importance of carbohydrates in various aspects of biology ranging from microbial pathogenesis to cell differentiation, the rapidly evolving area of glycomics has the potential to usher in a new understanding of biological recognition that will have a strong impact on diagnostic and therapeutic medicine.
Figure 12.
Acknowledgements
L.K.M. would like to acknowledge funding from the NIH (Grant: 1 DP2 OD004711-01).
Glossary
- Glycans
simple or complex polymers composed of monosaccharides
- Glycoconjugates
biomolecules which are modified with glycans such as lipids and proteins
- Glycome
the cohort of glycans and glycoconjugates found in a system
- Lectins
carbohydrate binding proteins that are not antibodies or enzymes
- Glycoproteomics
isolation and identification of glycosylated proteins from a complex mixture
- Microarrays
collection of probes immobilized on a solid support for high-throughput sample analysis
- Mass spectrometry
analytical technique that relies on unique mass signatures to identify components of a complex sample
- Biomarkers
proteins that are altered in certain disease states and could be used as diagnostic markers to indicate disease progression
References
- 1.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 2.Ajit Varki RDC, Esko JeffreyD, Freeze HudsonH, Stanley Pamela, Bertozzi CarolynR, Hart GeraldW, Etzler MarilynnE. Essentials of glycobiology. second ed. New York: Cold Spring Harbor Laboratory Press; 2008. [PubMed] [Google Scholar]
- 3.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Marth JD, Grewal PK. Mammalian glycosylation in immunity. Nat Rev Immunol. 2008 doi: 10.1038/nri2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu Rev Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 6.Sharon N. Carbohydrates as future anti-adhesion drugs for infectious diseases. Biochim Biophys Acta. 2006;1760:527–537. doi: 10.1016/j.bbagen.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varki A, Cummings RD, Esko JE, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of glycobiology. Second ed. New York: Cold Spring Harbor Laboratory Press; 2008. [PubMed] [Google Scholar]
- 10.Laine RA. A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05 × 10(12) structures for a reducing hexasaccharide: the Isomer Barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology. 1994;4:759–767. doi: 10.1093/glycob/4.6.759. [DOI] [PubMed] [Google Scholar]
- 11.Werz DB, Ranzinger R, Herget S, Adibekian A, von der Lieth CW, Seeberger PH. Exploring the structural diversity of mammalian carbohydrates ("glycospace") by statistical databank analysis. ACS Chem Biol. 2007;2:685–691. doi: 10.1021/cb700178s. [DOI] [PubMed] [Google Scholar]
- 12.Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nat Methods. 2005;2:817–824. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]
- 13.Taylor ME, Drickamer K. Introduction to glycobiology. Second ed. Oxford University Press; 2006. [Google Scholar]
- 14.Mahal LK. Glycomics: towards bioinformatic approaches to understanding glycosylation. Anticancer Agents Med Chem. 2008;8:37–51. doi: 10.2174/187152008783330806. [DOI] [PubMed] [Google Scholar]
- 15.Hossler P, Mulukutla BC, Hu WS. Systems analysis of N-glycan processing in mammalian cells. PLoS ONE. 2007;2:e713. doi: 10.1371/journal.pone.0000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budnik BA, Lee RS, Steen JA. Global methods for protein glycosylation analysis by mass spectrometry. Biochim Biophys Acta. 2006;1764:1870–1880. doi: 10.1016/j.bbapap.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Schachter H. The joys of HexNAc. The synthesis and function of N- and O-glycan branches. Glycoconj J. 2000;17:465–483. doi: 10.1023/a:1011010206774. [DOI] [PubMed] [Google Scholar]
- 18.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 19.Slawson C, Housley MP, Hart GW. O-GlcNAc cycling: how a single sugar post-translational modification is changing the way we think about signaling networks. J Cell Biochem. 2006;97:71–83. doi: 10.1002/jcb.20676. [DOI] [PubMed] [Google Scholar]
- 20.Yarema KJ, Bertozzi CR. Characterizing glycosylation pathways. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-5-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe JB, Marth JD. A genetic approach to Mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 22.Suga A, Yamanishi Y, Hashimoto K, Goto S, Kanehisa M. An improved scoring scheme for predicting glycan structures from gene expression data. Genome Inform. 2007;18:237–246. [PubMed] [Google Scholar]
- 23.Comelli EM, Head SR, Gilmartin T, Whisenant T, Haslam SM, North SJ, Wong NK, Kudo T, Narimatsu H, Esko JD, Drickamer K, Dell A, Paulson JC. A focused microarray approach to functional glycomics: transcriptional regulation of the glycome. Glycobiology. 2006;16:117–131. doi: 10.1093/glycob/cwj048. [DOI] [PubMed] [Google Scholar]
- 24.Nairn AV, York WS, Harris K, Hall EM, Pierce JM, Moremen KW. Regulation of glycan structures in animal tissues: transcript profiling of glycan-related genes. J Biol Chem. 2008;283:17298–17313. doi: 10.1074/jbc.M801964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundquist JJ, Toone EJ. The cluster glycoside effect. Chem Rev. 2002;102:555–578. doi: 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]
- 26.Lee RT, Lee YC. Affinity enhancement by multivalent lectin-carbohydrate interaction. Glycoconj J. 2000;17:543–551. doi: 10.1023/a:1011070425430. [DOI] [PubMed] [Google Scholar]
- 27.Dam TK, Roy R, Das SK, Oscarson S, Brewer CF. Binding of multivalent carbohydrates to concanavalin A and Dioclea grandiflora lectin. Thermodynamic analysis of the "multivalency effect". J Biol Chem. 2000;275:14223–14230. doi: 10.1074/jbc.275.19.14223. [DOI] [PubMed] [Google Scholar]
- 28.Sacchettini JC, Baum LG, Brewer CF. Multivalent protein-carbohydrate interactions. A new paradigm for supermolecular assembly and signal transduction. Biochemistry. 2001;40:3009–3015. doi: 10.1021/bi002544j. [DOI] [PubMed] [Google Scholar]
- 29.Hirabayashi J. Lectin-based structural glycomics: glycoproteomics and glycan profiling. Glycoconj J. 2004;21:35–40. doi: 10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- 30.Hirabayashi J, Kasai K. Separation technologies for glycomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;771:67–87. doi: 10.1016/s1570-0232(02)00057-0. [DOI] [PubMed] [Google Scholar]
- 31.Geyer H, Geyer R. Strategies for analysis of glycoprotein glycosylation. Biochim Biophys Acta. 2006;1764:1853–1869. doi: 10.1016/j.bbapap.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Sharon N, Lis H. History of lectins: from hemagglutinins to biological recognition molecules. Glycobiology. 2004;14:53R–62R. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- 33.Lis H, Sharon N. Lectins: Carbohydrate-Specific Proteins That Mediate Cellular Recognition. Chem Rev. 1998;98:637–674. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- 34.Xu Z, Zhou X, Lu H, Wu N, Zhao H, Zhang L, Zhang W, Liang YL, Wang L, Liu Y, Yang P, Zha X. Comparative glycoproteomics based on lectins affinity capture of N-linked glycoproteins from human Chang liver cells and MHCC97-H cells. Proteomics. 2007;7:2358–2370. doi: 10.1002/pmic.200600041. [DOI] [PubMed] [Google Scholar]
- 35.Vercoutter-Edouart AS, Slomianny MC, Dekeyzer-Beseme O, Haeuw JF, Michalski JC. Glycoproteomics and glycomics investigation of membrane N-glycosylproteins from human colon carcinoma cells. Proteomics. 2008;8:3236–3256. doi: 10.1002/pmic.200800151. [DOI] [PubMed] [Google Scholar]
- 36.Block TM, Comunale MA, Lowman M, Steel LF, Romano PR, Fimmel C, Tennant BC, London WT, Evans AA, Blumberg BS, Dwek RA, Mattu TS, Mehta AS. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc Natl Acad Sci U S A. 2005;102:779–784. doi: 10.1073/pnas.0408928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwientek T, Mandel U, Roth U, Muller S, Hanisch FG. A serial lectin approach to the mucin-type O-glycoproteome of Drosophila melanogaster S2 cells. Proteomics. 2007;7:3264–3277. doi: 10.1002/pmic.200600793. [DOI] [PubMed] [Google Scholar]
- 38.Durham M, Regnier FE. Targeted glycoproteomics: serial lectin affinity chromatography in the selection of O-glycosylation sites on proteins from the human blood proteome. J Chromatogr A. 2006;1132:165–173. doi: 10.1016/j.chroma.2006.07.070. [DOI] [PubMed] [Google Scholar]
- 39.Qiu R, Regnier FE. Comparative glycoproteomics of N-linked complex-type glycoforms containing sialic acid in human serum. Anal Chem. 2005;77:7225–7231. doi: 10.1021/ac050554q. [DOI] [PubMed] [Google Scholar]
- 40.Jung K, Cho W, Regnier FE. Glycoproteomics of plasma based on narrow selectivity lectin affinity chromatography. J Proteome Res. 2009;8:643–650. doi: 10.1021/pr8007495. [DOI] [PubMed] [Google Scholar]
- 41.Comunale MA, Wang M, Hafner J, Krakover J, Rodemich L, Kopenhaver B, Long RE, Junaidi O, Bisceglie AM, Block TM, Mehta AS. Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. J Proteome Res. 2009;8:595–602. doi: 10.1021/pr800752c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z, Hancock WS. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. J Chromatogr A. 2004;1053:79–88. [PubMed] [Google Scholar]
- 43.Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Multilectin affinity chromatography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin Chem. 2006;52:1897–1905. doi: 10.1373/clinchem.2005.065862. [DOI] [PubMed] [Google Scholar]
- 44.Heo SH, Lee SJ, Ryoo HM, Park JY, Cho JY. Identification of putative serum glycoprotein biomarkers for human lung adenocarcinoma by multilectin affinity chromatography and LC-MS/MS. Proteomics. 2007;7:4292–4302. doi: 10.1002/pmic.200700433. [DOI] [PubMed] [Google Scholar]
- 45.Zhao J, Simeone DM, Heidt D, Anderson MA, Lubman DM. Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: application to pancreatic cancer serum. J Proteome Res. 2006;5:1792–1802. doi: 10.1021/pr060034r. [DOI] [PubMed] [Google Scholar]
- 46.Sun Q, Kang X, Zhang Y, Zhou H, Dai Z, Lu W, Zhou X, Liu X, Yang P, Liu Y. DSA affinity glycoproteome of human liver tissue. Arch Biochem Biophys. 2009;484:24–29. doi: 10.1016/j.abb.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Dai Z, Fan J, Liu Y, Zhou J, Bai D, Tan C, Guo K, Zhang Y, Zhao Y, Yang P. Identification and analysis of alpha1,6-fucosylated proteins in human normal liver tissues by a target glycoproteomic approach. Electrophoresis. 2007;28:4382–4391. doi: 10.1002/elps.200700233. [DOI] [PubMed] [Google Scholar]
- 48.Wada Y, Tajiri M, Yoshida S. Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Anal Chem. 2004;76:6560–6565. doi: 10.1021/ac049062o. [DOI] [PubMed] [Google Scholar]
- 49.Tajiri M, Yoshida S, Wada Y. Differential analysis of site-specific glycans on plasma and cellular fibronectins: application of a hydrophilic affinity method for glycopeptide enrichment. Glycobiology. 2005;15:1332–1340. doi: 10.1093/glycob/cwj019. [DOI] [PubMed] [Google Scholar]
- 50.Hagglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J Proteome Res. 2004;3:556–566. doi: 10.1021/pr034112b. [DOI] [PubMed] [Google Scholar]
- 51.Wuhrer M, de Boer AR, Deelder AM. Structural glycomics using hydrophilic interaction chromatography (HILIC) with mass spectrometry. Mass Spectrom Rev. 2009;28:192–206. doi: 10.1002/mas.20195. [DOI] [PubMed] [Google Scholar]
- 52.Calvano CD, Zambonin CG, Jensen ON. Assessment of lectin and HILIC based enrichment protocols for characterization of serum glycoproteins by mass spectrometry. J Proteomics. 2008;71:304–317. doi: 10.1016/j.jprot.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 53.Sparbier K, Wenzel T, Kostrzewa M. Exploring the binding profiles of ConA, boronic acid and WGA by MALDI-TOF/TOF MS and magnetic particles. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;840:29–36. doi: 10.1016/j.jchromb.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 54.Xu Y, Wu Z, Zhang L, Lu H, Yang P, Webley PA, Zhao D. Highly specific enrichment of glycopeptides using boronic acid-functionalized mesoporous silica. Anal Chem. 2009;81:503–508. doi: 10.1021/ac801912t. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, Yi EC, Li XJ, Mallick P, Kelly-Spratt KS, Masselon CD, Camp DG, 2nd, Smith RD, Kemp CJ, Aebersold R. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol Cell Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat Protoc. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu T, Qian WJ, Gritsenko MA, Camp DG, 2nd, Monroe ME, Moore RJ, Smith RD. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J Proteome Res. 2009;8:651–661. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- 60.Wollscheid B, Bausch-Fluck D, Henderson C, O'Brien R, Bibel M, Schiess R, Aebersold R, Watts JD. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schiess R, Mueller LN, Schmidt A, Mueller M, Wollscheid B, Aebersold R. Analysis of cell surface proteome changes via label-free, quantitative mass spectrometry. Mol Cell Proteomics. 2009;8:624–638. doi: 10.1074/mcp.M800172-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McDonald CA, Yang JY, Marathe V, Yen TY, Macher BA. Combining results from lectin affinity chromatography and glycocapture approaches substantially improves the coverage of the glycoproteome. Mol Cell Proteomics. 2009;8:287–301. doi: 10.1074/mcp.M800272-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monzo A, Bonn GK, Guttman A. Boronic acid-lectin affinity chromatography. 1. Simultaneous glycoprotein binding with selective or combined elution. Anal Bioanal Chem. 2007;389:2097–2102. doi: 10.1007/s00216-007-1627-y. [DOI] [PubMed] [Google Scholar]
- 64.Cao J, Shen C, Wang H, Shen H, Chen Y, Nie A, Yan G, Lu H, Liu Y, Yang P. Identification of N-glycosylation sites on secreted proteins of human hepatocellular carcinoma cells with a complementary proteomics approach. J Proteome Res. 2009;8:662–672. doi: 10.1021/pr800826u. [DOI] [PubMed] [Google Scholar]
- 65.Laughlin ST, Bertozzi CR. Imaging the glycome. Proc Natl Acad Sci U S A. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 67.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 68.Hanson SR, Hsu TL, Weerapana E, Kishikawa K, Simon GM, Cravatt BF, Wong CH. Tailored glycoproteomics and glycan site mapping using saccharide-selective bioorthogonal probes. J Am Chem Soc. 2007;129:7266–7267. doi: 10.1021/ja0724083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci U S A. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nandi A, Sprung R, Barma DK, Zhao Y, Kim SC, Falck JR. Global identification of O-GlcNAc-modified proteins. Anal Chem. 2006;78:452–458. doi: 10.1021/ac051207j. [DOI] [PubMed] [Google Scholar]
- 71.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci U S A. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rexach JE, Clark PM, Hsieh-Wilson LC. Chemical approaches to understanding O-GlcNAc glycosylation in the brain. Nat Chem Biol. 2008;4:97–106. doi: 10.1038/nchembio.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 74.Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 75.Kaji H, Yamauchi Y, Takahashi N, Isobe T. Mass spectrometric identification of N-linked glycopeptides using lectin-mediated affinity capture and glycosylation site-specific stable isotope tagging. Nat Protoc. 2006;1:3019–3027. doi: 10.1038/nprot.2006.444. [DOI] [PubMed] [Google Scholar]
- 76.Hagglund P, Matthiesen R, Elortza F, Hojrup P, Roepstorff P, Jensen ON, Bunkenborg J. An enzymatic deglycosylation scheme enabling identification of core fucosylated N-glycans and O-glycosylation site mapping of human plasma proteins. J Proteome Res. 2007;6:3021–3031. doi: 10.1021/pr0700605. [DOI] [PubMed] [Google Scholar]
- 77.Nettleship JE, Aplin R, Aricescu AR, Evans EJ, Davis SJ, Crispin M, Owens RJ. Analysis of variable N-glycosylation site occupancy in glycoproteins by liquid chromatography electrospray ionization mass spectrometry. Anal Biochem. 2007;361:149–151. doi: 10.1016/j.ab.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 78.Lewandrowski U, Sickmann A. N-glycosylation site analysis of human platelet proteins by hydrazide affinity capturing and LC-MS/MS. Methods Mol Biol. 2009;534:225–238. doi: 10.1007/978-1-59745-022-5_17. [DOI] [PubMed] [Google Scholar]
- 79.Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 80.Zaia J. Mass spectrometry and the emerging field of glycomics. Chem Biol. 2008;15:881–892. doi: 10.1016/j.chembiol.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zaia J. Mass spectrometry of oligosaccharides. Mass Spectrom Rev. 2004;23:161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- 82.Morelle W, Michalski JC. Glycomics and mass spectrometry. Curr Pharm Des. 2005;11:2615–2645. doi: 10.2174/1381612054546897. [DOI] [PubMed] [Google Scholar]
- 83.Morelle W, Canis K, Chirat F, Faid V, Michalski JC. The use of mass spectrometry for the proteomic analysis of glycosylation. Proteomics. 2006;6:3993–4015. doi: 10.1002/pmic.200600129. [DOI] [PubMed] [Google Scholar]
- 84.Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science. 2001;291:2351–2356. doi: 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- 85.Haslam SM, North SJ, Dell A. Mass spectrometric analysis of N- and O-glycosylation of tissues and cells. Curr Opin Struct Biol. 2006;16:584–591. doi: 10.1016/j.sbi.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 86.Morelle W, Michalski JC. Analysis of protein glycosylation by mass spectrometry. Nat Protoc. 2007;2:1585–1602. doi: 10.1038/nprot.2007.227. [DOI] [PubMed] [Google Scholar]
- 87.North SJ, Hitchen PG, Haslam SM, Dell A. Mass spectrometry in the analysis of N-linked and O-linked glycans. Curr Opin Struct Biol. 2009 doi: 10.1016/j.sbi.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Go EP, Rebecchi KR, Dalpathado DS, Bandu ML, Zhang Y, Desaire H. GlycoPep DB: a tool for glycopeptide analysis using a "Smart Search". Anal Chem. 2007;79:1708–1713. doi: 10.1021/ac061548c. [DOI] [PubMed] [Google Scholar]
- 89.Vakhrushev SY, Dadimov D, Peter-Katalinic J. Software Platform for High-Throughput Glycomics. Anal Chem. 2009 doi: 10.1021/ac802408f. [DOI] [PubMed] [Google Scholar]
- 90.Goldberg D, Sutton-Smith M, Paulson J, Dell A. Automatic annotation of matrix-assisted laser desorption/ionization N-glycan spectra. Proteomics. 2005;5:865–875. doi: 10.1002/pmic.200401071. [DOI] [PubMed] [Google Scholar]
- 91.Barkauskas DA, An HJ, Kronewitter SR, de Leoz ML, Chew HK, de Vere White RW, Leiserowitz GS, Miyamoto S, Lebrilla CB, Rocke DM. Detecting glycan cancer biomarkers in serum samples using MALDI FT-ICR mass spectrometry data. Bioinformatics. 2009;25:251–257. doi: 10.1093/bioinformatics/btn610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ressom HW, Varghese RS, Goldman L, An Y, Loffredo CA, Abdel-Hamid M, Kyselova Z, Mechref Y, Novotny M, Drake SK, Goldman R. Analysis of MALDI-TOF mass spectrometry data for discovery of peptide and glycan biomarkers of hepatocellular carcinoma. J Proteome Res. 2008;7:603–610. doi: 10.1021/pr0705237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kyselova Z, Mechref Y, Kang P, Goetz JA, Dobrolecki LE, Sledge GW, Schnaper L, Hickey RJ, Malkas LH, Novotny MV. Breast cancer diagnosis and prognosis through quantitative measurements of serum glycan profiles. Clin Chem. 2008;54:1166–1175. doi: 10.1373/clinchem.2007.087148. [DOI] [PubMed] [Google Scholar]
- 94.Goldman R, Ressom HW, Varghese RS, Goldman L, Bascug G, Loffredo CA, Abdel-Hamid M, Gouda I, Ezzat S, Kyselova Z, Mechref Y, Novotny MV. Detection of Hepatocellular Carcinoma Using Glycomic Analysis. Clin Cancer Res. 2009 doi: 10.1158/1078-0432.CCR-07-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao J, Qiu W, Simeone DM, Lubman DM. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J Proteome Res. 2007;6:1126–1138. doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- 96.Kam RK, Poon TC, Chan HL, Wong N, Hui AY, Sung JJ. High-throughput quantitative profiling of serum N-glycome by MALDI-TOF mass spectrometry and N-glycomic fingerprint of liver fibrosis. Clin Chem. 2007;53:1254–1263. doi: 10.1373/clinchem.2007.085563. [DOI] [PubMed] [Google Scholar]
- 97.Mechref Y, Novotny MV. Mass spectrometric mapping and sequencing of N-linked oligosaccharides derived from submicrogram amounts of glycoproteins. Anal Chem. 1998;70:455–463. doi: 10.1021/ac970947s. [DOI] [PubMed] [Google Scholar]
- 98.Xie Y, Liu J, Zhang J, Hedrick JL, Lebrilla CB. Method for the comparative glycomic analyses of O-linked, mucin-type oligosaccharides. Anal Chem. 2004;76:5186–5197. doi: 10.1021/ac0496953. [DOI] [PubMed] [Google Scholar]
- 99.Xia B, Feasley CL, Sachdev GP, Smith DF, Cummings RD. Glycan reductive isotope labeling for quantitative glycomics. Anal Biochem. 2009;387:162–170. doi: 10.1016/j.ab.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Uematsu R, Furukawa J, Nakagawa H, Shinohara Y, Deguchi K, Monde K, Nishimura S. High throughput quantitative glycomics and glycoform-focused proteomics of murine dermis and epidermis. Mol Cell Proteomics. 2005;4:1977–1989. doi: 10.1074/mcp.M500203-MCP200. [DOI] [PubMed] [Google Scholar]
- 101.Atwood JA, 3rd, Cheng L, Alvarez-Manilla G, Warren NL, York WS, Orlando R. Quantitation by isobaric labeling: applications to glycomics. J Proteome Res. 2008;7:367–374. doi: 10.1021/pr070476i. [DOI] [PubMed] [Google Scholar]
- 102.Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem. 2007;282:9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- 103.Kang P, Mechref Y, Kyselova Z, Goetz JA, Novotny MV. Comparative glycomic mapping through quantitative permethylation and stable-isotope labeling. Anal Chem. 2007;79:6064–6073. doi: 10.1021/ac062098r. [DOI] [PubMed] [Google Scholar]
- 104.Xia B, Feasley CL, Sachdev GP, Smith DF, Cummings RD. Glycan reductive isotope labeling for quantitative glycomics. Anal Biochem. 2009;387:162–170. doi: 10.1016/j.ab.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bowman MJ, Zaia J. Tags for the stable isotopic labeling of carbohydrates and quantitative analysis by mass spectrometry. Anal Chem. 2007;79:5777–5784. doi: 10.1021/ac070581b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Orlando R, Lim JM, Atwood JA, Angel PM, Fang M, Aoki K, Alvarez-Manilla G, Moremen KW, York WS, Tiemeyer M, Pierce M, Dalton S, Wells L. IDAWG: Metabolic Incorporation of Stable Isotope Labels for Quantitative Glycomics of Cultured Cells. J Proteome Res. 2009 doi: 10.1021/pr8010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hitchcock AM, Yates KE, Costello CE, Zaia J. Comparative glycomics of connective tissue glycosaminoglycans. Proteomics. 2008;8:1384–1397. doi: 10.1002/pmic.200700787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Staples GO, Bowman MJ, Costello CE, Hitchcock AM, Lau JM, Leymarie N, Miller C, Naimy H, Shi X, Zaia J. A chip-based amide-HILIC LC/MS platform for glycosaminoglycan glycomics profiling. Proteomics. 2009;9:686–695. doi: 10.1002/pmic.200701008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsui ZC, Chen QR, Thomas MJ, Samuel M, Cui Z. A method for profiling gangliosides in animal tissues using electrospray ionization-tandem mass spectrometry. Anal Biochem. 2005;341:251–258. doi: 10.1016/j.ab.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 110.Parry S, Ledger V, Tissot B, Haslam SM, Scott J, Morris HR, Dell A. Integrated mass spectrometric strategy for characterizing the glycans from glycosphingolipids and glycoproteins: direct identification of sialyl Le(x) in mice. Glycobiology. 2007;17:646–654. doi: 10.1093/glycob/cwm024. [DOI] [PubMed] [Google Scholar]
- 111.Abd Hamid UM, Royle L, Saldova R, Radcliffe CM, Harvey DJ, Storr SJ, Pardo M, Antrobus R, Chapman CJ, Zitzmann N, Robertson JF, Dwek RA, Rudd PM. A strategy to reveal potential glycan markers from serum glycoproteins associated with breast cancer progression. Glycobiology. 2008;18:1105–1118. doi: 10.1093/glycob/cwn095. [DOI] [PubMed] [Google Scholar]
- 112.Knezevic A, Polasek O, Gornik O, Rudan I, Campbell H, Hayward C, Wright A, Kolcic I, O'Donoghue N, Bones J, Rudd PM, Lauc G. Variability, heritability and environmental determinants of human plasma N-glycome. J Proteome Res. 2009;8:694–701. doi: 10.1021/pr800737u. [DOI] [PubMed] [Google Scholar]
- 113.Campbell MP, Royle L, Radcliffe CM, Dwek RA, Rudd PM. GlycoBase and autoGU: tools for HPLC-based glycan analysis. Bioinformatics. 2008;24:1214–1216. doi: 10.1093/bioinformatics/btn090. [DOI] [PubMed] [Google Scholar]
- 114.Pilobello KT, Mahal LK. Lectin microarrays for glycoprotein analysis. Methods Mol Biol. 2007;385:193–203. doi: 10.1007/978-1-59745-426-1_14. [DOI] [PubMed] [Google Scholar]
- 115.Hsu KL, Mahal LK. A lectin microarray approach for the rapid analysis of bacterial glycans. Nat Protoc. 2006;1:543–549. doi: 10.1038/nprot.2006.76. [DOI] [PubMed] [Google Scholar]
- 116.Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nat Methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 117.Zheng T, Peelen D, Smith LM. Lectin arrays for profiling cell surface carbohydrate expression. J Am Chem Soc. 2005;127:9982–9983. doi: 10.1021/ja0505550. [DOI] [PubMed] [Google Scholar]
- 118.Angeloni S, Ridet JL, Kusy N, Gao H, Crevoisier F, Guinchard S, Kochhar S, Sigrist H, Sprenger N. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology. 2005;15:31–41. doi: 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- 119.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem. 2005;6:985–989. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 120.Pilobello KT, Slawek DE, Mahal LK. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proc Natl Acad Sci U S A. 2007;104:11534–11539. doi: 10.1073/pnas.0704954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen S, Zheng T, Shortreed MR, Alexander C, Smith LM. Analysis of cell surface carbohydrate expression patterns in normal and tumorigenic human breast cell lines using lectin arrays. Anal Chem. 2007;79:5698–5702. doi: 10.1021/ac070423k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tao SC, Li Y, Zhou J, Qian J, Schnaar RL, Zhang Y, Goldstein IJ, Zhu H, Schneck JP. Lectin microarrays identify cell-specific and functionally significant cell surface glycan markers. Glycobiology. 2008;18:761–769. doi: 10.1093/glycob/cwn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hsu KL, Pilobello KT, Mahal LK. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat Chem Biol. 2006;2:153–157. doi: 10.1038/nchembio767. [DOI] [PubMed] [Google Scholar]
- 124.Krishnamoorthy L, Bess JW, Jr, Preston AB, Nagashima K, Mahal LK. HIV-1 and microvesicles from T cells share a common glycome, arguing for a common origin. Nat Chem Biol. 2009 doi: 10.1038/nchembio.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tateno H, Uchiyama N, Kuno A, Togayachi A, Sato T, Narimatsu H, Hirabayashi J. A novel strategy for mammalian cell surface glycome profiling using lectin microarray. Glycobiology. 2007;17:1138–1146. doi: 10.1093/glycob/cwm084. [DOI] [PubMed] [Google Scholar]
- 126.Gemeiner P, Mislovicova D, Tkac J, Svitel J, Patoprsty V, Hrabarova E, Kogan G, Kozar T. Lectinomics II. A highway to biomedical/clinical diagnostics. Biotechnol Adv. 2009;27:1–15. doi: 10.1016/j.biotechadv.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 127.Van Damme EJM, Peumans WJAP, Bardocz S. Handbook of plant lectins: Properties and biomedical applications. Wiley; 1998. [Google Scholar]
- 128.Hsu KL, Gildersleeve JC, Mahal LK. A simple strategy for the creation of a recombinant lectin microarray. Mol Biosyst. 2008;4:654–662. doi: 10.1039/b800725j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zou Y, Broughton DL, Bicker KL, Thompson PR, Lavigne JJ. Peptide borono lectins (PBLs): a new tool for glycomics and cancer diagnostics. Chembiochem. 2007;8:2048–2051. doi: 10.1002/cbic.200700221. [DOI] [PubMed] [Google Scholar]
- 130.Yabe R, Suzuki R, Kuno A, Fujimoto Z, Jigami Y, Hirabayashi J. Tailoring a novel sialic acid-binding lectin from a ricin-B chain-like galactose-binding protein by natural evolution-mimicry. J Biochem. 2007;141:389–399. doi: 10.1093/jb/mvm043. [DOI] [PubMed] [Google Scholar]
- 131.Maenuma K, Yim M, Komatsu K, Hoshino M, Takahashi Y, Bovin N, Irimura T. Use of a library of mutated Maackia amurensis hemagglutinin for profiling the cell lineage and differentiation. Proteomics. 2008;8:3274–3283. doi: 10.1002/pmic.200800037. [DOI] [PubMed] [Google Scholar]
- 132.Chen S, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat Methods. 2007;4:437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 133.Li C, Simeone DM, Brenner DE, Anderson MA, Shedden KA, Ruffin MT, Lubman DM. Pancreatic Cancer Serum Detection Using a Lectin/Glyco-Antibody Array Method. J Proteome Res. 2009;8:483–492. doi: 10.1021/pr8007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Patwa TH, Zhao J, Anderson MA, Simeone DM, Lubman DM. Screening of glycosylation patterns in serum using natural glycoprotein microarrays and multi-lectin fluorescence detection. Anal Chem. 2006;78:6411–6421. doi: 10.1021/ac060726z. [DOI] [PubMed] [Google Scholar]
- 135.Zhao J, Patwa TH, Qiu W, Shedden K, Hinderer R, Misek DE, Anderson MA, Simeone DM, Lubman DM. Glycoprotein microarrays with multi-lectin detection: unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J Proteome Res. 2007;6:1864–1874. doi: 10.1021/pr070062p. [DOI] [PubMed] [Google Scholar]
- 136.Qiu Y, Patwa TH, Xu L, Shedden K, Misek DE, Tuck M, Jin G, Ruffin MT, Turgeon DK, Synal S, Bresalier R, Marcon N, Brenner DE, Lubman DM. Plasma glycoprotein profiling for colorectal cancer biomarker identification by lectin glycoarray and lectin blot. J Proteome Res. 2008;7:1693–1703. doi: 10.1021/pr700706s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Haseley SR, Talaga P, Kamerling JP, Vliegenthart JF. Characterization of the carbohydrate binding specificity and kinetic parameters of lectins by using surface plasmon resonance. Anal Biochem. 1999;274:203–210. doi: 10.1006/abio.1999.4277. [DOI] [PubMed] [Google Scholar]
- 138.Bryan MC, Plettenburg O, Sears P, Rabuka D, Wacowich-Sgarbi S, Wong CH. Saccharide display on microtiter plates. Chem Biol. 2002;9:713–720. doi: 10.1016/s1074-5521(02)00155-2. [DOI] [PubMed] [Google Scholar]
- 139.Duverger E, Frison N, Roche AC, Monsigny M. Carbohydrate-lectin interactions assessed by surface plasmon resonance. Biochimie. 2003;85:167–179. doi: 10.1016/s0300-9084(03)00060-9. [DOI] [PubMed] [Google Scholar]
- 140.Ratner DM, Adams EW, Su J, O'Keefe BR, Mrksich M, Seeberger PH. Probing protein-carbohydrate interactions with microarrays of synthetic oligosaccharides. Chembiochem. 2004;5:379–382. doi: 10.1002/cbic.200300804. [DOI] [PubMed] [Google Scholar]
- 141.Oyelaran O, Gildersleeve JC. Glycan arrays: recent advances and future challenges. Curr Opin Chem Biol. 2009 doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 2009;16:36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.de Boer AR, Hokke CH, Deelder AM, Wuhrer M. General microarray technique for immobilization and screening of natural glycans. Anal Chem. 2007;79:8107–8113. doi: 10.1021/ac071187g. [DOI] [PubMed] [Google Scholar]
- 144.Blixt O, Allin K, Bohorov O, Liu X, Andersson-Sand H, Hoffmann J, Razi N. Glycan microarrays for screening sialyltransferase specificities. Glycoconj J. 2008;25:59–68. doi: 10.1007/s10719-007-9062-z. [DOI] [PubMed] [Google Scholar]
- 145.Seeberger PH. Automated carbohydrate synthesis as platform to address fundamental aspects of glycobiology--current status and future challenges. Carbohydr Res. 2008;343:1889–1896. doi: 10.1016/j.carres.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 146.Seeberger PH, Werz DB. Synthesis and medical applications of oligosaccharides. Nature. 2007;446:1046–1051. doi: 10.1038/nature05819. [DOI] [PubMed] [Google Scholar]
- 147.Oyelaran O, McShane LM, Dodd L, Gildersleeve JC. Profiling human serum antibodies with a carbohydrate antigen microarray. J Proteome Res. 2009 doi: 10.1021/pr900515y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- 149.Wang D, Liu S, Trummer BJ, Deng C, Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol. 2002;20:275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- 150.Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray analysis of 24 lectins. Angew Chem Int Ed Engl. 2006;45:3607–3610. doi: 10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- 151.Park S, Shin I. Fabrication of carbohydrate chips for studying protein-carbohydrate interactions. Angew Chem Int Ed Engl. 2002;41:3180–3182. doi: 10.1002/1521-3773(20020902)41:17<3180::AID-ANIE3180>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 152.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Manimala JC, Li Z, Jain A, VedBrat S, Gildersleeve JC. Carbohydrate array analysis of anti-Tn antibodies and lectins reveals unexpected specificities: implications for diagnostic and vaccine development. Chembiochem. 2005;6:2229–2241. doi: 10.1002/cbic.200500165. [DOI] [PubMed] [Google Scholar]
- 154.Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology. 2007;17:17C–23C. doi: 10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- 155.Wang SK, Liang PH, Astronomo RD, Hsu TL, Hsieh SL, Burton DR, Wong CH. Targeting the carbohydrates on HIV-1: Interaction of oligomannose dendrons with human monoclonal antibody 2G12 and DC-SIGN. Proc Natl Acad Sci U S A. 2008;105:3690–3695. doi: 10.1073/pnas.0712326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ratner DM, Seeberger PH. Carbohydrate microarrays as tools in HIV glycobiology. Curr Pharm Des. 2007;13:173–183. doi: 10.2174/138161207779313650. [DOI] [PubMed] [Google Scholar]
- 157.Stevens J, Blixt O, Paulson JC, Wilson IA. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat Rev Microbiol. 2006;4:857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stevens J, Blixt O, Chen LM, Donis RO, Paulson JC, Wilson IA. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J Mol Biol. 2008;381:1382–1394. doi: 10.1016/j.jmb.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Liang PH, Wu CY, Greenberg WA, Wong CH. Glycan arrays: biological and medical applications. Curr Opin Chem Biol. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Horlacher T, Seeberger PH. Carbohydrate arrays as tools for research and diagnostics. Chem Soc Rev. 2008;37:1414–1422. doi: 10.1039/b708016f. [DOI] [PubMed] [Google Scholar]
- 161.Wu CY, Liang PH, Wong CH. New development of glycan arrays. Org Biomol Chem. 2009;7:2247–2254. doi: 10.1039/b902510n. [DOI] [PubMed] [Google Scholar]
- 162.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. Influencing receptor-ligand binding mechanisms with multivalent ligand architecture. J Am Chem Soc. 2002;124:14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 163.Wolfenden ML, Cloninger MJ. Mannose/glucose-functionalized dendrimers to investigate the predictable tunability of multivalent interactions. J Am Chem Soc. 2005;127:12168–12169. doi: 10.1021/ja053008n. [DOI] [PubMed] [Google Scholar]
- 164.Horan N, Yan L, Isobe H, Whitesides GM, Kahne D. Nonstatistical binding of a protein to clustered carbohydrates. Proc Natl Acad Sci U S A. 1999;96:11782–11786. doi: 10.1073/pnas.96.21.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Oyelaran O, Li Q, Farnsworth D, Gildersleeve JC. Microarrays with varying carbohydrate density reveal distinct subpopulations of serum antibodies. J Proteome Res. 2009;8:3529–3538. doi: 10.1021/pr9002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Seeberger PH. Chemical glycobiology: why now? Nat Chem Biol. 2009;5:368–372. doi: 10.1038/nchembio0609-368. [DOI] [PubMed] [Google Scholar]
- 167.Blow N. Glycobiology: A spoonful of sugar. Nature. 2009;457:617–620. doi: 10.1038/457617a. [DOI] [PubMed] [Google Scholar]
- 168.Raman R, Venkataraman M, Ramakrishnan S, Lang W, Raguram S, Sasisekharan R. Advancing glycomics: implementation strategies at the consortium for functional glycomics. Glycobiology. 2006;16:82R–90R. doi: 10.1093/glycob/cwj080. [DOI] [PubMed] [Google Scholar]
- 169.Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita KF, Ueda N, Hamajima M, Kawasaki T, Kanehisa M. KEGG as a glycome informatics resource. Glycobiology. 2006;16:63R–70R. doi: 10.1093/glycob/cwj010. [DOI] [PubMed] [Google Scholar]
- 170.Lutteke T, Bohne-Lang A, Loss A, Goetz T, Frank M, von der Lieth CW. GLYCOSCIENCES.de: an Internet portal to support glycomics and glycobiology research. Glycobiology. 2006;16:71R–81R. doi: 10.1093/glycob/cwj049. [DOI] [PubMed] [Google Scholar]