Abstract
The widespread use of zinc finger nucleases (ZFNs) for genome engineering is hampered by the fact that only a subset of sequences can be efficiently recognized using published finger archives. We describe a set of validated two-finger modules that complement existing finger archives and expand the range of ZFN-accessible sequences by three-fold. Using this archive, we successfully introduce lesions at 9 of 11 target sites in the zebrafish genome.
Zinc finger nucleases (ZFNs) are artificial restriction enzymes containing a zinc finger array (ZFA) engineered to recognize different DNA sequences, fused through a flexible linker to the nuclease domain of FokI1. These enzymes function as heterodimers to create site-specific double strand breaks in DNA. This technology has been successfully applied in a variety of cell lines and organisms that previously lacked efficient tools for targeted genome editing1. However, widespread use of ZFNs is hindered by the challenge of designing zinc finger arrays (ZFAs) with sufficient affinity and specificity for most DNA sequences within a genome.
Highly specific ZFAs can be selected from randomized finger libraries using phage or bacterial selection systems2–5, but this process is labor intensive. By contrast, modular assembly6–8, wherein pre-characterized single zinc finger (1F) modules that recognize three-base pair subsites are joined into ZFAs, rapidly yields ZFNs albeit with lower success rates9 presumably due to unfavorable “context-dependent” interactions at the finger-finger interface10. Efforts to generate more reliable tools for ZFA assembly (Supplementary Discussion 1) have focused on randomizing interface sequences for the selection of two-finger (2F) modules5, 11 or selecting compatible finger pairs2–4. Analogous 2F-modules have been used by Sangamo BioSciences to build highly specific ZFAs and active ZFNs1, 12, however their archive is proprietary, limiting its use to ZFNs purchased through Sigma-Aldrich. Recently, the Zinc Finger Consortium (ZFC) described a Context Dependent Assembly (CoDA) approach whereby 2F-modules selected from OPEN pools are assembled into three-finger ZFNs13. CoDA-derived ZFNs constructed from prescreened ZFAs displayed high success rates (~50%), but the assayed ZFNs were almost entirely constructed from 2F-modules that recognize ‘GNN-GNN’ 6bp sites (Supplementary Fig. 1). These ‘N-G’ type junctions are not the limiting factor in effective ZFN design; ZFAs composed of only GNN recognition modules generate the most reliable ZFNs when using a variety of single finger archives8, 9.
Here we describe an archive of 2F-modules that bind all possible 2bp-junctions at the finger-finger interface, for assembly with each other or with predefined 1F-modules to create active, multi-finger ZFNs. We isolated these 2F-modules from two orthogonal libraries containing randomized amino acids at the interface recognition positions, via bacterial one-hybrid (B1H) selection against all 16 ‘GAN-NCG’ target sites, where Adenine at the second base position is recognized by either Asparagine at +3 of finger 2 (Asn+3F2-library) or Histidine at +3 of finger 2 (His+3F2-library) (Figs. 1a and 1b). Analysis of clones recovered from selections for 30 of 32 library/target site combinations yielded a partial or full target consensus sequence, implying the recovery of motifs compatible with sequence-specific DNA recognition. Higher stringency yielded a more constrained consensus for seven of these selections (Supplementary Table 1).
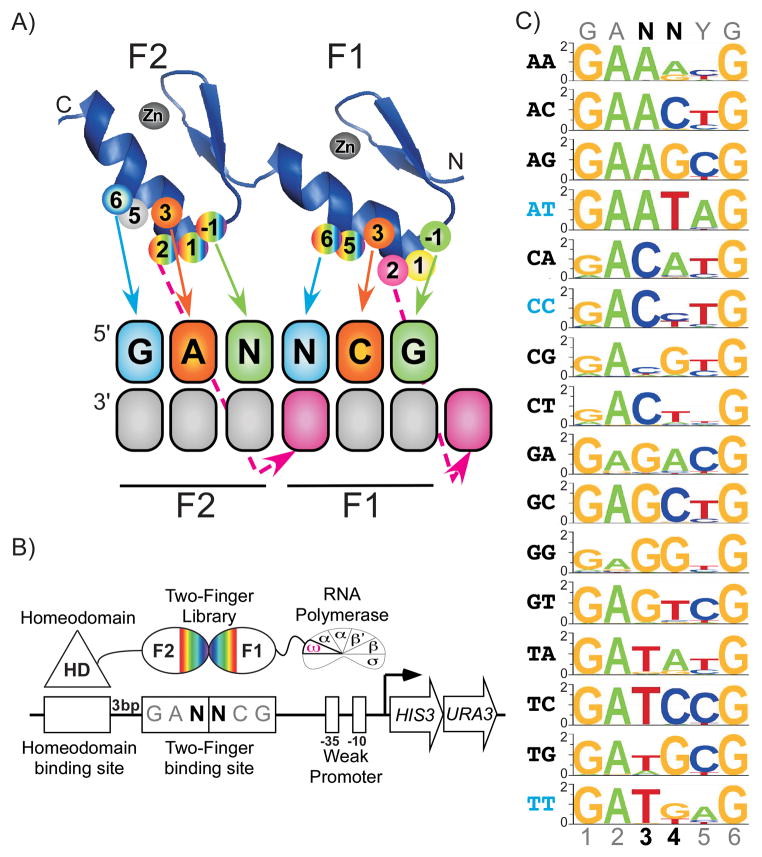
Figure 1.
Selection and characterization of 2F-modules recognizing GANNCG sequences. (a) Schematic representation of constructs encoded by the two-finger-ZFP library. Two orthogonal 2F-libraries were constructed each containing randomized amino acids at the finger-finger interface: at positions 5 and 6 of finger 1 (F1) and positions −1, 1 and 2 of finger 2 (F2). The residue at position 3 of F2 was either Asparagine (Asn+3F2-library) or Histidine (His+3F2-library). The arrows indicate canonical interactions between critical specificity determinants on the recognition helix and bases within each finger subsite (b) Schematic representation of bacterial one-hybrid (B1H) based selections to identify 2F-modules specific for a target site. The 2F-libraries were fused between the DNA-binding domain of the Engrailed homeodomain and the ωsubunit of the RNA polymerase. The 6 bp zinc finger target site is present on a His3/Ura3 reporter plasmid between the homeodomain binding site and the −35 box. (c) Montage of DNA-binding specificities for the most favorable 2F-modules with specificity for each GAN-NYG sequence where N-N represent the sixteen possible 2 bp junctions. The 2F-modules are designated as having ‘preferential specificity’ (black dinucleotide) or ‘compatible specificity’ (blue dinucleotide) for the desired target sequence. Further details on these clones are found in Supplementary Fig. 5
To confirm the target specificity of the B1H-selected 2F-modules, we determined the DNA-binding specificity of 87 2F-modules using our ‘constrained variation-B1H (CV-B1H)’ method14 (Supplementary Fig. 2). For 19 of 32 junctions, including 11 of 24 ‘non N-G’ junctions, we identified 2F-modules that preferred the desired binding site (Supplementary Fig. 3). For the seven selected 2F-modules that displayed a more constrained consensus at higher stringency, clones from the higher stringency displayed improved sequence selectivity (Supplementary Fig. 4).
Seven of the selected 2F-modules are compatible with the desired target site but preferentially recognize a different DNA sequence. To improve these 2F-modules and obtain specific modules for the other six junction sequences, we employed rational design, using principles of DNA recognition derived from our B1H-selections and from previous studies. This successfully expanded the number of 2F-modules that preferentially bind the desired junction sequence to 24, with an additional 6 junctions that can be recognized by 2F-modules with ‘compatible’ specificity (Fig. 1c and Supplementary Fig. 5; see online Methods for rating of specificity). Altogether, these 2F-modules can recognize a set of 60 ‘GRN-NYG’ 6bp sites, where R is A or G (recognized by Histidine at +3 of F2) and Y is C or T (recognized by Threonine at +3 of F1).
While some of our 2F-modules contain previously observed residues at the finger-finger interface11, 13, many contain novel combinations of residues. Based on these interface sequences, some CoDA 2F-modules previously described as recognizing ‘N-A’ junctions might prefer alternate junction sequences13. We assessed five of these CoDA 2F-modules to investigate their sequence preferences using B1H binding site selections and activity assays. We observed that the CoDA modules prefer ‘N-G’ to ‘N-A’ junctions, highlighting the advantage of explicit optimization of the finger-finger interface for the generation of highly specific ZFAs (Supplementary Fig. 6).
To generate additional modules targeting new hexameric target sites, we employed focused substitutions within our selected modules. In many instances, desired alterations in specificity could be obtained through substitutions at specific determinant positions (e.g. Supplementary Fig. 7a) or through substitution of sets of residues (−1, 1 and 2) at the N-terminus of finger 1 (Supplementary Fig. 7b). These modifications expanded the archive of 2F-modules to encompass a total of 162 unique 6bp sites including 132 ‘non-N-G’ junction-containing sites (Supplementary Table 2).
To demonstrate the utility of these 2F-modules for gene disruption, we combined them with each other or with published 1F-modules8 to create ZFNs (3 pairs of 3-finger ZFNs and 8 pairs of 4-finger ZFNs) targeting 11 sites in the zebrafish genome, where each site contains at least one ‘non-N-G’ junction (Supplementary Table 3). We determined the DNA-binding specificities of these assembled ZFAs using our B1H system and a 28 bp randomized library, and found that the incorporated 2F modules display the desired DNA-binding specificity (Supplementary Fig. 8). We observed that the majority of the resulting ZFNs (9 of 11) were active in a yeast-based chromosomal reporter assay12 (Supplementary Fig. 9). For some 4F-ZFNs constructs, we found that the presence of a non-canonical linker (TGSQKP) between the second and the third finger could both increase ZFN activity and reduce its toxicity, presumably by moderating its activity at non-target sequences (Supplementary Fig. 10). Finally, we injected mRNAs encoding these ZFN into zebrafish embryos and evaluated the induction of insertions and deletions (InDels) at the target site. 9 of 11 ZFNs induced InDels (>1bp) at the target sites at >0.5% frequency (Table 1 and Supplementary Table 4). Consistent with the yeast assay, lesion frequency in zebrafish was also improved for some ZFNs when incorporating a non-canonical linker (Supplementary Table 5 and Supplementary Discussion 2). We assayed ZFN-injected adults for germline transmission of mutant alleles for four targets and in all cases identified founders from a small number of screened animals (Supplementary Table 6).
Table 1.
Analysis of ZFN-induced lesions in zebrafish
| Gene | 5p ZFP binding site | 3p ZFP binding site | Spacer Length (bp) | Lesion Frequency (%) |
|---|---|---|---|---|
| dab2ip | GACTCGgac | GACATGgac | 6 | 8.0 |
| hey2 | GGTATGgtt | gggGAACTG | 6 | 0.6 |
| rock1 | gctGGCACG | GACTCGgcc | 6 | 0.0 |
| zgc77041* | GAACTGGAGTTG | GACATGGGTACG | 6 | 15.7 |
| dclk2* | GAGCCGGAATGG | GAAACGGGATCG | 5 | 1.1 |
| mc4r | gctgcagatgaa | gtagcaGACTTG | 6 | 12.9 |
| lrp8 | gtcggggcaggc | GAACTGGGCCCG | 6 | 7.3 |
| mc3r | GACGTGGAGCTG | gctgcatgaagg | 6 | 3.1 |
| apoeb | GGTGCGtaggtt | ggaGGGCCGtgt | 5 | 2.8 |
| lepr* | aagGAATTGgat | gcagtggaaggt | 6 | 0.9 |
| irs2* | aggGGCATGgta | cagGAAAAGgta | 6 | 0.4 |
ZFNL and ZFNR sites are shown wherein the 6bp subsites for the 2F-modules are represented in uppercase. In some cases two 2F-modules abut, leading to a 12bp element. An asterisk indicates targets where a non-canonical linker (TGSQKP) between the second and the third finger was employed to increase ZFN activity; the position of the non-canonical linker is underlined in each half-site where it is present.
In this study we report a unique set of 87 validated 2F-modules that recognize 162 6-bp target sites with high specificity. These 2F-modules can be readily combined together or with available single finger modules8 to rapidly create active ZFNs that target sequences containing ‘non-N-G’-junctions in vivo. We determined the number of potential ZFN target sites in protein-coding exons within the zebrafish and human genomes and compared this with two other recently described two-finger archives for ZFN construction (CoDA13 and the two-finger archive from Kim and colleagues15; Supplementary Table 7 and Supplementary Discussion 1). Our combined archive allows targeting of ~95% of the protein-coding genes (exons, Zv9) in the zebrafish genome, with an average density of one unique ZFN site every ~140 bp, which is a ~5-fold higher density than available through the CoDA archive. The Kim archive has the highest targeting density of the three archives with an average of one unique ZFN site every 10 bp, albeit with a lower overall success rate for assayed ZFNs15. To facilitate public use of our archive, we have developed a web interface that allows users to search for potential ZFN sites within an input sequence (http://pgfe.umassmed.edu/ZFPmodularsearchV2.html). The website ranks the quality of each ZFN site within the input sequence and provides information for the assembly or direct synthesis of the requisite ZFAs (Supplementary Discussion 3).
Although our archive is the largest set of ‘non-N-G’-junction recognizing 2F-modules described to date, it comprises only 132 of the possible 3072 non-N-G junction sites. Additional archives of ‘non-N-G’-junction 2F-modules exist; for example, sixty-one are found in the CoDA archive, but we note that only 3 of 10 tested ZFAs containing these modules were active13. Thus, there is a need to expand the set of high-quality 2F-modules covering these junctions to further increase the targeting resolution of ZFNs.
As observed in our study, selections alone may not always be sufficient to obtain highly specific modules for a given target sequence since both affinity and specificity play a role in module activity. The continued development of more accurate predictive models of DNA recognition for zinc fingers are likely to be needed to inform design efforts. Ultimately, these efforts should lead to important advances in nuclease precision and activity not only for engineering model systems, but also for creating therapeutic reagents for the treatment of disease.
Online Methods
ZFN website scoring function
Our new ZFN site identification tool (http://pgfe.umassmed.edu/ZFPmodularsearchV2.html) uses 2F-modules from this study and 1F-modules from our previous archive8 to define favorable combinations of these modules for constructing active ZFNs. These ZFNs are designed to target sequences with 5, 6 or 7 bp gaps between the monomer recognition sequences, where each ZFN monomer can contain three or four fingers. ZFNs with higher scores are more likely to be active, where the current 2F-modules are scored based on their DNA-binding specificity (as determined in the B1H system) where good, fair and poor represent 4, 3 and 2 points respectively. If the modules utilize an A-cap (QRG at the N-terminus of the 2F-module) instead of the standard RSD sequence for G-recognition one point is subtracted from the score. The 1F-modules are scored as previously described8. ZFNs containing 2F-modules are readily identified in the output from the website by the presence of lowercase triplet sequences in the site breakdown, and by the presence of “2FM-#” in the output Module ID information.
Animal husbandry
Zebrafish were handled according to established protocols16 and in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines of the University of Massachusetts Medical School.
2F-Library construction
2F-libraries were constructed in two stages. First, individual F1 and F2 libraries were independently constructed via cassette mutagenesis of annealed randomized oligonucleotides into pBluescript vector containing the appropriate zinc finger backbone derived from Zif268. The sequences for the randomized oligonucleotides are listed in Supplementary Table 8 where lowercase letters denote the randomized bases. Individual finger library diversity greatly exceeded the theoretical library size; ~1×105 transformed cells were obtained for the F1 library (>100 times theoretical size) and ~1×106 transformed cells for the F2 library (30 times theoretical size of the library). Constructed libraries were grown at low density on 2xYT plates containing 100μg/ml carbenicillin at 37 °C for 14 hours. Individual F1 and F2 libraries in pBluescript were harvested from pooled cells from these surviving colonies.
The 2F-library was constructed from the single finger libraries by PCR assembly, individual F1 and F2 libraries were separately amplified from the pooled pBluescript clones by PCR and then joined via overlapping PCR, where the number of amplification cycles in both steps was minimized by employing high concentrations of template DNA. This 2F-library was then cloned into the B1H expression vector 1352-omega-UV2 between unique BssHII and Acc65I restriction enzyme sites such that the ω-subunit of the RNA polymerase is fused at the N-terminus of the zinc fingers and the Engrailed homeodomain at the C-terminus. Following electroporation into bacterial cells, 1×108 cells (5 times the theoretical size of the library) were plated on 10 2xYT-carbenicillin plates (150 × 15 mm) and grown at 37 °C for 14 hours. 1352-omega-UV2 plasmids containing the 2F-library were isolated from pooled surviving colonies and used for selections.
Zinc finger Binding site cloning
The 16 GANNCG zinc finger binding sites (ggccTAATTACCTGANNCGGacg) were cloned between the EcoRI and NotI sites in the pH3U3-mcs reporter vector. The Homeodomain (Engrailed) binding site TAATTA (underlined) is present 3 bp away and on the strand opposite to the zinc finger binding site to minimize any interference between the Homeodomain and the zinc fingers. For selecting 2F-modules from the Asn+3F2 library that recognize the ‘G-G’ interface, sufficient stringency could not be obtained to narrow the selected clones merely through increased 3-aminotriazole (3-AT) concentration or reduced inducer (isopropyl-b-D-thiogalactoside; IPTG) levels. To reduce the activity of the ZFP-HD construct the homeodomain site was mutated to TAAAGG to increase the dependence on zinc finger binding.
2F B1H Selections
Selections for 2F-modules were performed as described previously4. The zinc finger library (20 ng) and the reporter vector (1 μg) containing the zinc finger target site were cotransformed via electroporation in the selection strain that lacks endogenous expression of the ω-subunit of RNA polymerase (US0ΔhisBΔpyrFΔrpoZ). 2×107 cotransformed cells were plated on selective NM minimal medium plates (where stringency was controlled via 3-AT and IPTG concentration) and grown at 37 °C until moderate number of colonies (typically 100s) were visible. Post-selection, 2F-modules from 6–10 surviving colonies were sequenced to identify functional amino acid sequences for further evaluation. The success of the selection was judged on the diversity of sequences obtained from these selections, with the expectation that successful selections will converge on a small number of functional residues at the critical recognition positions.
Cloning B1H-selected 2F modules into 3F F1-GCG constructs
To determine the binding specificities of 2F-modules a ‘GCG’ binding anchor zinc finger (recognition helix: RSDTLAR) was fused at the N-terminus of the 2F-module via overlapping PCR (Supplementary Table 8). Following overlapping PCR, the 3F-ZFA was cloned into 1352-omega-UV2 vector between the Acc65I and BamHI sites for expression as an omega fusion.
CV-B1H method
To determine binding site specificities of 2F-modules, the CV-B1H assay was performed as described before14. Post-transformations into the selection strain, 1×106 cells containing the zinc finger plasmid (1352-omega-UV2-ZFP) and the randomized binding site library plasmid (pH3U3) were plated on selective NM minimal medium plates (100 × 15 mm) containing 50 μM IPTG and 1 or 2 mM 3-AT and grown at 37 °C for 22–30 hrs. The surviving colonies were pooled and the binding site plasmid was isolated for identification of the functional DNA sequences. The binding site region was PCR amplified and Sanger sequenced to rapidly obtain binding site profiles for each 2F-module. For quantitative modeling, the binding site pools for multiple 2F-modules were barcoded and sequenced via Illumina sequencing, and then binding specificities were modeled from this data using both W log-odds and GRaMSc methods (Supplementary Methods).
Rating of 2F modules
For every 2F-module, the frequency of each of the 16 possible 2bp-junctions was determined in the binding sites that were recovered by Illumina sequencing. The 2F-modules for which the frequency of the desired 2bp-junction was the highest among all 16 2bp-junctions were designated as possessing ‘preferential specificity’. If the frequency of the desired 2bp-junction was the second highest and represented more than 20% of the dinucleotide population, the 2F-module was designated as having ‘compatible specificity’. The remaining 2F-modules were designated as having ‘poor specificity’.
Comparison of CoDA-2F modules and B1H-selected 2F-modules
The CoDA 2F-modules were created using overlapping PCR where the desired recognition helix sequences were introduced into the Zif268 finger 2 backbone. The 2F-modules were fused to the N-terminal ‘GCG’ binding finger and CV-B1H assay was performed followed by binding site modeling using the W log-odds and GRaMS methods as described above. B1H-based activity assay were performed as described previously17.
Rational design of 2F modules with improved specificity
The archive of selected 2F-modules was expanded through rational design. For improved recognition of junction sequences, the specificity determinants at the interface positions were altered based on recognition trends that were observed in our selected modules or prior interface selection studies11. Changing the residue at position 3 of finger 2 or finger 1, respectively, created modules with altered specificity at the 2nd or 5th position of the six base pair recognition sequence. Substituting the three N-terminal cap residues in finger 1 (RSD at positions −1, 1 and 2) produced alterations in sequence preference at the 6th position, where substitution of a QRG cap reliably produced an alteration in sequence preference from G to A.
Creating ZFAs
Three Finger (3F) and Four Finger (4F) ZFAs for use in ZFNs were assembled from the 2F-module archive described herein and a 1F-module archive that we recently described8 using overlapping PCR. The primer sequences used for these different assemblies are listed in Supplementary Table 9. If desired these ZFAs can also be synthesized from the DNA sequence output from our website application.
For amplifying individual 1F and 2F modules, the following PCR conditions were used: 10 ng DNA template, 1 μM each of forward and reverse primer, 200 μM dNTPs and 0.5 unit of Phusion High Fidelity DNA polymerase (New England Biolabs) in 25 μl reaction volume. PCR cycles: 98 °C 3 min, [98 °C 15 sec, 50 °C 15 sec, 72 °C 30 sec] 6 repeats, [98 °C 15 sec, 56 °C 15 sec, 72 °C 30 sec] 24 repeats, 72 °C 5 min, 4 °C.
For ZFA assembly from the individual 1F and 2F module amplicons was mediated by overlapping PCR under the following conditions: 1–5ng DNA for each component, 200 μM dNTPs and 0.5 unit of Phusion High Fidelity DNA polymerase (New England Biolabs) in 25 μl reaction volume. PCR cycles: 98 °C 3 min, [98 °C 15 sec, 50 °C 15 sec, 72 °C 30 sec] 6 repeats 72 °C 5 min. Following this initial assembly step the forward and reverse primers (final concentration of 1 μM each) were added to the reaction and PCR amplification proceeded using the following cycles: 98 °C 3 min, [98 °C 15 sec, 56 °C 15 sec,72 °C 30 sec] 25 repeats, 72 °C 5 min. Post-amplification, the 3F/4F PCR products were digested with Acc65I and BamHI enzymes and cloned into appropriate vectors.
Note
The QRG cap is introduced into the 2F module using a special QRG(X) primer set that substitutes the RSD cap with the QRG cap in F1. When Thr is present at position 3 of F1 use the QRG(T) primer, when Asn is present at position 3 of F1 use the QRG(N) primer, and when His is present at position 3 of F1 use the QRG(H) primer. For ZFNs recognizing a seven base pair gap utilize the F3RnTGPGAAGS or 2FM-F3RnTGPGAAGS instead of the F3RnLRGS or F3RnLRGS primers to incorporate the longer linker associated with increased activity (Supplementary Discussion). To incorporate the non-canonical linker (TGSQKP) between F1 and F2 fingers of a 4F construct, the sequences for F2-forward primer and F1-reverse primer were modified to introduce the additional Serine in the linker.
3F-ZFAs assemblies from F1, F2 and F3 1F-modules
The single fingers are amplified individually and then assembled. F1 was amplified using the F1(noF0)Fn and F1Rn primers. F2 was amplified using the F2Fn and F2Rn primers. F3 was amplified using F3Fn and F3RnLRGS primers. The amplified DNA was gel purified using a Qiagen gel purification kit. For finger assembly, 5ng of the F1, F2 and F3 amplicons were combined and assembled as described above, where the F1(noF0)Fn and F3RnLRGS primers were added to the PCR reaction for the final amplification.
3F-ZFAs assemblies from F1 1F-module and 2F-module
F1 was amplified using the F1(noF0)Fn and F1Rn primers. 2F-module was amplified using 2FM-F2Fn and 2FM-F3RnLRGS primer. The amplified DNA was gel purified and the finger amplicons were assembled as described above, where the F1(noF0)Fn and 2FM-F3RnLRGS primers were added to the PCR reaction for the final amplification.
3F-ZFAs assemblies from a 2F-module and F3 1F-module
2F-module was amplified using the 2FM-F1(noF0)Fn and 2FM-F2Rn primers and F3 was amplified using the F3Fn and F3RnLRGS primers. The amplified DNA was gel purified and the finger amplicons were assembled as described above, where the 2FM-F1(noF0)Fn and F3RnLRGS primers were added to the PCR reaction for the final amplification.
3F-ZFAs assemblies from a F1 1F-modules and 2F-module-QRG cap
F1 was amplified using the F1(noF0)Fn and F1Rn primers. 2F-module was first amplified with 2FM-F1Fn and 2FM-F2Rn primers, gel purified and then 1–5ng of gel purified DNA was used as template for amplification with 2FM-F2-QRG(X)Fn and 2FM-F3RnLRGS primers to substitute the RSD N-terminal cap with the QRG-N-terminal cap. The amplified 2F-module-QRG and F1 module were gel purified and the finger amplicons were assembled as described above, where the F1(noF0)Fn and 2FM-F3RnLRGS primers were added to the PCR reaction for the final amplification.
3F-ZFAs assemblies from a 2F-module-QRG cap and F3 1F-modules
2F-module was first amplified with 2FM-F1Fn and 2FM-F2Rn primers, gel purified and then 1–5ng of gel purified DNA was used as template for amplification with 2FM-F1(noF0)-QRG(X)Fn and 2FM-F2Rn primers. F3 was amplified using the F3(noF0)Fn and F3RnLRGS primers. The amplified 2F-module-QRG and F3 module were gel purified and the finger amplicons were assembled as described above, where the 2FM-F1(noF0)Fn and F3RnLRGS primers were added to the PCR reaction for the final amplification.
4F-ZFAs assemblies from F0, F1, F2 and F3 1F-modules
F0 was amplified using the F0Fn and F0Rn primers. F1 was amplified using the F1Fn and F1Rn primers. F2 was amplified using the F2Fn and F2Rn primers. F3 was amplified using F3Fn and F3RnLRGS primers. The amplified DNA was gel purified and the finger amplicons were assembled as described above, where the sF0Fn and F3RnLRGS primers were added to the PCR reaction for the final amplification.
4F-ZFAs assemblies from F0 and F1 1F-modules, and 2F-module
F0 was amplified using the F0Fn and F0Rn primers. F1 was amplified using the F1Fn and F1Rn primers. The 2F-module was amplified using 2FM-F2Fn and 2FM-F3RnLRGS primer. The amplified DNA was gel purified and the finger amplicons were assembled as described above, where the F0Fn and 2FM-F3RnLRGS primers were added to the PCR reaction for the final amplification.
4F-ZFAs assemblies from F0, 2F-module and F3
F0 was amplified using the F0Fn and F0Rn primers. 2F-module was amplified using the 2FM-F1Fn and 2FM-F2Rn primers and F3 was amplified using the F3Fn and F3Rn primers. The amplified DNA was gel purified and the finger amplicons were assembled as described above, where the F0Fn and F3RnLRGS primers were added to the PCR reaction for the final amplification.
4F-ZFAs assemblies from 2F-module, F2 and F3
The 2F-module was amplified using the 2FM-F0Fn and 2FM-F1Rn primers. F2 was amplified using the F2Fn and F2Rn primers. F3 was amplified using the F3Fn and F3Rn primers. The amplified DNA was gel purified and the finger amplicons were assembled as described above, where the 2FM-F0Fn and F3RnLRGS primers were added to the PCR reaction for the final amplification.
4F-ZFAs assemblies from N-terminal-2F-module, C-terminal-2F-module
The N-terminal 2F-module was amplified with the 2FM-NT-in-Fn and 2FM-F1Rn primers and the C-terminal 2F-module was amplified with the 2FM-F2Fn and 2FM-F3RnLRGS primers. The amplified products were gel purified and the finger amplicons were assembled as described above, where the 2FM-NT-out-Fn and 2FM-CT-out-Rn primers were added to the PCR reaction for the final amplification.
4F-ZFAs assemblies from F0, F1 and 2F-module-QRG
F0 was amplified using the F0Fn and F0Rn primers. F1 was amplified using the F1Fn and F1Rn primers. 2F-module was first amplified with 2FM-F1Fn and 2FM-F2Rn primers, gel purified and then 1–5ng of gel purified DNA was used as template for amplification with 2FM-F2-QRG(X)Fn and 2FM-F3RnLRGS primers to substitute the RSD N-terminal cap with the QRG-N-terminal cap. The amplified 2F-module-QRG, F0 and F1 modules were gel purified and the finger amplicons were assembled as described above, where the F0Fn and 2FM-F3RnLRGS primers were added to the PCR reaction for the final amplification.
4F-ZFAs assemblies from F0, 2F-module-QRG and F3
F0 was amplified using the F0Fn and F0Rn primers. 2F-module was first amplified with 2FM-F1Fn and 2FM-F2Rn primers, gel purified and then 1–5ng of gel purified DNA was used as template for amplification with 2FM-F1-QRG(X)Fn and 2FM-F2Rn primers. F3 was amplified using the F3Fn and F3Rn primers. The amplified the F0, 2F-module-QRG and F3 modules were gel purified and amplicons were assembled as described above, where the F0Fn and F3RnLRGS primers were added to the PCR reactions for the final amplification
4F-ZFAs assemblies from 2F-module-QRG, F2 and F3
The 2F-module was first amplified with 2FM-F1Fn and 2FM-F2Rn primers, gel purified and then 1–5ng of gel purified DNA was used as template for amplification with 2FM-F0-QRG(X)Fn and 2FM-F1Rn primers. F2 was amplified using the F2Fn and F2Rn primers. F3 was amplified using the F3Fn and F3Rn primers. The amplified DNA was gel purified and the finger amplicons were assembled as described above, where the 2FM-F0Fn and F3RnLRGS primers were added to the PCR reaction for the final amplification.
4F-ZFAs assemblies from N-terminal-2F-module-QRG, C-terminal-2F-module
The N-terminal 2F-module was amplified with 2FM-F1Fn and 2FM-F2Rn, gel purified and then 1–5ng of gel purified DNA was used as template for amplification with 2FM-F0-QRG(X)Fn and 2FM-F1Rn primers. This amplified 2F-module-QRG was again gel purified and PCR amplified with 2FM-NT-in-Fn and 2FM-F1Rn primers. The C-terminal 2F-module was amplified with 2FM-F2Fn and 2FM-F3RnLRGS. The amplified N-terminal and C-terminal 2F-modules were gel purified and the finger amplicons were assembled as described above, where the 2FM-NT-out-Fn and 2FM-CT-out-Rn primers were added to the PCR reaction for the final amplification.
4F-ZFAs assemblies from N-terminal-2F-module, C-terminal-2F-module-QRG
The N-terminal 2F-module was amplified with 2FM-NT-in-Fn and 2FM-F1Rn. The C-terminal 2F-module was amplified with 2FM-F1Fn and 2FM-F2Rn, gel purified and then 1–5ng of gel purified DNA was used as template for amplification with 2FM-F2-QRG(X)Fn and 2FM-F3RnLRGS primers. The amplified N-terminal and C-terminal 2F-modules were gel purified and the finger amplicons were assembled as described above, where the 2FM-NT-out-Fn and 2FM-CT-out-Rn primers were added to the PCR reaction for the final amplification.
4F-ZFAs assemblies from N-terminal-2F-module-QRG, C-terminal-2F-module-QRG
The N-terminal 2F-module was amplified with 2FM-F1Fn and 2FM-F2Rn, gel purified and then 1–5ng of gel purified DNA was used as template for amplification with 2FM-F0-QRG(X)Fn and 2FM-F1Rn primers. This amplified 2F-module-QRG was again gel purified and PCR amplified with 2FM-NT-in-Fn and 2FM-F1Rn primers. The C-terminal 2F-module was amplified with 2FM-F1Fn and 2FM-F2Rn, gel purified and then 1–5ng of gel purified DNA was used as template for amplification with 2FM-F2-QRG(X)Fn and 2FM-F3RnLRGS primers. The amplified N-terminal and C-terminal 2F-modules were gel purified and the finger amplicons were assembled as described above, where the 2FM-NT-out-Fn and 2FM-CT-out-Rn primers were added to the PCR reaction for the final amplification.
B1H-binding site selections using the 28bp library
The selections for 3F and 4F ZFAs were performed as previously described17. 1–5×107 selection strain cells transformed with the 1352-omega-UV2 ZFA expression plasmid and the 28 bp pH3U3 library plasmid were plated on NM minimal medium selective plates lacking uracil and containing 3-AT (2.5, 5 or 10 mM) as the competitor and grown at 37 °C for 36–72 hours. The number of surviving bacterial colonies on each plate was estimated and then these colonies were pooled and the population of recovered DNA sequences was determined via Illumina sequencing. Unique sequences were ranked based on the number of recovered reads. From this list an overrepresented sequence motif was determined with MEME18 using as input the number of unique sequences from the top of the list that correspond to the estimated number of colonies on the selection plate (typically >1000). The aligned sequences were then used to generate Sequence logos using Weblogo19.
Yeast-based ZFN activity assay
To assess the activity of our ZFNs in an independent system we employed a Mel1-based yeast activity assay12. The target sites for test ZFNs and the positive control ZFN were cloned in the modified ySSA vector and then integrated into the yeast genome (BY4741 strain) at the HO locus. The ZFAs were cloned at the N-terminus of the wild type FokI nuclease domain in the pYHis3 and pYLeu2 vectors in between the Acc651 and BamH1 sites. Test ZFNs, positive control ZFN and negative control (EGFP containing pYHis3 and pYLeu2 vectors) were transformed in the yeast strain containing the ZFN target site. ZFN expression was induced by 2% galactose treatment for 30 min. The activity assay was performed ~16 hours post-induction as previously described20. In brief, yeast cultures were diluted to an OD600 of 0.4–0.6. 950 μl of diluted cells were centrifuged and the pellet was resuspended in 200 μl of 20 mM HEPES (pH 7.5)–10 mM dithiothreitol–0.002% sodium dodecyl sulfate. 10 μl of chloroform was added to cells and vortexed for 10 s. After a 5 min preequilibration at 30 °C, 800 μl of a 7 mM solution of PNPG (4-Nitrophenyl α-D-galacto-pyranoside; Sigma) in 61 mM citric acid–77 mM Na2HPO4 (pH 4) was added and incubated at 30 °C for 30 min. Following incubation, 100 μl aliquot was added to 900 μl of 0.1 M Na2CO3 to stop the reaction and the OD405 was recorded.α-galactosidase units were calculated as follows: α-gal = (OD405*1000)/(OD600*tpnpg) where tpnpg is the time of incubation with PNPG. Relative activity for test ZFNs was calculated as follows: (100*α-galtest ZFN)/ α-galpositive control.
ZFN injections and lesion analysis
For gene targeting in zebrafish, ZFAs were cloned in pCS2 vectors containing the DD/RR obligate heterodimer version of the FokI nuclease domain21, 22. pCS2-ZFN constructs were linearized with NotI and mRNA was transcribed using the mMesagemMachine SP6 kit from Ambion. ZFN mRNAs were injected into the blastomere of one-cell-stage zebrafish embryos as previously described4. ZFN-injected embryos with normal appearance (8–30) and uninjected embryos were collected 24 hpf and incubated in 50 mM NaOH (15μl/embryo) for 15 min at 95 C to isolate genomic DNA and then neutralized with 0.5 M Tris-HCl (4 μl/embryo). The DNA solution was centrifuged for 1 min at 13,000 rpm and supernatant was taken for lesion analysis. For initial validation of ZFN activity, the region flanking the ZFN target site was amplified using the Phire Hot Start DNA Polymerase (Finnzymes) and RFLP analysis or Cel I nuclease assay (Transgenomics) was performed as described previously4. For Illumina sequencing, the region flanking the ZFN sites was amplified using the primers and then digested with the appropriate restriction enzyme (Supplementary Table 10). The ends for the digested DNA were polished using Klenow exo−enzyme (New England Biolabs) or T4 DNA polymerase (New England Biolabs) and A-tailed using Klenow exo− enzyme (New England Biolabs). The barcoded adapters (Supplementary Table 10) were ligated to each DNA pool and then PCR amplified with the Illumina genomic primers 1.1 and 1.2. Following sequencing, identification of InDels was performed as described previously17. Briefly, two tags unique to a ZFN target site were employed, a 5’ tag and a 3’ tag (Supplementary Table 10) and the distance between the tags was used to distinguish wild type sequence from the InDel containing sequence. Lesion frequency was calculated as follows: Lesion frequency = (100*NInDels)/Ntotal where, NInDels represents number of sequences containing InDels that are >1bp in length and Ntotal represents number of total sequences.
Genomic analysis of ZFN target sites
The targeting density and overlap of ZFN sites were determined for three archives (Gupta 1/2FM, CoDA 2FM13 & Kim 1/2FM15) on the unique protein-coding exons zebrafish (Zv9) and human (GRCh37.p5) Ensembl genes 64. Target sites for each finger archive were determined using custom perl scripts, where only ZFN sites that map to a single unique gene were counted in this analysis. This analysis provides information on the fraction of genes that can be targeted and the density of the sites per base pair.
Germline Transmission Analysis
ZFNs were injected at optimal doses in wild type zebrafish embryos. Injected embryos were grown to maturity and crossed with wild type zebrafish to identify carriers. PCR products spanning the target loci in F1 embryos were screened using Cel1 surveyor nuclease assay for presence of lesions4. The compositions of these lesions were characterized through cloning and sequencing PCR products spanning the ZFN target site for each gene (Supplementary Table 10).
Supplementary Material
Supplementary Figure 1. Comparison of target site composition for CoDA-ZFNs and our ZFNs.
Supplementary Figure 2. Identification of DNA binding specificity for 2Fmodules using the CV-B1H method.
Supplementary Figure 3. Montage showing the binding site specificities of the best 2F-modules selected from the Asn+3F2 and the His+3F2 library for each 2bp junction.
Supplementary Figure 4. Influence of stringency on the specificity of the recovered 2F-module.
Supplementary Figure 5. DNA-binding site specificities for 2F modules that bind GAN-NYG and GGN-NYG sequences
Supplementary Figure 6. Comparison of specificities of CoDA-2F modules and our 2F modules
Supplementary Figure 7. Examples of module alterations designed to expand the archive of targetable sequences through rational design.
Supplementary Figure 8. Binding site specificities of ZFAs incorporated into each ZFN pair
Supplementary Figure 9. Assessment of ZFN activity using the yeast based chromosomal reporter assay
Supplementary Figure 10. Influence of non-canonical linker on ZFN specificity and activity
Supplementary Table 1. List of all 2F-modules obtained after B1H-selections from the Asn+3F2-library and the His+3F2-library. Recognition helix sequences (−1, 1, 2, 3, 4, 5, 6) for F1 and F2 for selected 2F-modules are shown for 2F-modules selected for zinc finger binding sites (gaNNcg where NN represents the 2bp-interface). The amino acid at position 3 of F2 can either be Asn or His depending on the zinc finger library used. The selection conditions (3-AT, IPTG and Uracil concentrations) are given. For selection of 2F-modules that bind the G-G interface with the Asn+3F2-library library, a mutant homeodomain binding site (TAAAGG) was used.
Supplementary Table 2. List of all 2F-modules and target sites for the assembly of ZFNs. Recognition helix sequences for F1 and F2 of each 2F-module are shown.
Supplementary Table 3. List of all ZFNs and their target sites.
Supplementary Table 4. Analysis of ZFN-induced lesions in zebrafish.
Supplementary Table 5. Influence of non-canonical linker on ZFN activity in zebrafish.
Supplementary Table 6. Founder rates for four ZFN target genes.
Supplementary Table 7. Metrics for comparison of different ZFN assembly systems.
Supplementary Table 8. Primer sequences for 2F-module library construction and specificity analysis.
Supplementary Table 9. Primer sequences for ZFN assembly.
Supplementary Table 10. Sequences of the genotyping primers used for lesions detection in zebrafish embryos.
Supplementary Discussion 1. Comparison to previously described Finger Archives.
Supplementary Discussion 2. Modifications to our ZFNs to increase their activity in vivo.
Supplementary Discussion 3. Description of web interface for identification of ZFN sites within query sequences
Supplementary Methods. Description of GRaMSc and W log-odds motif construction algorithms used in this study.
Acknowledgments
This research was supported by the US National Institutes of Health (NIH) R01GM068110 (S.A.W.), R24GM078369, R01HL093766 (N. Lawson & S.A.W.) and R01HG00249 (G.D.S). We thank N. Lawson and his laboratory for their insightful advice and zebrafish husbandry training. We thank J. Zhu for her assistance with the website construction.
Footnotes
Author contributions:
S.A.W. conceived the study; A.G. and A.L.R. carried out the selection experiments. R.G.C. and G.D.S. developed the computational platform for motif analysis. A.L. performed the analysis of ZFN sites in multiple genomes. A.G. and S.A.W. wrote the manuscript with input from all authors.
References
- 1.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 2.Greisman HA, Pabo CO. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 3.Maeder ML, et al. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Nature biotechnology. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isalan M, Klug A, Choo Y. Nature biotechnology. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Nature protocols. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 7.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu C, et al. Development. 2011;138:4555–4564. doi: 10.1242/dev.066779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramirez CL, et al. Nature methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Nucleic Acids Res. 2009;37:506–515. doi: 10.1093/nar/gkn962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isalan M, Klug A, Choo Y. Biochemistry. 1998;37:12026–12033. doi: 10.1021/bi981358z. [DOI] [PubMed] [Google Scholar]
- 12.Doyon Y, et al. Nature biotechnology. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sander JD, et al. Nature methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen RG, et al. Nucleic Acids Res. 2011;39:e83. doi: 10.1093/nar/gkr239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S, Lee MJ, Kim H, Kang M, Kim JS. Nature methods. 2011;8:7. doi: 10.1038/nmeth0111-7a. [DOI] [PubMed] [Google Scholar]
- 16.Westerfield M. The Zebrafish Book. University of Oregon Press; Eugene, Oregon: 1993. [Google Scholar]
- 17.Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Nucleic Acids Res. 2010;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey TL, Elkan C. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 19.Crooks GE, Hon G, Chandonia JM, Brenner SE. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan MP, Jones R, Morse RH. Mol Cell Biol. 1998;18:1774–1782. doi: 10.1128/mcb.18.4.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller JC. Nature biotechnology. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 22.Szczepek M, et al. Nature biotechnology. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Comparison of target site composition for CoDA-ZFNs and our ZFNs.
Supplementary Figure 2. Identification of DNA binding specificity for 2Fmodules using the CV-B1H method.
Supplementary Figure 3. Montage showing the binding site specificities of the best 2F-modules selected from the Asn+3F2 and the His+3F2 library for each 2bp junction.
Supplementary Figure 4. Influence of stringency on the specificity of the recovered 2F-module.
Supplementary Figure 5. DNA-binding site specificities for 2F modules that bind GAN-NYG and GGN-NYG sequences
Supplementary Figure 6. Comparison of specificities of CoDA-2F modules and our 2F modules
Supplementary Figure 7. Examples of module alterations designed to expand the archive of targetable sequences through rational design.
Supplementary Figure 8. Binding site specificities of ZFAs incorporated into each ZFN pair
Supplementary Figure 9. Assessment of ZFN activity using the yeast based chromosomal reporter assay
Supplementary Figure 10. Influence of non-canonical linker on ZFN specificity and activity
Supplementary Table 1. List of all 2F-modules obtained after B1H-selections from the Asn+3F2-library and the His+3F2-library. Recognition helix sequences (−1, 1, 2, 3, 4, 5, 6) for F1 and F2 for selected 2F-modules are shown for 2F-modules selected for zinc finger binding sites (gaNNcg where NN represents the 2bp-interface). The amino acid at position 3 of F2 can either be Asn or His depending on the zinc finger library used. The selection conditions (3-AT, IPTG and Uracil concentrations) are given. For selection of 2F-modules that bind the G-G interface with the Asn+3F2-library library, a mutant homeodomain binding site (TAAAGG) was used.
Supplementary Table 2. List of all 2F-modules and target sites for the assembly of ZFNs. Recognition helix sequences for F1 and F2 of each 2F-module are shown.
Supplementary Table 3. List of all ZFNs and their target sites.
Supplementary Table 4. Analysis of ZFN-induced lesions in zebrafish.
Supplementary Table 5. Influence of non-canonical linker on ZFN activity in zebrafish.
Supplementary Table 6. Founder rates for four ZFN target genes.
Supplementary Table 7. Metrics for comparison of different ZFN assembly systems.
Supplementary Table 8. Primer sequences for 2F-module library construction and specificity analysis.
Supplementary Table 9. Primer sequences for ZFN assembly.
Supplementary Table 10. Sequences of the genotyping primers used for lesions detection in zebrafish embryos.
Supplementary Discussion 1. Comparison to previously described Finger Archives.
Supplementary Discussion 2. Modifications to our ZFNs to increase their activity in vivo.
Supplementary Discussion 3. Description of web interface for identification of ZFN sites within query sequences
Supplementary Methods. Description of GRaMSc and W log-odds motif construction algorithms used in this study.