Abstract
Introduction
Retroviral vectors have been developed for hematopoietic stem cell (HSC) gene therapy and have successfully cured X-linked severe combined immunodeficiency (SCID-X1), adenosine deaminase deficiency (ADA-SCID), adrenoleukodystrophy, and Wiskott-Aldrich syndrome. However, in HSC gene therapy clinical trials, genotoxicity mediated by integrated vector proviruses has led to clonal expansion, and in some cases frank leukemia. Numerous studies have been performed to understand the molecular basis of vector-mediated genotoxicity with the aim of developing safer vectors and safer gene therapy protocols. These genotoxicity studies are critical to advancing HSC gene therapy.
Areas covered
This review provides an introduction to the mechanisms of retroviral vector genotoxicity. It also covers advances over the last 20 years in designing safer gene therapy vectors, and in integration site analysis in clinical trials and large animal models. Mechanisms of retroviral-mediated genotoxicity, and the risk factors that contribute to clonal expansion and leukemia in HSC gene therapy are introduced.
Expert opinion
Continued research on virus–host interactions and next-generation vectors should further improve the safety of future HSC gene therapy vectors and protocols.
Keywords: gene therapy, genotoxicity, leukemia, retroviral vector
1. Introduction: retroviral vectors as therapeutic agents and insertional mutagens
The ability of retroviral vectors to efficiently integrate into the host genome, thereby permanently delivering a therapeutic transgene has led to their use for hematopoietic stem cell (HSC) gene therapy. In this approach patient hematopoietic cells are collected, enriched for repopulating cells using the CD34 surface marker, exposed to retroviral vector preparations ex vivo, and then re-infused into the patient. If the procedure is successful, enough gene-modified repopulating cells engraft and produce sufficient numbers of corrected mature hematopoietic cells to correct a disease phenotype. To date several genetic immunodeficiencies have been corrected using this approach, including X-linked severe combined immunodeficiency (SCID-X1) [1,2], adenine deaminase deficiency (ADA-SCID) [3–6], chronic granulomatous disease (CGD) [7], X-linked adrenoleukodysrophy [8] and Wiskott-Aldrich syndrome [9]. A major advantage of using retroviral vectors including gammaretroviral, and more recently lentiviral, vectors, is that the genetic modification is permanent. The differentiation and massive expansion of long-term repopulating cells during hematopoiesis requires that any gene delivery platform allow for efficient transmission of transgenes to daughter cells. When repopulating cells that contain the integrated retroviral vector provirus divide, transmission of the therapeutic transgene to both daughter cells is ensured via mitosis. To date other integrating or episomal vectors have not allowed for efficient gene delivery to HSCs in large animal models, or in patients. The ability to permanently modify the genome of hematopoietic repopulating cells has allowed for sustained correction of ADA-SCID, SCID-X1, adrenoleukodystrophy and Wiskott-Aldrich syndrome in patients. Thus, a lasting cure is effected by somatic mutation. However, the ability of retroviruses to permanently modify the host genome of repopulating cells is also a disadvantage for HSC gene therapy; retroviral vectors are insertional mutagens.
1.1 Success and adverse events in the French SCID-X1 trial
The report of successful treatment of SCID-X1 patients with a gammaretroviral vector in 2000 generated great enthusiasm and optimism in the gene therapy field [2]. SCID-X1 is an inherited disease caused by inactivating mutations of the γC cytokine receptor common subunit (IL2RG locus on the X chromosome) which delivers growth, survival and differentiation signals to early lymphoid progenitors. SCID-X1 patients do not have mature T cells or NK cells and have dysfunctional B cells. This disease is fatal early in life if left untreated due to uncontrolled infections. Years of development of retroviral vectors and ex vivo transduction protocols with testing in small and large animal models paved the way for this and other HSC gene therapy successes. However, a few years after the gene therapy protocol, two patients were diagnosed with lymphoproliferative leukemias. It was established that the transformed cells from these two patients contained integrated vector proviruses near LIM domain only 2 (LMO2), a known proto-oncogene, suggesting the leukemias were vector-mediated [10]. Leukemia in this trial came as a relative surprise. It was well understood that replicating retroviruses can cause leukemia, but it was thought that replication-incompetent vectors had a low probability of transforming cells. Assays for contaminating replication-competent virus were negative in these patients, and the field was left to accept that replication-incompetent vectors do in fact pose a significant risk for leukemia via insertional mutagenesis, at least in some disease settings. Additional patients in this trial, and also in a SCID-X1 gene therapy trial in the UK developed vector-mediated leukemias [11,12].
Following these reports, major efforts went into better understanding the cause of these events. In these trials, vector-mediated dysregulation of nearby genes by integrated vector proviruses led to clonal expansion, and it appears that additional mutations were necessary to cause frank leukemia [11,12]. What these studies showed is that there is a significant risk of severe adverse events based on vector-mediated dysregulation of proto-oncogenes, at least for immunodeficiencies. Thus, like other drugs, retroviral vectors can cause severe side-effects. Understanding the mechanisms of retroviral vector genotoxicity is thus a relatively new field, and one that is already starting to provide insight into how to design safer viral vectors. This review covers mechanisms of retroviral vector genotoxicity and recent advances in designing safer gene therapy vectors.
1.2 Retroviruses as insertional mutagens
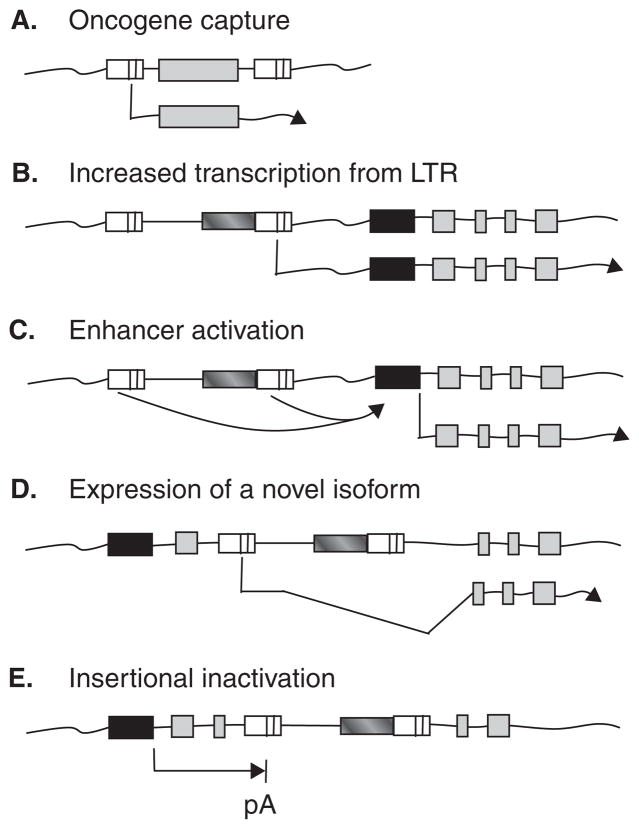
Before their development as vectors, retroviruses were invaluable tools to identify mechanisms of oncogenesis. In 1911 Peyton Rous discovered the first known tumor virus, Rous sarcoma virus (RSV), an avian retrovirus that induces tumors in chickens [13]. This discovery resulted in a Nobel prize many years later in 1966. The subsequent identification of mammalian tumor viruses, and the discovery by Bishop and Varmus that retroviruses can capture and deliver cellular proto-oncogenes [14], led to the identification of numerous additional proto-oncogenes. Retroviral mutagenesis has thus provided major advances in our understanding of cancer. There are several mechanisms whereby retroviruses can transform cells (Figure 1). It is important to note that oncogene capture (Figure 1A) is specific to replicating retroviruses, whereas several additional mechanisms (Figure 1B – E) can also occur with replication-incompetent retroviral vectors.
Figure 1. Retroviral mechanisms of oncogenesis.
A. Acute-transforming replication-competent retroviruses capture a cellular proto-oncogene that mediates transformation. This mechanism does not occur in replication-incompetent vectors. B. The provirus 3′ long terminal repeat (LTR) triggers transcription of a cellular proto-oncogene at increased levels. C. Enhancers in the provirus LTRs activate transcription from a nearby cellular proto-oncogene promoter. D. Transcription from the provirus 5′ LTR creates a novel truncated isoform of a cellular proto-oncogene via splicing. E. A provirus disrupts transcription by causing premature polyadenylation (pA). The integrated provirus is indicated by two LTRs. Cellular proto-oncogene promoter and exons regions are indicated by black and grey boxes respectively.
1.3 Replication-incompetent vector design reduces, but does not eliminate, oncogenic potential
The discovery of retroviral reverse transcription and provirus integration led to the development of retroviral vectors as gene transfer vehicles [15,16]. The ability of retroviruses to efficiently enter cells, migrate to the nucleus and insert their reverse-transcribed DNA genome into a host chromosome were co-opted to efficiently deliver a transgene. Early replication-competent vectors carried selectable markers in addition to their full complement of retroviral genes. Replication-incompetent vectors were then designed where only the ‘cis-acting’ sequences required for efficient gene delivery are retained in the vector. In this design the structural genes, gag, pol and env are replaced by a transgene in the vector (Figure 2A). The Gag, Pol and Env proteins necessary for virion formation are provided in ‘trans’ either from an engineered packaging cell line, or from plasmid DNAs that are co-transfected with the vector plasmid into a cell line. The resulting replication-incompetent vector is able to deliver the therapeutic transgene, but does not express any viral genes in the target cell. It is therefore a replicative dead-end. Unlike acute transforming retroviruses, replication-incompetent vectors do not capture and deliver cellular proto-oncogenes at a detectable frequency. However, the integrated vector provirus is not inert. Once integrated, the vector provirus can still influence the expression of nearby cellular genes via several mechanisms (Figure 1B – E). To date the primary mechanism of oncogenicity in gene therapy trials appears to be via enhancer-mediated activation of nearby genes including LMO2 in the SCID-X1 gene therapy trial, although transcriptional activation has also been observed [17]. The development of leukemia in the SCID-X1 gene therapy trial stimulated research to identify where different types of retroviral vectors integrate in the genome and to better understand vector provirus–host gene interactions.
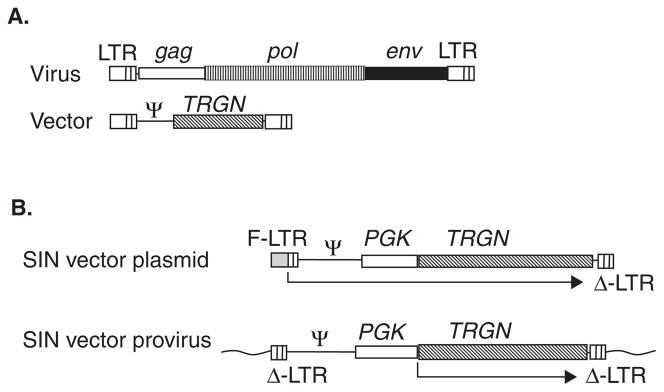
Figure 2. Retroviral vectors.
A. Replication-incompetent vector design. The viral gag, pol and env genes are removed and a transgene (TRGN) is inserted. Cis-acting regions including the Psi packaging region are retained. B. Self-inactivating (SIN) vector design. During vector production, vector transcripts are driven by a 5′ fusion long terminal repeat (F-LTR) promoter that contains a heterologous enhancer (grey box) fused with the viral LTR. The 3′ LTR contains a deletion in the U3 region. During reverse transcription the deleted U3 region of the 3′ LTR is copied to the 5′ LTR of the vector provirus, resulting in a provirus with flanking deleted LTRs that are essentially inactive. An internal promoter such as phosphoglycerate kinase (PGK) drives transcription.
1.4 Where retroviral vectors integrate and why
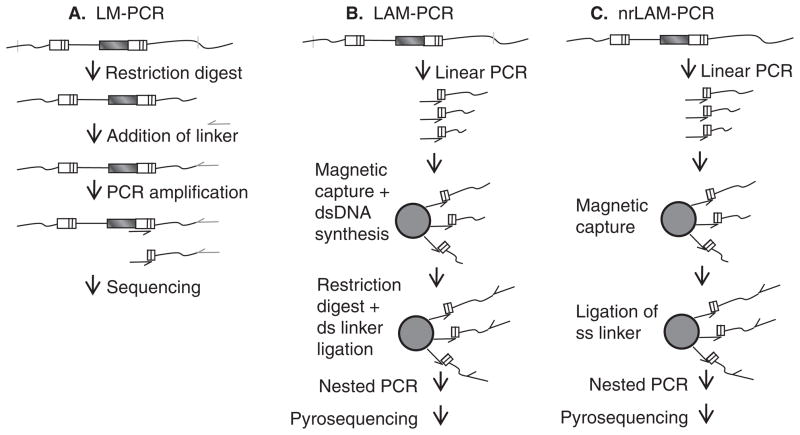
One important factor that affects the genotoxicity of vector proviruses is where they integrate in the host genome. Early studies that sought to understand the integration profiles of different viruses were limited by relatively small numbers of proviruses, and an incomplete genome sequence with which to map these integrants [18,19]. With the sequencing of the human genome, and also the genomes of model organisms such as mouse, dog and rhesus macaque, large-scale identification of retroviral vector integration site distributions became possible. The first large-scale analysis of HIV (lentivirus) integration sites was performed by infecting a human T cell line with HIV or HIV vectors [20]. Provirus–chromosome junctions were detected by cleaving genomic DNA with restriction enzymes, ligating a linker, and PCR amplifying the provirus–chromosome junctions as shown in Figure 3A. Using this approach 524 unique integration sites were identified and then mapped on the draft human genome to determine the distribution of integration sites. Improved techniques have been developed since this study including a method (Figure 3C) that does not require restriction enzyme digestion and thereby overcomes restriction enzyme site-mediated biases [21]. HIV integration was strongly favored within genes, and active genes were preferentially targeted [20]. This study documented unexpectedly strong biases in integration site distribution, and also showed that there were integration hotspots. Large-scale integration studies followed for the murine leukemia virus and avian sarcoma-leukosis virus gammaretroviruses [22–24], and for foamy retroviruses [25]. Integration sites were mapped relative to several genomic features including transcription start sites, transcription units, and also CpG islands. CpG islands are genomic regions that contain a high frequency of CpG dinucleotides and are associated with promoters [26,27]. Remarkably, each type of virus was found to have a unique integration site profile. Table 1 summarizes the findings in these studies. From these and subsequent high-definition studies [28,29] the following observations were made. i) Different types of retroviral vectors have distinct integration profiles. ii) The route of entry does not appear to strongly affect integration distribution. HIV vectors pseudotyped with vesicular stomatitis virus (VSV)-G have a similar integration profile to HIV virions with the native HIV envelope [20,30] despite differences in route of entry [31]. iii) The unique integration profile for each virus type is largely independent of target cell type [23–25], although the transcriptional program and epigenetic status of the target cell can influence integration site selection [28,32]. iv) For lentiviruses, which can integrate independently of mitosis, the cell cycle status of the target cell has only a modest effect on integration site distribution [33].
Figure 3. Identification of provirus-chromosome junctions.
A. Ligation-mediated PCR (LM-PCR). Genomic DNA with integrated vector proviruses is digested with frequent-cutting restriction enzymes. A linker is ligated to the cleaved DNA, and after PCR amplification the products are sequenced with a long terminal repeat (LTR)-specific primer. B. Linear-amplification-mediated PCR (LAM-PCR). Linear PCR with only a LTR-specific primer is performed on genomic DNA with integrated vector proviruses to amplify provirus–chromosome junctions. Following amplification, provirus–chromosome junctions of various lengths are produced. The products are enriched on magnetic beads converted to dsDNA, digested with a restriction enzyme (s), typically frequent-cutting enzymes with a 4 bp recognition site, and a double-stranded linker is ligated to the ends. Nested PCR is followed by direct sequencing, shotgun cloning into bacterial plasmids and sequencing or pyrosequencing. C. Non-restrictive linear-amplification-mediated PCR (nrLAM-PCR). Linear PCR with only a LTR-specific primer is performed on genomic DNA with integrated vector proviruses to amplify provirus–chromosome junctions. The products are enriched on magnetic beads and a single-stranded linker is ligated to the ends. Nested PCR is followed by pyrosequencing. For all methods aligning the chromosomal sequence immediately next to the LTR to a published genome allows identification of the provirus integration site.
Table 1.
Retroviral integration site distributions.
| Virus/vector | Percentage of integration sites
|
Description | |
|---|---|---|---|
| In genes* | In CpG islands‡ | ||
| MLV§ | 43.3 | 17.7 | §§ Strong preference for transcript start sites |
| HIV¶ | 65.2 | 1.5 | ¶¶ Strong preference for transcription units |
| ASLV# | 42.9 | 2.7 | Weak preference for transcription units |
| Foamy** | 28.6 | 9.9 | Modest preference for transcript start sites |
| Random‡‡ | 34.1 | 2.6 | NA |
Within the transcribed region of Refseq genes, human genome build 35 (hg17).
In a CpG island or within 1 kilobase.
1757 HIV integrations in primary lung fibroblasts (426), PBMCs (487), SupT1 cells (504), H9 cells (215) and HeLa (125) cells from [20,22,24], as analyzed in [25].
480 Avian sarcoma-leukosis virus (ASLV) integrations in HeLa (181) and 293T-TVA cells (299) from [23,24], as analyzed in [25].
2829 Foamy integrations in normal human fibroblasts (1008) and CD34+ hematopoietic cells (1,821) [25].
10,000 randomly generated sites as previously described [25].
Strong as defined by > 15% and modest as defined by between 10 and 12% of integrants within 2.5 kb of transcript start sites respectively.
Strong as defined by > 60% and weak as defined by < 45% of integrants within transcription units respectively.
NA: Not applicable.
Our understanding of the factors that determine this integration profile is incomplete, but it is partially determined by the types of host proteins the retroviral pre-integration complex (PIC) associates with. The PIC is a nucleoprotein complex including the viral genome, integrase and additional viral proteins. During infection the PIC must migrate to the nucleus. A study that investigated the contribution of the viral integrase to integration site distribution by creating murine leukemia virus MLV-HIV integrase and Gag chimeras showed that the MLV integrase is a major determinant of integration site specificity, but that MLV Gag proteins also can play a role [34]. One well-characterized example of a host protein that affects this process for HIV is lens-epithelium-derived growth factor (LEDGF), which binds tightly to HIV integrase [35]. LEDGF protects the HIV integrase from proteasomal degredation [36], and integrase mutants that cannot bind LEDGF are defective for tethering to chromosomes and replication [37]. In cells depleted for LEDGF, not only is HIV integration drastically reduced, but the distribution of HIV integration sites is altered [38]. Integration in LEDGF-depleted cells was less frequent in transcription units, less frequent in genes regulated by LEDGF and more frequent in GC-rich DNA [38]. Thus, interaction of the viral PIC with host proteins that facilitate tethering is currently considered to be one of the main factors responsible for retroviral integration site distribution. There are also weak preferences for small palindromic sequences at integration sites [39]. RNA interference screens have identified numerous other host cell proteins that affect viral replication [40,41]. As we better characterize and understand host cell–virus interactions we will undoubtedly uncover additional viral PIC–host protein interactions that influence viral integration patterns.
2. Clonal expansion following HSC gene therapy
Following integration, a vector provirus can influence the expression of nearby genes and confer an altered phenotype on the gene-modified target cell. In HSC gene therapy this can lead to an over-representation of repopulating clones that harbor a particular vector provirus. In some cases clonal expansion with additional mutations has led to frank leukemia [10–12] or myelodysplasia [42]. Conveniently, the vector provirus acts as a molecular tag with a unique vector provirus–chromosome junction. Tracking the frequency of individual clones by tracking the presence and relative frequency of unique provirus–chromosome junctions in animal models and in patients can provide information on clonal expansion. Understanding this process, and designing improved vectors that reduce the potential for clonal expansion is key to improving the safety of HSC gene therapy.
It is important to note that although improvements have been made in the methodology to detect integration sites as described above and outlined in Figure 3, our ability to accurately track clonality still relies on exponential PCR amplification of DNA templates. It is well recognized that the identification of clonality is biased, and the frequency of re-capture of sites has been used to investigate and correct for this bias [43,44]. Shuttle vector rescue is an alternative proven approach [25,45] that does not require exponential amplification of DNA templates prior to sequencing, and could be used in future pre-clinical studies to further investigate the extent of PCR-based bias.
The potential for clonal expansion introduces an important consideration when evaluating integration sites in hematopoietic repopulating cells. Following the initial integration site distribution of a given vector type in a given cell, the vector provirus may alter the phenotype of the transduced cell. If the insertion sites are evaluated a significant amount of time after infection, the observed integration profile is a combination of i) the initial integration site distribution and ii) the effects of clonal expansion of gene-modified cells during culture. For HSC gene therapy, the ex vivo culture conditions may play a significant role in enriching specific clones, even prior to infusion of gene-modified cells. For example, in one study evaluating the effects of prolonged ex vivo culture in a macaque HSC gene therapy model, significant enrichment of gammaretroviral vector integration sites near myelodysplasia syndrome 1 homolog/ecotropic viral integration site 1 (MDS1/EVI1) occurred during only 6 additional days in culture [46]. It should be noted that 6 days in culture is longer than gene therapy trials where cells have been kept in culture for a maximum of 3 – 4 days.
Following infusion of gene-modified cells, further selective pressures can influence the observed integration site distribution. The ability of transduced cells to avoid apoptosis, home to the bone marrow, engraft and provide long term hematopoiesis can all potentially be influenced by host genes dysregulated by integrated vector proviruses. It is important to consider the length of time in which the enrichment of specific clones can occur in vivo. In several studies in patients and in large animal models, analysis of integration sites in long-term repopulating cells was performed up to several years after provirus integration and infusion [8,47–52]. Thus even very subtle changes in the phenotype of individual cells within a polyclonal population of infused cells could affect their relative fitness. This in turn may have significant effects on their clonal contribution over long time periods. Integration site analysis in repopulating cells therefore measures not only vector-specific integration site distribution, but also potentially the final effect on clonality of provirus dysregulation of host genes that affect apoptosis, homing, engraftment and long-term hematopoiesis. Because of this, integration site analysis in repopulating cells can provide information on vector genotoxicity that is a combination of the initial integration site distribution, and the effects of host gene dysregulation on clonality. Identifying vector provirus locations in repopulating cells can therefore potentially identify disease setting-specific genotoxic effects.
2.1 Clonal expansion in HSC gene therapy clinical trials
In the French and UK SCID-X1 trials, analysis of integration sites in repopulating cells has now identified common integration sites (CISs) near several proto-oncogenes including B lymphoma Moloney murine leukemia virus insertion region 1 (BMI1), cyclin D2 (CCND2), LMO2, high mobility group AT-hook 2 (HMGA2), runt related transcription factor (RUNX)2, RUNX3, septin 9 (SEPT9) and zinc finger protein 217 (ZNF217) in long-term repopulating cells [10–12,43,53]. Additionally, the genes observed near vector proviruses were enriched for growth-promoting genes and genes that affect survival. Integration sites were also compared between gene modified cells prior to infusion, and in long term mature repopulating cells for SCID-X1 [53] and ADA-SCID gene therapy [54]. In the ADA-SCID study, significant differences were not observed between pre-infusion and repopulating cells. However, as noted by the authors, this data is only suggestive, since the integration site analysis performed in the pre-infusion samples came from a cell population where only a minority of the gene-modified cells were long-term repopulating cells. Currently it is not possible to purify human or large animal long-term repopulating cells to homogeneity, so a clear comparison of integration sites in long-term repopulating cells pre- and post-infusion is not possible. The integration pattern in the ADA-SCID study did correlate with the expression profile and epigenetic state in the CD34+ target cells [32,54]. This finding supported a previous observation of an enrichment for gammaretroviral vector proviruses in non-human primate long-term repopulating cells near genes expressed in primitive CD34+ cells [51]. In the SCID-X1 study the authors concluded that preferential vector integration into genes expressed in the target CD34+ cell population was followed by the vector provirus actively influencing the fate of corrected cell clones in vivo [53].
CGD is a rare inherited life-threatening immunodeficiency where 70% of affected individuals have mutations in the X-linked gene encoding glycoprotein gp91phox, resulting in a lack of antimicrobial activity of phagocytes. Unlike SCID-X1, the therapeutic transgene gp91phox is not known to confer a selective advantage to corrected cells in vivo. Thus, a relatively high percentage of repopulating cells must be corrected to attain a therapeutic outcome. Because of this a reduced intensity conditioning with liposomal busulfan was used to condition patients prior to infusion of gene-modified cells in a CGD clinical trial conducted by Ott and colleagues [7]. No conditioning was used in the SCID-X1 trials described above. Following infusion, polyclonal reconstitution occurred. However, starting approximately 3 months after the therapy, clones began to emerge in these patients, and predominant clones were observed with CISs in the MDS1/EVI1, PR domain containing 16 (PRDM16) and SET binding protein 1 (SETBP1) loci [7]. In these patients repopulating clones with insertions near these genes became predominant with over 80% of detected vector provirus integration sites in these three CISs. In this study repopulating cells over-expressed MDS1/EVI1 and SETBP1 in one patient and MDS1/EVI1 and PRDM16 in the second patient. This clonal expansion, presumably as a result of dysregulation of these and possibly additional genes by vector proviruses, contributed to the success of the therapy by increasing the percentage of corrected cells [7]. The authors concluded that the vector proviruses affected the fate of repopulating cells, possibly in part due to the inclusion of a strong spleen-focus forming virus (SFFV) long-terminal repeat (LTR). Unfortunately following this initially successful treatment, silencing of the transgene occurred in both patients due to methylation of the viral promoter. One patient died from sepsis and the other was treated with an alternative therapy, allogeneic transplantation. Additional analysis of these patients showed that vector-mediated dysregulation of EVI1 led to genomic instability and clonal progression toward myelodysplasia [42].
Clonal expansion was also observed in a lentiviral HSC gene therapy trial for β-thalassemia, as result of transcriptional activation of HMGA2 [17]. In this study most of the therapeutic benefit resulted from a dominant clone, in which the integrated vector activated HMGA2. Integration created a novel HMGA2 transcript that was resistant to degredation and may have played a role in expansion in vivo. Thus, in several high-profile clinical gene therapy studies, the vector provirus appears to have played a role in enhancing the survival and/or growth of repopulating clones. In both CGD and β-thalassemia transgene-mediated clonal expansion played a role in therapeutic benefit.
HSC gene therapy has also been performed for Wiskott-Aldrich syndrome, a severe X-linked recessive immunodeficiency associated with thrombocytopenia, eczema and autoimmunity. In this trial the results from two boys who received transplants of gammaretroviral transduced CD34+ cells after conditioning with busulfan have been reported [9]. Stable chimerism with polyclonal repopulation and resolution of disease symptoms occurred in both patients. Linear-amplification-mediated (LAM)-PCR analysis with pyrosequencing showed that despite the absence of dominant clones, clonal skewing had occurred with enrichment of genes near vector proviruses that are involved in cellular development, growth, proliferation and hematopoiesis. Interestingly, there was an enrichment of vector insertions in or near genes regulating immune and autoimmune responses in both patients [9]. Clones containing vector proviruses near CCND2 and MDS1/EVI1 were also observed. Analysis of sorted lymphoid and myeloid fractions showed that the over-represented genes near vector proviruses contributing to long-term repopulation were distinct in both lineages. This suggests that distinct lineage-specific proliferative advantages mediated by vector proviruses contributed to over-representation of specific clones in each lineage. Unfortunately, a severe adverse event of treatment-related T cell leukemia has recently been reported in this trial, but details on the molecular cause have not yet been published.
2.2 Over-representation of repopulating clones without clonal expansion or leukemia in large animal studies
Large-animal models of HSC gene therapy allow analysis of integration sites and clonality in long-term repopulating cells. Two studies of gammaretroviral and lentiviral vector integration sites in non-human primates reported the distribution of integration sites in long-term repopulating cells [50,51]. In these studies there was no evidence of clonal expansion or leukemia. When data from these two studies were pooled, a statistically significant increase in the frequency of vector proviruses near proto-oncogenes was observed for either MLV-based gammaretroviral vectors, or HIV and SIV-based lentiviral vectors. A total of 8.8% of lentiviral, and 8.2% of gammaretroviral proviruses were observed within proto-oncogenes in repopulating cells, whereas in a random dataset only 2.5% of integrations occurred [51]. Highly significant over-representation of specific gene classes near vector provi-ruses occurred in both studies although the over-represented gene classes differed. Interestingly, in these two primate studies integrants were observed near LMO2, MDS1/EVI1, RUNX2, RUNX3 and HMGA2. All of which are proto-oncogenes near CISs in the SCID-X1 trials. Thus, even when clonal expansion is not observed, vector proviruses are over-represented near genes that may have influenced the survival of that repopulating clone. One CIS occurred in repopulating cells from three different baboons in a very small 664 bp window within a CpG island between two genes, leupaxin (LPXN) and zinc finger protein 91 (ZFP91) [55]. Repopulating cells with these integrants were tracked over time using allele-specific PCR, and clonal expansion was not detected. As mentioned above, it is not possible to directly compare repopulating cells pre- and post-infusion to formally show that these integrants were enriched during engraftment. However, it can be speculated that dysregulation of these genes may have enhanced this process. This can be directly tested in future studies where these genes are over-expressed or knocked down in a competitive repopulation assay [56,57]. Thus, vector integration data from long-term repopulating cells in large-animal models and also patients may identify novel genes and gene pathways that contribute to homing, engraftment and hematopoiesis. Gene therapy studies that analyze integration sites in repopulating cells can thus provide novel and important information on genes involved in engraftment. The integration site data from studies on long term repopulating cells may thus complement retroviral mutagenesis studies in mice that identify genes involved in oncogenesis, stem cell renewal or competitive fitness [58,59].
3. Risk factors for clonal expansion
One of the reasons that the finding of leukemia in the SCID-X1 trials came as a relative surprise is that a number of gene therapy studies in large-animal models had suggested that gene transfer with gammaretroviral vectors was safe [60]. Additionally a number of T cell gene therapy trials had also suggested that gammaretroviral gene transfer was safe [61,62]. However these studies differed from the SCID-X1 gene therapy trials in that in the animal studies, the recipients were not immunodeficient, and in the T cell studies, the target cells were T lymphocytes, not CD34+ cells. Following the finding of leukemia in the SCID-X1 studies, a number of studies in vitro and in animal models sought to better understand the relative contribution of different disease settings, cell target types, and retroviral vector design. These findings can be summarized as follows; i) the specific transgene and disease setting may play a role in the potential for adverse events, ii) different target cell types such as T cells and CD34+ cells appear to differ in their potential for transformation, and iii) vector type and design can play a major role in the propensity for transformation in vivo.
3.1 Effects of the transgene on genotoxicity
In the case of SCID-X1 the γC therapeutic transgene confers a selective advantage to T cells in vivo. In the SCID-X1 patients treated to date, the percentage of gene-corrected cells in the myeloid lineage is much lower than in lymphoid cells, where the transgene promotes expansion. At long-term follow-up gene-modified cells in the B cell and myeloid lineage are essentially undetectable, in part due to a lack of conditioning [63]. The γC subunit is part of several cytokine receptors including the IL-7 cytokine receptor which is involved in cell growth and plays a role in inhibiting apoptosis in T cells [64]. IL-7 and the γC therapeutic transgene may play a role in genotoxicity for SCID-X1 [65–67]. IL-7 increases the viability and proliferation of acute lymphoblastic leukemia and induces the activation of Jak/signal transducer and activator of transcription (STAT), MAPK kinase (MEK)/extracellular signal-regulated kinase (Erk) and PI3K/Akt signaling pathways in these cells. The PI3K/Akt is the dominant pathway that mediates these effects and PI3K signaling is required for the induction of B cell leukemia/lympoma associated gene 2 (Bcl-2), the downregulation of p27 kinase inhibitor protein 1 (kip1) and cell cycle progression [65]. In mice, leukemias occurred with retroviral insertions near Lmo2 and the γC gene (Il2rg) in the same cell implying that these two genes were both dysregulated and cooperated to induce leukemia [56]. However, some caution must be made when translating findings from mouse models to patients [68]. Integration site analyses in long term repopulating cells in different disease settings have suggested that different therapeutic trans-genes have different specific risks. Because of this, pre-clinical studies with animal models should attempt to replicate the specific disease-setting and therapeutic protocol as closely as possible. The merits and also limitations of mouse and large animal models for genotoxicity studies have been discussed elsewhere [69,70].
3.2 The contribution of target cell type to genotoxicity
Several studies have sought to better understand why leukemia was not observed in several T cell gene therapy studies performed prior to the SCID-X1 HSC gene therapy trials. Analysis of integration sites in T cells modified with gammaretroviral vectors encoding a suicide gene showed that there was a preference for integration near genes that were transcriptionally active in T cells [71]. Furthermore, transcript analysis in individual clones indicated that almost one in five promoter-proximal integrations resulted in gene activation [71]. However, unlike the HSC gene therapy trials, in these T cell studies the infused cells showed no evidence of clonal selection for up to nine years post infusion. Comparison of the vector integration site distribution in T cells before infusion and after engraftment suggests that integrations in growth-controlling genes were counter-selected in vivo [71]. In ADA-SCID a comparison of gammaretroviral vector integration sites between HSCs and T cell targets showed cell-specific integration preferences related to the epigenetic and expression profile at the time of transduction [32]. A direct comparison of mouse HSCs and mature T cells as cell targets for gene transfer leading to T cell transformation also showed that T cells are resistant to transformation [72]. The authors proposed that one mechanism restricting T cell clonal expansion may be that clones with the same T cell receptor compete for the same stimulatory MHC-peptide niche. A direct comparison of the same gammaretroviral ADA-SCID-encoding vector in the same genetic disease background but in different HSC or T cell targets showed a different integration site distribution [32]. Regardless of the mechanisms, these studies highlight the fact that the target cell type can play a major role in the genotoxic risk.
3.3 Impact of vector design on genotoxicity
The contribution of the vector type and design has also been shown to affect genotoxicity. As discussed above, different retroviral vectors have distinct integration site distributions which can affect their genotoxicity. However when the percentage of proviruses observed in or near genes or proto-oncogenes are compared for different vector types in the absence of major selection pressure, only relatively modest twofold to threefold differences are seen [25]. Thus, a major component of the potential genotoxicity will instead be determined by the propensity of a given vector provirus to dysregulate nearby genes. This is influenced both by the vector type and the vector design. A comparison of gamma- and lentiviral vectors with different vector designs showed that lentiviral self-inactivating (SIN) vectors with an internal housekeeping promoter were the least likely to cause tumors in a tumor-prone mouse model and that a lentiviral integration pattern was safer than a gammaretroviral pattern [73,74]. SIN vector design is described in Figure 2B.
In another study the propensity of gamma-, lenti- and foamy retrovirus vectors to transactivate a nearby reporter was directly compared using a plasmid-based transfection assay [75]. Both enhancer activation and transcriptional read-through were evaluated. In this study, MLV-based vectors similar to the ones used in the SCID-X1 trial mediated strong enhancer-based, and also read-through transcription of a nearby gene. HIV vectors also showed significant transcriptional read-through [75]. Unlike MLV or HIV-based vectors, foamy vectors did not show read-through transcription. Additionally, the use of an internal housekeeping phosphoglycerate kinase (PGK) promoter significantly reduced enhancer activation relative to the use of a strong viral promoter.
4. Next-generation vectors designed to reduce the potential for leukemogenesis
It is clear from the above studies that like other drugs, retroviral vectors can be modified to reduce their toxicity and improve their safety profile. The choice of vector type can affect both the initial integration site distribution, and the probability that nearby genes will be dysregulated leading to clonal expansion and possibly leukemia. The use of SIN vectors with internal housekeeping promoters can reduce genotoxicity [73]. However, additional vector modifications are likely to further increase safety. Another approach is to avoid semi-random integration using non-integrating vectors that effect homologous recombination at specific ‘safe harbor’ locations in the genome in order to deliver transgenes [76]. This approach has recently been reviewed elsewhere [77,78].
4.1 Chromatin insulators as a means to reduce genotoxicity
One promising approach to reduce vector-mediated genotoxicity is to incorporate enhancer-blocking insulators into retroviral vectors. Insulators, also called boundary elements, are DNA sequences that block the activity of enhancers on promoters when placed between them [79]. The chicken hypersensitive site-4 (cHS4) insulator is a well characterized insulator that has been incorporated into retroviral vectors [80–85]. When inserted into the U3 region of the 3′ LTR of a vector, the insulator is copied to the 5′ LTR during reverse transcription, thereby bracketing the transgene cassette with insulators. The insulator can potentially provide three functions; i) insulators can protect nearby host genes from enhancer activation mediated by the vector internal enhancer, ii) they can reduce the variable expression observed from vectors integrated in different chromosomal locations, referred to as clonal variegation, and iii) they can also potentially protect the vector transgene cassette from silencing. One challenge has been to develop insulated vectors with high titers [86]. A 650 bp cHS4 insulator element containing the 250 bp ‘core’ element combined with an additional distal 400 bp cHS4 element was found to provide efficient insulating properties [86]. Incorporation of this 650 bp cHS4 element into the LTR only had a modest effect on lentiviral vector titers, reducing titers approximately 2.5-fold [86]. A direct comparison of insulated versus non-insulated lentiviral vectors showed that in a T cell line, incorporation of an cHS4 insulator element reduced the frequency at which dominant clones were observed [80]. In a β-thalassemia clinical study a lentiviral vector with cHS4 insulator did not provide protection from clonal dominance [17]. In this study a dominant clone arose that had only one 250-bp cHS4 insulator core element in each LTR, whereas the lentiviral vector used in the study was designed with two copies of this core element to improve insulator activity [17]. It is currently unclear if clonal dominance was related to loss of one core element, and more studies are needed to establish the potential of insulated vectors.
4.2 Incorporation of cell-type-specific control elements
Strong constitutive viral promoters such as the SFFV promoter have been very useful for pre-clinical studies to evaluate the ability of different vector types and designs to deliver transgenes to multi-lineage long-term repopulating cells [87,88]. However, restricting transgene expression to the therapeutic target cell can potentially reduce genotoxicity by eliminating or reducing enhancer activation in more primitive progenitor cells [89]. Thus, lineage-restricted promoters are an attractive approach to reduce gentoxicity for hematopoietic diseases such as hemoglobinopathies where the disease phenotype is restricted to one lineage. In this example vector proviruses with erythroid-specific promoters would transduce HSCs capable of long term repopulation, but these promoters would not be active in HSCs, multi-potential progenitors or potentially even erythroid progenitors. Only during erythroid differentiation would the enhancer become active, expressing the therapeutic transgene in erythrocytes. This would reduce the potential for enhancer-mediated transactivation of cellular proto-oncogenes in stem and progenitor cells. In the β-thalassemia clinical trial, activation of HMGA2 and clonal expansion occurred in erythroblasts in one patient, consistent with the erythroid specificity of the β-locus-control-region-derived promoter used. In this patient bone marrow cytology and immunotyping were normal with the exception of erythroid hyperplasia. Bone marrow karyotype and high-resolution array-comparative-genomic-hybridization analysis of chromosomes were also normal, consistent with erythroid-lineage-specific clonal amplification. B cell lineage-specific promoters have also been developed and show promise [90–92].
4.3 Targeting retroviral vector proviruses to specific chromosomal locations
Another approach to improve the safety of retroviruses is to develop means to target integration to specific chromosomal locations, or at least to reduce the frequency with which they integrate near proto-oncogenes. The semi-random integration distribution of retroviruses suggests that this will be a challenge. However, there are examples of highly specific targeted integration. Yeast retrotransposons have evolved mechanisms to integrate into a specific location in their host genome. Unlike retroviruses which may actually benefit from genotoxicity by allowing for increased growth, yeast Ty element LTR retrotransposons are intracellular parasites, which are dependent on the long-term survival of their unicellular hosts. The yeast Ty1 and Ty3 elements integrate upstream of polymerase III-transcribed genes. The Ty3 element has a particularly restricted integration profile, integrating within one or two nucleotides of a tRNA transcription site [93]. Attempts to re-target retroviral vectors have shown some potential [94,95], but have not as yet led to safer, high-titer vectors. Recently one report showed that HIV integration could be re-targeted to regions outside of genes and into heterochromatin by modifying LEDGF [96]. The authors created target cells in which LEDGF was modified to contain the chromatin-interacting domain of chromobox homolog 1 (CBX1). CBX1 binds di- or tri-methylated histone H3 which is associated with pericentric heterochromatin and intergenic regions. In this study the HIV vectors expressed an enhanced green fluorecent protein (EGFP) transgene, despite being integrated in genomic regions that are considered unfavorable for transcription. Additional work will be needed to develop a method to target lentiviral integration in cells that express wild-type LEDGF [96]. However the demonstration that significant re-targeting can occur in living cells when an appropriate tether is provided is very encouraging. The findings of this study support future studies to engineer vectors with modified integration site preferences, and also studies to better understand virus–host interactions.
5. Expert opinion and conclusions
Retroviral vectors can be thought of as a new class of drugs with great therapeutic potential, but with incompletely characterized side-effects. As we gain a better understanding of the factors that influence the safety of HSC gene therapy, we should be able to develop safer vectors, and also safer protocols. Key to the development of safer vectors is the development of appropriate models for genotoxicity testing. In vitro models can be used for initial vector design, allowing for rapid development and comparison of novel vectors. Mouse models are appropriate for directly comparing the genotoxicity of promising high-titer vectors. Large-animal models are the most appropriate for identifying disease-specific genotoxicity considerations using therapeutic vectors to predict the effects of vector design on clonal over-representation and expansion in long-term repopulating cells. However, while there are large animal models for some hematopoietic diseases including SCID-X1, leukocyte adhesion deficiency and pyruvate kinase deficiency (reviewed in [70]), many other diseases cannot currently be modeled in large animals.
The enormous potential of HSC gene therapy will provide the impetus for continued development and refinement of retroviral vectors to reduce their genotoxicity. In conjunction with improved vector design, studies are needed to better define and understand virus–host interactions. Understanding these interactions may allow us to develop novel approaches to control integration towards developing safer vectors. This field, while challenging, has the potential to greatly reduce the genotoxicity of retroviral vectors. It should also provide insight into the biology of retroviral infection, and is likely to have benefits outside of HSC gene therapy. For severe life-threatening diseases such as SCID-X1, SIN vectors that use internal housekeeping promoters such as PGK or the elongation factor alpha 1 promoter are a compelling therapeutic option for patients who do not have HLA-matched donors for allogeneic transplantation. As we move forward we should expect that there will be additional therapeutic successes for HSC gene therapy. However, like other drugs there will also probably and unfortunately be some adverse events. We need to continue to identify the molecular mechanisms responsible for retroviral vector genotoxicity, and to study virus–host interactions with the aim of developing safer vectors.
Article highlights.
Hematopoietic stem cell gene therapy with retroviral vectors has great promise but vector-mediated genotoxicity currently limits its potential.
Dysregulation of nearby proto-oncogenes by integrated vector proviruses has led to clonal expansion and frank leukemia in some gene therapy clinical trials.
There are several risk factors for vector-mediated genotoxicity including the therapeutic transgene, the target cell type and the vector design.
Improvements in vector design including the use of self-inactivating vectors, non-viral promoters and chromatin insulators should improve the safety of future gene therapy trials.
Additional work to is needed to characterize the molecular mechanisms of vector-mediated genotoxicity, and to better understand virus–host interactions to further improve the safety of gene therapy.
This box summarizes key points contained in the article.
Footnotes
Declaration of interest
The author was sponsored by an NIH grant and by a New Faculty Seed Grant from Washington State University.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Gaspar HB, Parsley KL, Howe S, et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364:2181–7. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- 2••.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–72. doi: 10.1126/science.288.5466.669. Pioneering study showing unequivocal therapeutic benefit of HSC gene therapy. [DOI] [PubMed] [Google Scholar]
- 3.Aiuti A, Slavin S, Aker M, et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–3. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 4.Aiuti A, Vai S, Mortellaro A, et al. Immune reconstitution in ADA-SCID after PBL gene therapy and discontinuation of enzyme replacement. Nat Med. 2002;8:423–5. doi: 10.1038/nm0502-423. [DOI] [PubMed] [Google Scholar]
- 5.Gaspar HB, Bjorkegren E, Parsley K, et al. Successful reconstitution of immunity in ADA-SCID by stem cell gene therapy following cessation of PEG-ADA and use of mild preconditioning. Mol Ther. 2006;14:505–13. doi: 10.1016/j.ymthe.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Aiuti A, Cattaneo F, Galimberti S, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–58. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- 7•.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–9. doi: 10.1038/nm1393. This research demonstrates clonal expansion of gene-modified cells in vivo. [DOI] [PubMed] [Google Scholar]
- 8•.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–23. doi: 10.1126/science.1171242. Pioneering study using lentiviral vector for HSC gene therapy. [DOI] [PubMed] [Google Scholar]
- 9.Boztug K, Schmidt M, Schwarzer A, et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N Engl J Med. 2010;363:1918–27. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–9. doi: 10.1126/science.1088547. Detailed description of vector-mediated leukemia in patients. [DOI] [PubMed] [Google Scholar]
- 11.Howe SJ, Mansour MR, Schwarzwaelder K, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–50. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–42. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rous R. Transmission of a malignant new growth by means of a cell-free filtrate. JAMA. 1911;56:198. [PubMed] [Google Scholar]
- 14.Varmus HE, Weiss RA, Friis RR, et al. Detection of avian tumor virus-specific nucleotide sequences in avian cell DNAs (reassociation kinetics-RNA tumor viruses-gas antigen-Rous sarcoma virus, chick cells) Proc Natl Acad Sci USA. 1972;69:20–4. doi: 10.1073/pnas.69.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller AD, Jolly DJ, Friedmann T, Verma IM. A transmissible retrovirus expressing human hypoxanthine phosphoribosyltransferase (HPRT): gene transfer into cells obtained from humans deficient in HPRT. Proc Natl Acad Sci USA. 1983;80:4709–13. doi: 10.1073/pnas.80.15.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joyner A, Keller G, Phillips RA, Bernstein A. Retrovirus transfer of a bacterial gene into mouse haematopoietic progenitor cells. Nature. 1983;305:556–8. doi: 10.1038/305556a0. [DOI] [PubMed] [Google Scholar]
- 17•.Cavazzana-Calvo M, Payen E, Negre O, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–22. doi: 10.1038/nature09328. Significant progress in treatment of a difficult disease that requires high levels of corrected cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mooslehner K, Karls U, Harbers K. Retroviral integration sites in transgenic Mov mice frequently map in the vicinity of transcribed DNA regions. J Virol. 1990;64:3056–8. doi: 10.1128/jvi.64.6.3056-3058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scherdin U, Rhodes K, Breindl M. Transcriptionally active genome regions are preferred targets for retrovirus integration. J Virol. 1990;64:907–12. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Schroder AR, Shinn P, Chen H, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–9. doi: 10.1016/s0092-8674(02)00864-4. A major breakthrough in understanding retroviral integration preferences. [DOI] [PubMed] [Google Scholar]
- 21.Gabriel R, Eckenberg R, Paruzynski A, et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med. 2009;15:1431–6. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–51. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 23.Narezkina A, Taganov KD, Litwin S, et al. Genome-wide analyses of avian sarcoma virus integration sites. J Virol. 2004;78:11656–63. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell RS, Beitzel BF, Schroder AR, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trobridge GD, Miller DG, Jacobs MA, et al. Foamy virus vector integration sites in normal human cells. Proc Natl Acad Sci USA. 2006;103:1498–503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–13. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 27.Antequera F, Bird A. CpG islands. EXS. 1993;64:169–85. doi: 10.1007/978-3-0348-9118-9_8. [DOI] [PubMed] [Google Scholar]
- 28.Wang GP, Ciuffi A, Leipzig J, et al. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–94. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cattoglio C, Pellin D, Rizzi E, et al. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood. 2010;116:5507–17. doi: 10.1182/blood-2010-05-283523. [DOI] [PubMed] [Google Scholar]
- 30.Barr SD, Ciuffi A, Leipzig J, et al. HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Mol Ther. 2006;14:218–25. doi: 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 31.Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol. 1997;71:5871–7. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biasco A, Ambrosi A, Pellin D, et al. Integration profile of retroviral vector in gene therapy treated patients is cell-specific according to gene expression and chromatin conformation of target cell. EMBO Mol Med. 2011;3:89–101. doi: 10.1002/emmm.201000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brady T, Agosto LM, Malani N, et al. HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS. 2009;23:1461–71. doi: 10.1097/QAD.0b013e32832caf28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewinski MK, Yamashita M, Emerman M, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherepanov P, Maertens G, Proost P, et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–81. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 36.Llano M, Delgado S, Vanegas M, Poeschla EM. Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J Biol Chem. 2004;279:55570–7. doi: 10.1074/jbc.M408508200. [DOI] [PubMed] [Google Scholar]
- 37.Emiliani S, Mousnier A, Busschots K, et al. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J Biol Chem. 2005;280:25517–23. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- 38.Ciuffi A, Llano M, Poeschla E, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–9. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 39.Wu X, Li Y, Crise B, et al. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J Virol. 2005;79:5211–4. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brass AL, Dykxhoorn DM, Benita Y, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 41.Zhou H, Xu M, Huang Q, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Stein S, Ott MG, Schultze-Strasser S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 43.Wang GP, Berry CC, Malani N, et al. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood. 2010;115:4356–66. doi: 10.1182/blood-2009-12-257352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang GP, Garrigue A, Ciuffi A, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cepko CL, Roberts BE, Mulligan RC. Construction and applications of a highly transmissible murine retrovirus shuttle vector. Cell. 1984;37:1053–62. doi: 10.1016/0092-8674(84)90440-9. [DOI] [PubMed] [Google Scholar]
- 46.Sellers S, Gomes TJ, Larochelle A, et al. Ex vivo expansion of retrovirally transduced primate CD34+ cells results in overrepresentation of clones with MDS1/EVI1 insertion sites in the myeloid lineage after transplantation. Mol Ther. 2010;18:1633–9. doi: 10.1038/mt.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt M, Carbonaro DA, Speckmann C, et al. Clonality analysis after retroviral-mediated gene transfer to CD34+ cells from the cord blood of ADA-deficient SCID neonates. Nat Med. 2003;9:463–8. doi: 10.1038/nm844. [DOI] [PubMed] [Google Scholar]
- 48.Calmels B, Ferguson C, Laukkanen MO, et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–3. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deichmann A, Hacein-Bey-Abina S, Schmidt M, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–32. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hematti P, Hong BK, Ferguson C, et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beard BC, Dickerson D, Beebe K, et al. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol Ther. 2007;15:1356–65. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- 52.Beard BC, Keyser KA, Trobridge GD, et al. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, or foamy virus. Hum Gene Ther. 2007;18:423–34. doi: 10.1089/hum.2007.011. [DOI] [PubMed] [Google Scholar]
- 53.Schwarzwaelder K, Howe SJ, Schmidt M, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–9. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aiuti A, Cassani B, Andolfi G, et al. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J Clin Invest. 2007;117:2233–40. doi: 10.1172/JCI31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiem HP, Ironside C, Beard BC, Trobridge GD. A retroviral vector common integration site between leupaxin and zinc finger protein 91 (ZFP91) observed in baboon hematopoietic repopulating cells. Exp Hematol. 2010;38:819–22. doi: 10.1016/j.exphem.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrison DE. Competitive repopulation: a new assay for long-term stem cell functional capacity. Blood. 1980;55:77–81. [PubMed] [Google Scholar]
- 57.Kiem HP, Heyward S, Winkler A, et al. Gene transfer into marrow repopulating cells: comparison between amphotropic and gibbon ape leukemia virus pseudotyped retroviral vectors in a competitive repopulation assay in baboons. Blood. 1997;90:4638–45. [PubMed] [Google Scholar]
- 58.Kustikova OS, Geiger H, Li Z, et al. Retroviral vector insertion sites associated with dominant hematopoietic clones mark “stemness” pathways. Blood. 2007;109:1897–907. doi: 10.1182/blood-2006-08-044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akagi K, Suzuki T, Stephens RM, et al. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32(Database issue):D523–7. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kiem HP, Sellers S, Thomasson B, et al. Long-term clinical and molecular follow-up of large animals receiving retrovirally transduced stem and progenitor cells: no progression to clonal hematopoiesis or leukemia. Mol Ther. 2004;9:389–95. doi: 10.1016/j.ymthe.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 61.Blaese RM, Culver KW, Miller AD, et al. T lymphocyte-directed gene therapy for ADA–SCID: initial trial results after 4 years. Science. 1995;270:475–80. doi: 10.1126/science.270.5235.475. [DOI] [PubMed] [Google Scholar]
- 62.Bordignon C, Notarangelo LD, Nobili N, et al. Gene therapy in peripheral blood lymphocytes and bone marrow for ADA-immunodeficient patients. Science. 1995;270:470–5. doi: 10.1126/science.270.5235.470. [DOI] [PubMed] [Google Scholar]
- 63.Hacein-Bey-Abina S, Hauer J, Lim A, et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2010;363:355–64. doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barata JT, Cardoso AA, Nadler LM, Boussiotis VA. Interleukin-7 promotes survival and cell cycle progression of T-cell acute lymphoblastic leukemia cells by down-regulating the cyclin-dependent kinase inhibitor p27(kip1) Blood. 2001;98:1524–31. doi: 10.1182/blood.v98.5.1524. [DOI] [PubMed] [Google Scholar]
- 65.Barata JT, Cardoso AA, Boussiotis VA. Interleukin-7 in T-cell acute lymphoblastic leukemia: an extrinsic factor supporting leukemogenesis? Leuk Lymphoma. 2005;46:483–95. doi: 10.1080/10428190400027852. [DOI] [PubMed] [Google Scholar]
- 66.Dave UP, Akagi K, Tripathi R, et al. Murine leukemias with retroviral insertions at Lmo2 are predictive of the leukemias induced in SCID-X1 patients following retroviral gene therapy. PLoS Genet. 2009;5:e1000491. doi: 10.1371/journal.pgen.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woods NB, Bottero V, Schmidt M, et al. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440(7088):1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- 68.Thrasher AJ, Gaspar HB, Baum C, et al. Gene therapy: X-SCID transgene leukaemogenicity. Nature. 2006;443:E5–6. doi: 10.1038/nature05219. discussion E7. [DOI] [PubMed] [Google Scholar]
- 69.Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–49. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 70.Trobridge GD, Kiem HP. Large animal models of hematopoietic stem cell gene therapy. Gene Ther. 2010;17:939–48. doi: 10.1038/gt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Recchia A, Bonini C, Magnani Z, et al. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc Natl Acad Sci USA. 2006;103:1457–62. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Newrzela S, Cornils K, Li Z, et al. Resistance of mature T cells to oncogene transformation. Blood. 2008;112:2278–86. doi: 10.1182/blood-2007-12-128751. [DOI] [PubMed] [Google Scholar]
- 73•.Montini E, Cesana D, Schmidt M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119:964–75. doi: 10.1172/JCI37630. A direct demonstration of the effects of vector design on genotoxicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Modlich U, Navarro S, Zychlinski D, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther. 2009;17:1919–28. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hendrie PC, Huo Y, Stolitenko RB, Russell DW. A rapid and quantitative assay for measuring neighboring gene activation by vector proviruses. Mol Ther. 2008;16:534–40. doi: 10.1038/sj.mt.6300398. [DOI] [PubMed] [Google Scholar]
- 76.Lombardo A, Genovese P, Beausejour CM, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 77.Wanisch K, Yanez-Munoz RJ. Integration-deficient lentiviral vectors: a slow coming of age. Mol Ther. 2009;17:1316–32. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Handel EM, Cathomen T. Zinc-finger nuclease based genome surgery: it’s all about specificity. Curr Gene Ther. 2011;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- 79.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Evans-Galea MV, Wielgosz MM, Hanawa H, et al. Suppression of clonal dominance in cultured human lymphoid cells by addition of the cHS4 insulator to a lentiviral vector. Mol Ther. 2007;15:801–9. doi: 10.1038/sj.mt.6300103. [DOI] [PubMed] [Google Scholar]
- 81.Li CL, Xiong D, Stamatoyannopoulos G, Emery DW. Genomic and functional assays demonstrate reduced gammaretroviral vector genotoxicity associated with use of the cHS4 chromatin insulator. Mol Ther. 2009;17:716–24. doi: 10.1038/mt.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li CL, Emery DW. The cHS4 chromatin insulator reduces gammaretroviral vector silencing by epigenetic modifications of integrated provirus. Gene Ther. 2008;15:49–53. doi: 10.1038/sj.gt.3303009. [DOI] [PubMed] [Google Scholar]
- 83.Aker M, Tubb J, Groth AC, et al. Extended core sequences from the cHS4 insulator are necessary for protecting retroviral vectors from silencing position effects. Hum Gene Ther. 2007;18:333–43. doi: 10.1089/hum.2007.021. [DOI] [PubMed] [Google Scholar]
- 84.Arumugam PI, Scholes J, Perelman N, et al. Improved human beta-globin expression from self-inactivating lentiviral vectors carrying the chicken hypersensitive site-4 (cHS4) insulator element. Mol Ther. 2007;15:1863–71. doi: 10.1038/sj.mt.6300259. [DOI] [PubMed] [Google Scholar]
- 85.Malik P, Arumugam PI, Yee JK, Puthenveetil G. Successful correction of the human Cooley’s anemia beta-thalassemia major phenotype using a lentiviral vector flanked by the chicken hypersensitive site 4 chromatin insulator. Ann NY Acad Sci. 2005;1054:238–49. doi: 10.1196/annals.1345.030. [DOI] [PubMed] [Google Scholar]
- 86.Arumugam PI, Urbinati F, Velu CS, et al. The 3′ region of the chicken hypersensitive site-4 insulator has properties similar to its core and is required for full insulator activity. PloS One. 2009;4(9):e6995. doi: 10.1371/journal.pone.0006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trobridge GD, Wu RA, Beard BC, et al. Protection of stem cell-derived lymphocytes in a primate AIDS gene therapy model after in vivo selection. PloS One. 2009;4:e7693. doi: 10.1371/journal.pone.0007693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trobridge GD, Beard BC, Gooch C, et al. Efficient transduction of pigtailed macaque hematopoietic repopulating cells with HIV-based lentiviral vectors. Blood. 2008;111:5537–43. doi: 10.1182/blood-2007-09-115022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang AH, Sadelain M. The genetic engineering of hematopoietic stem cells: the rise of lentiviral vectors, the conundrum of the LTR, and the promise of lineage-restricted vectors. Mol Ther. 2007;15:445–56. doi: 10.1038/sj.mt.6300060. [DOI] [PubMed] [Google Scholar]
- 90.Luo XM, Maarschalk E, O’Connell RM, et al. Engineering human hematopoietic stem/progenitor cells to produce a broadly neutralizing anti-HIV antibody after in vitro maturation to human B lymphocytes. Blood. 2009;113:1422–31. doi: 10.1182/blood-2008-09-177139. [DOI] [PubMed] [Google Scholar]
- 91.Sather BD, Ryu BY, Stirling BV, et al. Development of B-lineage predominant lentiviral vectors for use in genetic therapies for B cell disorders. Mol Ther. 2010 doi: 10.1038/mt.2010.259. published online 7 December 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kerns HM, Ryu BY, Stirling BV, et al. B cell-specific lentiviral gene therapy leads to sustained B-cell functional recovery in a murine model of X-linked agammaglobulinemia. Blood. 2010;115:2146–55. doi: 10.1182/blood-2009-09-241869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sandmeyer S. Integration by design. Proc Natl Acad Sci USA. 2003;100:5586–8. doi: 10.1073/pnas.1031802100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bushman FD, Miller MD. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J Virol. 1997;71:458–64. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tan W, Zhu K, Segal DJ, et al. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J Virol. 2004;78:1301–13. doi: 10.1128/JVI.78.3.1301-1313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96••.Gijsbers R, Ronen K, Vets S, et al. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol Ther. 2010;18:552–60. doi: 10.1038/mt.2010.36. Description of significant retroviral re-targeting in living cells. [DOI] [PMC free article] [PubMed] [Google Scholar]