Abstract
Elucidating the forces responsible for genomic variation is critical for understanding evolution. Under standard conditions, X-linked diversity is expected to be three-quarters the level of autosomal diversity. Empirical data often deviate from this prediction, but the reasons for these departures are unclear. We demonstrate that population size changes can greatly alter relative levels of X-linked and autosomal variation: population size reductions lead to particularly low X-linked diversity, while growth elevates X-linked relative to autosomal diversity. Genetic variation from a diverse array of taxa supports an important role for this effect in accounting for population differences in the ratio of X-linked to autosomal diversity. Consideration of this effect may improve the inference of population history and other evolutionary processes.
Keywords: Population history, X-linked variation, autosomal variation, population bottleneck, human genome diversity
A fundamental enigma in evolutionary biology concerns the relative levels of X chromosome and autosome diversity. Since X chromosomes are present in two copies in females but only one copy in males, the effective population size (and level of diversity) for X chromosomes is expected to be ¾ of that of the autosomes. However, departures from this ratio are frequently observed in natural populations. A number of processes have been suggested to account for such deviations, including sex-biased mutation (reviewed in Li et al. 2002), sex-specific variance in reproductive success (Caballero 1995; Charlesworth 2001), sex-biased migration (Laporte and Charlesworth 2002), the effect of linked negative selection (Charlesworth 1996), and positive selection (e.g. Aquadro et al. 1994).
The quest to detect molecular signatures of adaptation has been a strong motivation for comparing X-linked and autosomal variation, particularly in Drosophila. Recessive beneficial mutations at low frequency can be “seen” by selection in males if the locus is X-linked, while similar autosomal alleles must first drift to a high enough frequency to be present as homozygotes. If beneficial mutations tend to be recessive on average, hitchhiking may be more frequent on the X chromosome (Charlesworth et al. 1987) and the X chromosome may have disproportionately lower variation than the autosomes (Aquadro et al. 1994).
In Drosophila melanogaster, interest has centered on the comparison between ancestral range populations from sub-Saharan Africa and more recently founded populations from elsewhere in the world. Andolfatto (2001) examined X-linked and autosomal sequence data in D. melanogaster, finding that cosmopolitan populations had a considerably lower X-to-autosome (X/A) diversity ratio than sub-Saharan populations. Kauer et al. (2002; 2003) confirmed this pattern with a large microsatellite data set, arguing that a disproportionate reduction of cosmopolitan X-linked variation should not result from a founder event bottleneck, but could reflect a higher rate of hitchhiking as cosmopolitan populations adapted to new environments outside Africa.
It is commonly assumed that historical changes in population size (such as founder event bottlenecks) should have similar effects on X-linked and autosomal variation, and thus should have little or no impact on the X-to-autosome (X/A) diversity ratio. However, a few studies have provided clues that population size changes may have contrasting effects on chromosomes with different modes of inheritance. For example, Fay and Wu (1999) and Hey and Harris (1999) found that mitochondrial and autosomal loci can differ in their allele frequency distributions after a population bottleneck. Wall et al. (2002) simulated the effect of a population bottleneck on X-linked and autosomal diversity, focusing primarily on linkage disequilibrium. While in this case the pre-bottleneck X/A diversity ratio was assumed to be less than 0.75, the post-bottleneck X/A ratio was found to be slightly lower yet. Lastly, Lawson-Handley et al. (2006) simulated haplotype diversity for mitochondrial, Y-linked and X-linked loci (in the absence of intragenic recombination), finding that haplotype diversity recovered more quickly for the uniparentally inherited chromosomes than for the X chromosome.
Although the studies cited above have suggested that demographic history may have distinct effects on genetic markers with differing modes of inheritance, the magnitude of this effect is unclear, and its potential influence on chromosomal diversity differences is often ignored. Here we use theoretical predictions to show that population size changes can profoundly alter the X/A diversity ratio. Due to the X chromosome’s smaller effective population size, X-linked variation will converge faster to its new equilibrium after a size change. Therefore, we find that reductions in population size lead to lower X/A diversity ratios, while population growth yields the opposite effect. We show that empirical data from diverse taxa consistently support an important role for this process. Finally, we suggest that jointly considering patterns of X-linked and autosomal variation may improve demographic inference and assist in differentiating population history from other processes, such as positive selection.
Theoretical models
The effect of changing population sizes on the distribution of coalescence times has been extensively treated in the literature (e.g. Slatkin and Hudson 1991; Rogers and Harpending 1992; Polanski et al. 1998) and is well-understood. Here we re-derive expressions for the expected coalescent time for a pair of sequences with a specific inheritance factor (h). Assuming equal effective male and female population sizes, h equals 1 for autosomal markers, ¾ for X-linked markers and ¼ for mtDNA or Y-linked markers. These inheritance factors will allow us to compare the expected nucleotide diversity in different types of markers.
We first derive results based on a discrete time population model with non-overlapping generations, although the results are applicable to a much larger class of population genetic models. Assume a discrete change in population size from 2Nh to 2Nhf, g generations ago. The probability of coalescence in a single generation is then 1/(2Nhf), and we have the following recursion for the expected pairwise coalescent time given a population size change g generations ago:
| (1) |
and initial condition C0 = 2Nh, the familiar result for Wright-Fisher model. The solution to this recursion is
| (2) |
The expected number of mutations separating two alleles is 2μn, where μ is the mutation rate. 2μCn then gives the expected nucleotide diversity (π), or average number of pairwise differences, under an infinite sites model (Kimura 1969).
Now consider two types of genetic markers, with inheritance factors h1 and h2, and mutation rates μ1 and μ2. Without a change in population size, the diversity ratio is
| (3) |
Using Equation 2, we find that with a change in population size, we have
| (4) |
which follows from the result first obtained by Li (1977). Notice that the relative effect of a population size change does not depend on the mutation rates, which only alter diversity ratios as simple scalars.
In the diffusion limit of large population sizes, a similar result can easily be derived using coalescent theory (Kingman 1982a; Kingman 1982b; Husdon 1993). Two lineages coalesce at rate 1/2Nf and 1/2Nhf, before and after the change in population size, respectively. The expected coalescence time for a pair of sequences is then
| (5) |
which agrees with Equation 2 in the limit of large N, and an equation similar to (4) follows trivially.
Similar results can be obtained for more complicated demographic models. For example, consider a bottleneck model in which the population size is reduced by a factor f, g1 generations ago, returning to the original size g1 + g2 generations ago. The expected coalescence time, using the standard coalescent model, is
| (6) |
and the diversity ratio becomes
| (7) |
Derivations similar to Equation 5 and Equation 6 can be found in much existing work (e.g. Polanski et al. 1998; Jesus et al. 2006).
Predictions of the models
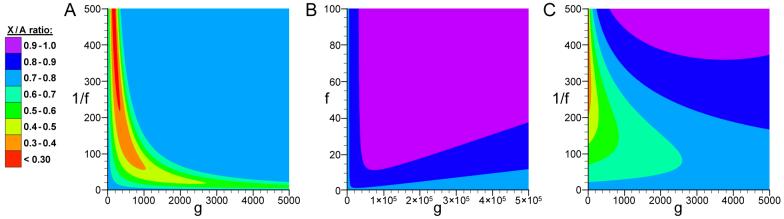
From the model given in Equation 4 (using h1 = ¾, h2 = 1, and μ1 = μ2), we find that reductions in population size lead to disproportionately reduced X-linked variation (Figure 1A). Although weaker population size reductions can lead to prolonged decreases in X/A diversity ratios, the strongest departures are caused by recent, severe size reductions. For example, if a population of initial size 10,000 suffered a 500-fold reduction in size 163 generations ago, the expected X/A diversity ratio is 0.248. Thus, population size reductions compound the difference in effective population size between X chromosomes and autosomes, as genetic drift causes X-linked variation to be lost more quickly. Conversely, population growth can generate X/A diversity ratios closer to unity (but not exceeding 1.0; Figure 1B). Following growth, new X-linked and autosomal variation accumulates at more similar rates, and this effect can be quite long-lasting.
Figure 1. Predictions of the population size change model for X/A diversity ratios.
Predicted ratio of X chromosome to autosome diversity for a population with initial N = 10,000 following (A) population reduction, (B) population growth, or (C) population bottleneck lasting 100 generations. In each plot, the x-axis indicates the number of generations since this event (g), the y-axis indicates the magnitude of the size change (in terms of f for population growth, or 1/f for population reductions and bottlenecks; see equations for details), and color depicts the X/A diversity ratio (see scale).
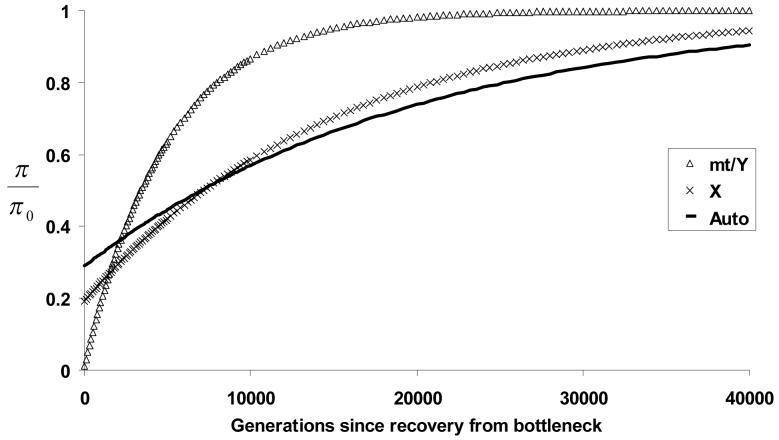
Using Equation 7, we find that population bottlenecks (reductions with subsequent recovery) initially reduce X/A diversity ratios, but with time they may produce X/A ratios greater than the initial value (Figure 1C). This pattern can also be seen in Figure 2, which depicts the recovery of variation after a bottleneck (with initial size N = 10,000, reduction factor f = 0.04, and bottleneck duration g2 = 100 generations): the X chromosome initially loses more of its variation after the bottleneck, but then recovers more quickly than the autosomes.
Figure 2. Recovery of variation after a population bottleneck.
Expected diversity through time following a population bottleneck (relative to pre-bottleneck levels) is shown separately for autosomal, X-linked, and uniparentally inherited chromosomes (mt/Y). In this bottleneck, a population of N = 10,000 experiences a population size reduction to f = 0.004 times this size, lasting for g2 = 100 generations until recovery to the initial size.
Figure 2 also shows the recovery of variation for uniparentally inherited markers (such as mitochondria and the Y chromosome), which have an inheritance factor of h = 0.25. The pattern observed for these haploid markers, as compared to the autosomes, is even more dramatic than for the X chromosome: nearly all variation is lost during the bottleneck, but recovery occurs much more quickly. Importantly, this more rapid recovery of variation is entirely due to the lower effective population sizes of the uniparentally inherited chromosomes, and would not be altered by mutation rate differences between chromosomes.
Comparing predictions to empirical data
We obtained published data from every species that was found to have X-linked and autosomal sequence polymorphism data from at least two populations with differing levels of nucleotide diversity. Data of this type were found for humans (Yu et al. 2002), chimpanzees and orangutans (Kaessmann et al. 2001; Fischer et al. 2006), three subspecies of the house mouse Mus musculus (Baines and Harr 2007), Drosophila melanogaster (Andolfatto 2001; Haddrill et al. 2005; Ometto et al. 2005), and Drosophila simulans (Andolfatto 2001). Additional details concerning the data can be found in the Supplemental Text.
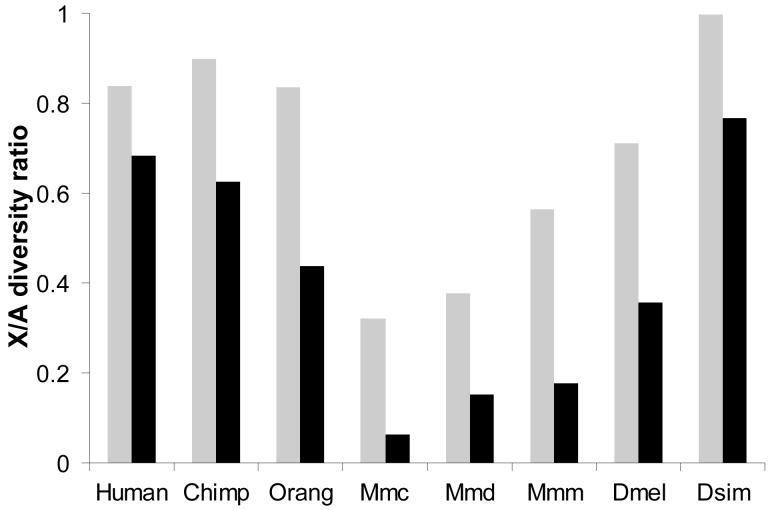
To test whether the empirical data is consistent with our model, we compared X/A diversity ratios between populations for each species/subspecies. To the extent that differences in variability between these populations reflect population size changes subsequent to their divergence (such as founder event bottlenecks leading to the formation of new populations), our model predicts that the less variable population should have a reduced X/A diversity ratio. Indeed, this pattern was observed in every case (Figure 3). For example, as the ancestors of Asian and European human populations migrated out of Africa, they experienced a population bottleneck which reduced genetic variation, and as predicted by the size change model, the X/A diversity ratio is lower in non-African (0.68) than in African (0.84) populations.
Figure 3. Reduced X/A diversity ratios in less variable populations.
Bars indicate the X/A diversity ratios of more genetically diverse (gray) and less genetically diverse (black) populations of Homo sapiens (Human), Pan Troglodytes (Chimp), Pongo pygmaeus (Orang), Mus musculus castaneus (Mmc), Mus musculus domesticus (Mmd), Mus musculus musculus (Mmm), Drosophila melanogaster (Dmel), and Drosophila simulans (Dsim). Information regarding the data can be found in the Supplemental Text.
In addition to the taxa shown in Figure 3, we are aware of SNP diversity data showing reduced X/A diversity ratios in the less variable population of rhesus macaque (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007), along with microsatellite polymorphism data showing lower X/A ratios in less diverse, recently founded populations of Drosophila pseudoobscura (Reiland et al. 2002), and Drosophila subobscura (Pascual et al. 2007). The probability that the less variable population of all eleven of the above taxa would have the lower X/A diversity ratio by chance is less than 0.0005.
Discussion
Because the size change model invokes no phenomena beyond founder event bottlenecks or other population size changes, it represents a parsimonious explanation for the observed population differences in X/A diversity ratios (Figure 3). Clearly, we can not exclude the influence of other mutational, demographic, and selective processes in contributing to these X/A diversity ratios, particularly with regard to the differences between species. For example, male-biased mutation rates may influence X/A diversity ratios in mammalian species to differing degrees (Li et al. 2002), while no such effect has been detected in Drosophila (e.g. Bauer and Aquadro 1997). However, as shown in Equations 4 and 7, mutation rate differences between X-linked and autosomal loci only affect X/A diversity ratios as constant factors, independently of the size change effect. Therefore, male-biased mutation can not account for the population differences shown in Figure 3.
By comparing X/A diversity ratios between closely related populations, we restrict our focus to evolutionary processes that might differ between a given pair of populations. For example, if sex-specific variance in reproductive success is higher for males in the more variable population, but higher for females in the less variable population, a reduced X/A diversity ratio for the latter population would be expected (Charlesworth 2001). However, it seems unlikely that such a shift in reproductive variances in this direction would have occurred in all or most of the species examined here.
It has also been suggested that accelerated rates of adaptive evolution for recently founded populations in new environments may lead to reduced X/A diversity ratios, under the logic that the X chromosome’s hemizygosity in males renders selection more efficient. However, the X chromosome will actually have a lower rate of adaptation than the autosomes if most beneficial alleles come from standing variation (Orr and Betancourt 2001), as may often be the case for populations expanding into new environments. Also, at least in the case of Drosophila-like recombination (none in males), hitchhiking should not reduce X/A diversity ratios unless most beneficial mutations are recessive (Betancourt et al. 2004). The fact that Thornton et al. (2006) detected no “faster-X” effect in Drosophila (i.e. X-linked loci do not have higher rates of protein evolution) might indicate that the majority of beneficial mutations do not fit the above criteria (at least in Drosophila).
While three of the species examined here have expanded from tropical Africa into temperate habitats (i.e. humans, D. melanogaster, and D. simulans), for the remaining species we are comparing populations from less drastically different environments. While such environments may still have very important ecological differences, it is difficult to imagine why, for example, Bornean orangutans should have a vastly higher rate of adaptation than Sumatran orangutans. It is also noteworthy that a reduced X/A diversity ratio was observed in North American D. subobscura (Pascual et al. 2007), given that this population was established only about 25 years prior to the study (roughly 125 generations; Prevosti et al. 1988), which is perhaps too short an interval for a genome-wide effect of hitchhiking to be expected.
Further studies will be needed to determine whether a demographic model of historical size changes can adequately account for patterns of genetic variation in any given species. One example of such an approach is given by Baines and Harr (2007), who fit demographic models for Mus musculus populations using autosomal data, and then tested whether the best-fitting model could account for X-linked polymorphism as well. While this strategy may not formally exclude the possibility that other demographic scenarios are consistent with both X-linked and autosomal diversity, it certainly represents a step toward fully accounting for the influence of population history.
For simplicity, we have assumed throughout this study that X/A diversity ratios before a population size change were equal to 0.75, but clearly the models presented above are not limited to this case. Different mutation rates for X-linked and autosomal loci can easily be substituted into the equations given here. If additional factors are suspected to influence X/A diversity ratios in a particular species, the X-linked and autosomal inheritance factors from our equations could be adjusted to account for the predicted impact of processes such as sex-specific variance in reproductive success (Charlesworth 2001) or background selection (Charlesworth 1996) on the effective population size of X-linked and autosomal loci. Under some scenarios, e.g. if males have an extremely low probability of reproductive success, the effective population size of X-linked loci may even exceed that of autosomal loci (Charlesworth 2001), and the population size change effect would then be in the opposite direction from that presented above. As an alternative to inferring such processes, if the pre- size change X/A diversity ratio can be estimated directly (e.g. by using data from a population that recently diverged from the study population but is not thought to have undergone a recent size change, or from ancient DNA samples), one can focus on the predicted change from initial X-linked and autosomal diversity levels by multiplying these values by the size change terms of Equations 2 and 6 (the terms in parentheses).
We have stated our findings mainly in terms of X-linked and autosomal diversity, but the models given here are equally applicable to comparisons involving mitochondrial or Y-linked variation (e.g. Hey 1997; Fay and Wu 1999; Hey and Harris 1999; Lawson Handley et al. 2006). At first glance, the predictions shown in Figure 2 might suggest that comparisons involving uniparentally inherited chromosomes would be very powerful for detecting population size changes. We caution, however, that since non-recombining markers represent only a single realization of the evolutionary process, they may be particularly sensitive to stochastic variation (and the effects of positive and negative selection). Comparisons involving these markers must account for such uncertainty before demographic inferences can be made.
In conclusion, we suggest that population history may be an important determinant of chromosomal variability in many species. Joint consideration of the diversity level of chromosomes with differing modes of inheritance - along with chromosomal differences in allele frequency spectra (Fay and Wu 1999; Hey and Harris 1999) and linkage disequilibrium (Wall et al. 1999) - should offer new insights into the relative importance of demographic, mutational, and selective forces in shaping genetic diversity.
Acknowledgments
We thank B. Charlesworth and two anonymous reviewers for helpful comments on earlier versions of this manuscript. This research was supported by an N.I.H. Kirschstein-NRSA postdoctoral fellowship (1 F32 HG004182-01) to J.E.P., and by grants from Danmarks Grundforskningsfond and the N.I.H. (U01HL084706) to R.N.
References
- Andolfatto P. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 2001;18:279–290. doi: 10.1093/oxfordjournals.molbev.a003804. [DOI] [PubMed] [Google Scholar]
- Aquadro CF, Begun DJ, Kindahl EC. Selection, recombination and DNA polymorphism in Drosophila. In: Golding B, editor. Non-Neutral Evolution. Chapman & Hall; New York: 1994. pp. 46–56. [Google Scholar]
- Baines JF, Harr B. Reduced X-linked diversity in derived populations of house mice. Genetics. 2007;175:1911–1921. doi: 10.1534/genetics.106.069419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry E, Derome N, Huet M, Veuille M. Contrasted polymorphism patterns in a large sample of populations from the evolutionary genetics model Drosophila simulans. Genetics. 2006;173:759–767. doi: 10.1534/genetics.105.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer VL, Aquadro CF. Rates of DNA sequence evolution are not sex-biased in Drosophila melanogaster and D. simulans. Mol. Biol. Evol. 1997;14:1252–1257. doi: 10.1093/oxfordjournals.molbev.a025734. [DOI] [PubMed] [Google Scholar]
- Betancourt AJ, Kim Y, Orr HA. A pseudohitchhiking model of X vs. autosomal diversity. Genetics. 2004;168:2261–2269. doi: 10.1534/genetics.104.030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A. On the effective size of populations with separate sexes, with particular reference to sex-linked genes. Genetics. 1995;139:1007–1011. doi: 10.1093/genetics/139.2.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Background selection and patterns of genetic diversity in Drosophila melanogaster. Genet. Res. 1996;134:1289–1303. doi: 10.1017/s0016672300034029. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The effect of life-history and mode of inheritance on neutral genetic variability. Genet. Res., Camb. 2001;77:153–166. doi: 10.1017/s0016672301004979. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 1987;130:113–146. [Google Scholar]
- Fay JC, Wu C-I. A human population bottleneck can account for the discordance between patterns of mitochondrial versus nuclear DNA variation. Mol. Biol. Evol. 1999;16:1003–1005. doi: 10.1093/oxfordjournals.molbev.a026175. [DOI] [PubMed] [Google Scholar]
- Fischer A, Pollack J, Thalmann O, Nikel B, Pääbo S. Demographic history and genetic differentiation in apes. Curr. Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Haddrill P, Thornton KR, Charlesworth B, Andolfatto P. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 2005;15:790–799. doi: 10.1101/gr.3541005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. Mitochondrial and nuclear genes present conflicting portraits of human origins. Mol. Bio. Evol. 1997;14:166–172. doi: 10.1093/oxfordjournals.molbev.a025749. [DOI] [PubMed] [Google Scholar]
- Hey J, Harris E. Population bottlenecks and patterns of human polymorphism. Mol. Biol. Evol. 1999;16:1423–1426. doi: 10.1093/oxfordjournals.molbev.a026054. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 1983;23:183–201. doi: 10.1016/0040-5809(83)90013-8. [DOI] [PubMed] [Google Scholar]
- Jesus FF, Wilkins JF, Solferini VN, Wakeley J. Expected coalescence times and segregating sites in a model of glacial cycles. Genet. Mol. Res. 2006;5:466–474. [PubMed] [Google Scholar]
- Kaessmann H, Wiebe V, Weiss G, Pääbo S. Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nature Genetics. 2001;27:155–156. doi: 10.1038/84773. [DOI] [PubMed] [Google Scholar]
- Kauer M, Zangerl B, Dieringer D, Schlötterer C. Chromosomal patterns of microsatellite variability contrast sharply in African and non-African populations of Drosophila melanogaster. Genetics. 2002;160:247–256. doi: 10.1093/genetics/160.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer M, Dieringer D, Schlötterer C. A microsatellite variability screen for positive selection associated with the “out of Africa” habitat expansion of Drosophila melanogaster. Genetics. 2003;165:1137–1148. doi: 10.1093/genetics/165.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutation. Genetics. 1969;61:893–903. doi: 10.1093/genetics/61.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman JFC. The coalescent. Stochastic Process. Appl. 1982;13:235–248. [Google Scholar]
- Kingman JFC. On the genealogy of large populations. J. Appl. Probab. 1982;19A:27–43. [Google Scholar]
- Laporte V, Charlesworth B. Effective population size and population subdivision in demographically structured populations. Genetics. 2002;162:501–519. doi: 10.1093/genetics/162.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson Handley LJ, Berset-Brändli L, Perrin N. Disentangling reasons for low Y chromosome variation in the greater white-toothed shrew Crocidura russula. Genetics. 2006;173:935–942. doi: 10.1534/genetics.105.050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. Distribution of nucleotide differences between two randomly chosen cistrons in a finite population. Genetics. 1977;85:331–337. doi: 10.1093/genetics/85.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H, Yi S, Makova K. Male-driven evolution. Curr. Op. Gen. Dev. 2002;12:650–656. doi: 10.1016/s0959-437x(02)00354-4. [DOI] [PubMed] [Google Scholar]
- Ometto L, Glinka S, De Lorenzo D, Stephan W. Inferring the effects of demography and selection on Drosophila melanogaster populations from a chromosome-wide scan of DNA variation. Mol. Biol. Evol. 2005;22:2219–2230. doi: 10.1093/molbev/msi207. [DOI] [PubMed] [Google Scholar]
- Orr HA, Betancourt AJ. Haldane’s sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Chapuis MP, Mestres F, Balanya J, Huey RB, Gilchrist GW, Serra L, Estoup A. Introduction history of Drosophila subobscura in the New World: a microsatellite-based survey using ABC methods. Mol. Ecol. 2007;16:3069–3083. doi: 10.1111/j.1365-294X.2007.03336.x. [DOI] [PubMed] [Google Scholar]
- Polanski A, Kimmel M, Chakraborty R. Application of a time-dependent coalescence process for inferring the history of population size changes from DNA sequence data. Proc. Natl. Acad. Sci. 1998;95:5456–5461. doi: 10.1073/pnas.95.10.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevosti A, Ribo G, Serra L, Aguade M, Balana J, Monclus M, Mestres F. Colonization of America by Drosophila subobscura: Experiment in Natural Populations that Supports the Adaptive Role of Chromosomal-Inversion Polymorphism. Proc. Natl. Acad. Sci. 1988;85:5597–5600. doi: 10.1073/pnas.85.15.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland J, Hodge S, Noor MAF. Strong founder effect in Drosophila pseudoobscura colonizing New Zealand from North America. J. Heredity. 2002;93:415–420. doi: 10.1093/jhered/93.6.415. [DOI] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Rogers AR, Harpending HC. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129:555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K, Bachtrog D, Andolfatto P. X chromosomes and autosomes evolve at similar rates in Drosophila: No evidence for faster-X protein evolution. Genome Res. 2006;16:498–504. doi: 10.1101/gr.4447906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Andolfatto P, Przeworski M. Testing models of selection and demography in Drosophila simulans. Genetics. 2002;162:203–216. doi: 10.1093/genetics/162.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Chen F-C, Ota S, Jorde LB, Pamilo P, Patthy L, Ramsey M, Jenkins T, Shyue S-K, Li W-H. Larger genetic differences within Africans than between Africans and Eurasians. Genetics. 2002;161:269–274. doi: 10.1093/genetics/161.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]