Abstract
A method involving electron paramagnetic resonance spectroscopy of a site-selectively spin-labeled peripheral membrane protein in the presence and absence of membranes and of a water-soluble spin relaxant (chromium oxalate) has been developed to determine how bee venom phospholipase A2 sits on the membrane. Theory based on the Poisson-Boltzmann equation shows that the rate of spin relaxation of a protein-bound nitroxide by a membrane-impermeant spin relaxant depends on the distance (up to tens of angstroms) from the spin probe to the membrane. The measurements define the interfacial binding surface of this secreted phospholipase A2.
Many interfacial enzymes such as phospholipases are water-soluble and must bind to the membrane-water interface in order to hydrolyze components of the membrane. Although the high-resolution structures of aqueous forms of several phospholipases and lipases are known (1), there are no reports that reveal the positioning of an interfacial enzyme at the membrane-water interface. The same can be said for most membrane-bound proteins. In the case of 14-kD secreted phospholipases A2 (sPLA2s), such as bee venom phospholipase A2 (bvPLA2), the interfacial recognition surface is thought to surround the active site slot; the latter is a deep cavity into which a single phospholipid molecule enters to reach the catalytic residues (2) (Fig. 1). Here we describe a high-resolution structure determination tool based on electron paramagnetic resonance (EPR) spectroscopy that allows peripheral membrane proteins such as sPLA2s to be oriented with respect to the membrane-aqueous interface.
Fig. 1.
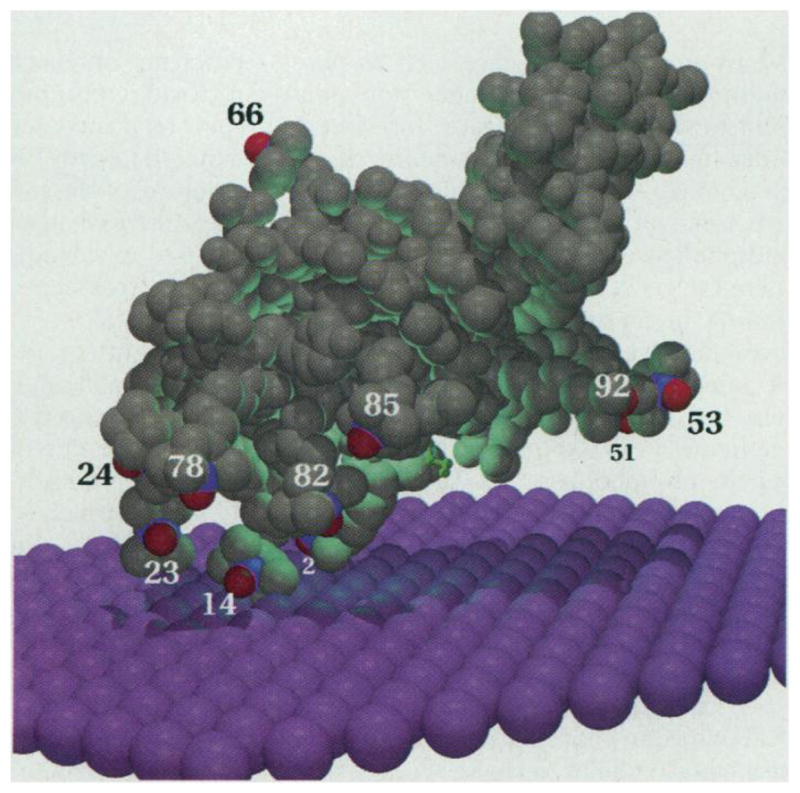
bvPLA2 (gray) positioned on the membrane surface (purple) with the use of the EPR data in Table 1 and the theory developed in this study. The nitrogen and oxygen of each spin label (N-O•) are colored blue and red, respectively. The distance from each spin label to the membrane is as shown in Fig. 3. Spin labels 13 and 15 are hidden from view. A short-chain phospholipid analog inhibitor in the active site slot, as seen in the x-ray structure (14), is shown in green (1). This inhibitor is replaced by a DTPM molecule in these studies. Each membrane sphere has a radius of 2.2 Å, and thus there are about three spheres per phospholipid. The image was created with MOL-SCRIPT and Raster3D (25).
EPR methods have been developed that make use of protein site-specific spin labeling and spin relaxants for probing the membrane penetration depth of segments of integral membrane proteins that pass through the membrane (3). In theory developed below, it will be shown that the efficiency of relaxation of a protein-bound nitroxide spin probe by a water-soluble spin relaxant such as tris(oxalato)chromate(III) (Crox) is dependent on the positioning of the membrane with respect to the spin probe, even when the probe is exposed to the aqueous phase. By measuring the Crox-dependent relaxation of several nitroxides placed at defined locations on the surface of bvPLA2, both in the presence and absence of membranes to which the enzyme binds, it is possible to position the enzyme on the membrane.
In order to apply this method to bvPLA2, 13 site-selectively spin-labeled enzymes were prepared (4), 12 with the spin label located on or near the putative interfacial recognition surface (1,2) and 1 with the probe on the opposite side. The ability of Crox to relax the spin label of each bvPLA2 mutant can be quantified by obtaining the continuous-wave EPR spectra as a function of microwave irradiation power. This series of experiments was carried out in the presence and absence of 10 mM Crox for the enzyme in the aqueous phase or bound to small unilamellar vesicles of the nonhydrolyzable, anionic phospholipid 1,2-dimyristoyl-snglycero-3-phosphomethanol (DTPM) (5). bvPLA2 binds tightly to such vesicles (6). For each data set, the power dependence of the peak to peak height of the central line of the first derivative EPR spectrum, ΔY, was fit by least squares to the power saturation rollover equation (3,7)
| (1) |
where is the microwave amplitude in gauss, P0 is the power incident on the sample, and α is the conversion efficiency factor for the resonator (5) (4.5 G/W1/2). The quantities c, ε, and P2 were allowed to vary during curve fitting. The parameter c is a scaling factor, and P2 is a power parameter that depends only on the properties of the nitroxide (7):
| (2) |
Here, R1 and R2 are the spin lattice and spin-spin relaxation rates (in gauss) and are related by the electron gyromagnetic ratio γe to the relaxation times T1 and T2 as shown (7). The parameter ε is a measure of the curvature of the power dependence and is 3/2 for a homogenous line and 1/2 for a completely inhomogeneous line shape (7). This parameter enables us to obtain very high-quality fits to the data, as it absorbs the effects of inhomogeneous broadening and partially slowed rotational tumbling of the nitroxide.
The change in the fitted value of P2 on addition of Crox is taken as a measure of the effect of this metal on relaxation. The presence of Crox increases both R1 and R2 as follows (8):
| (3) |
where χ is the relaxivity of Crox (9) and the superscript zero refers to the absence of Crox. The quantity ΔP2 is defined as the difference in P2 values in the presence and absence ( ) of Crox:
| (4) |
Thus, ΔP2 is directly proportional to the concentration of Crox in the vicinity of the spin probe; because , the term that is quadratic in [Crox] is small and neglected (8). ΔP2 is measured in the presence and absence of DTPM vesicles, and these two quantities are used to obtain the exposure factor (Φ) as follows:
| (5) |
The superscript “local” refers to the effective concentration of Crox near the spin label and the presence of the membrane reduces this concentration. The equality on the right side of Eq. 5 follows directly from Eq. 4. The quantity 1 – Φ is a measure of the ability of the membrane to shield the protein-bound nitroxide from Crox in the aqueous phase (there is negligible Crox in membranes).
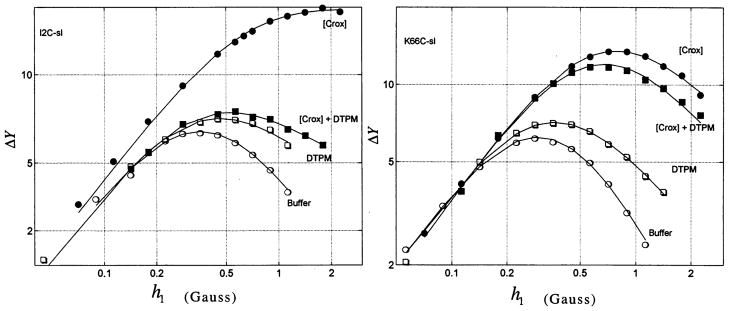
Power saturation rollover curves for the mutant in which isoleucine 2 is replaced with spin-labeled cysteine (I2C-sl) and K66C-sl are shown in Fig. 2 along with the fit to Eq. 1. Values of ε and P2 for all mutants are listed in (10), and values of Φ are listed in Table 1. Ideally, one would expect to be independent of the presence of the membrane, but it is not (10, 11). K66C-sl has its spin label on the face of bvPLA2 that is opposite the putative interfacial recognition surface, and, as expected, Φ for this mutant is close to unity (maximum exposure; Fig. 2 and Table 1). At the other extreme are I2C-sl, K14C-sl, and I78C-sl, which display values of Φ close to zero (Fig. 2 and Table 1), and thus the membrane confers nearly complete protection from Crox relaxation on these nitroxides. The other nine mutants display Φ values of intermediate magnitude (Table 1).
Fig. 2.
Power saturation rollover curves for I2C-Sl and K66C-sl. Curves are shown for bvPLA2 mutant in buffer with and without Crox ([Crox] and buffer) and bound to membranes with and without Crox ([Crox] + DTPM and DTPM). The fit to Eq. 1 is shown by the solid lines. The units of ΔY are arbitrary.
Table 1.
Exposure factor Φ for spin-labeled bvPLA2s.
The key to understanding the data in Table 1 is that the highly negative surface electrostatic potential of DTPM vesicles reduces the concentration of anions in solution near the membrane relative to their bulk concentrations. This results from the Boltzmann equation, which says that the concentration of Crox is a function of the electrostatic potential due to the membrane
| (6) |
Here CCrox (r) is the molar concentration of Crox at a normal distance r from the membrane; ψ(r) is the electrostatic potential; zCrox is the charge on Crox; and F, R, and T have their usual meanings Poisson’s equation describes the electrostatic potential around any set of charges, and for a planar membrane surface of uniform charge density, the potential depends only on r. The final result is the Poisson-Boltzmann equation appropriate for a planar charged membrane (12), which can be written as a first-order differential equation as follows:
| (7) |
Here Ci is the bulk molar concentration of each electrolyte of charge zi in solution, and ε is the dielectric of bulk water (a value of 78). Given the experimental value of ψ(0) = − 77 ± 3 mV for our system (13), Eq. 7 can be solved numerically to obtain ψ(r), and CCrox (r) is obtained using Eq. 6.
Theoretical exposure factors Φ (r) can be calculated as
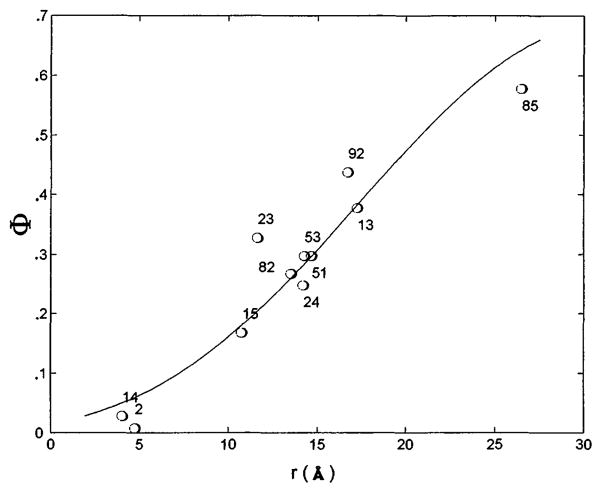
(by analogy to Eq. 5) and compared with experimental Φ values (Table 1) to obtain the normal distance of each spin label to the membrane. To do this, it was assumed that the x-ray structure determined for bvPLA2 in solution (14) is maintained for the enzyme at the interface and that the membrane that contacts the enzyme is a plane. Marquardt-Levenberg regression analysis (15) was carried out by varying the protein-to-membrane distance and the Euler angles for the rotation of bvPLA2 about its center. Several trials were executed with systematic variation of the initial conditions. In all cases, the analysis converged to a single bvPLA2-membrane orientation. Figure 3 shows the remarkably good fit of experimental Φ to calculated Φ (16–18) and Fig. 1 shows the derived structure. The data in Table 1 and (10) also show that values of Φ are significantly larger when the neutral spin relaxant nickel(ethylenediaminediacetic acid) is used instead of Crox, proving that there is a significant electrostatic component to Φ.
Fig. 3.
Regression analysis of the bvPLA2-membrane orientation. The solid line shows calculated values of Φ as a function of the distance from the spin label to the membrane (r), and the circles are the experimental Φ as a function of the modeled distance from the spin label to the membrane for each residue.
Because the effect of Crox on the EPR parameters was measured for bvPLA2 in solution and bound to membranes, to a first approximation the effect of the electrostatic potential at each nitroxide due to the protein alone is removed from the problem because Φ is the ratio (ΔP2)+membrane/ (ΔP2)−membrane Strictly speaking, this is true if the electrostatic potential at each spin label of the protein-membrane complex is equal to the sum of the potentials from the membrane and protein alone. To examine this in more detail, we numerically solved the nonlinear Poisson-Boltzmann equation for bvPLA2 bound to DTPM vesicles as given by Fig. 1 in 50 mM monovalent salt solution and for enzyme and vesicles alone (19). The electrostatic potential of the complex was generally similar to the sum of the potentials due to enzyme and vesicles alone. Very close to the membrane [near spin labels at positions 2 and 14, for which values of Φ near zero were measured (Table 1)], however, the low-dielectric enzyme enhances the negative electrostatic potential of DTPM vesicles by causing the Faraday electric field lines to bend around it and increase in density (20). However, this does not affect the conclusion that these residues are closest to the membrane. Overall, the results suggest that the first-order approach of simply ignoring the nonlinear electrostatic effects is valid.
A clear result of the present study is that bvPLA2 sits on the membrane surface rather than digging into the membrane. This is consistent with monolayer pressure studies showing poor penetration of sPLA2 into an anionic phospholipid monolayer at the air-water interface (21). The opening to the active site slot of bvPLA2 faces the membrane (Fig. 1); however, this opening is not firmly against the membrane. This result is unequivocal as several diagnostic spin labels (at positions 51, 53, 82, 85, and 92) are clearly not as close to the membrane as those at positions 2 and 14 (Table 1). This implies that the alkyl chains of a long-chain phospholipid bound in the active site slot of the enzyme at the interface are partly in contact with the interior of the bilayer, with the hydrophobic walls of the active site slot, and with solvent water [because these experiments were done in the presence of CaCl2, a molecule of DTPM occupies the active site of bvPLA2 at the interface (22)].
The surface of bvPLA2 that contains the opening to the active site slot contains eight cationic residues and only one anionic residue. bvPLA2 and other sPLA2s bind more tightly by orders of magnitude to anionic vesicles than to zwitterionic ones, and it has been hypothesized that these surface cations drive interfacial binding by means of electrostatics. However, our recent study shows that these cationic residues, individually and collectively, are not very important for interfacial binding, because mutating them to glutamates has virtually no effect on the binding of bvPLA2 to anionic vesicles (23). The present study shows that the membrane contact surface of bvPLA2 corresponds to a prominent patch of hydrophobic residues found on all sPLA2s and that all basic residues except K14 are not in close contact with the membrane [see figure 1 of (23)]. The hydrophobic residues are not deeply inserted into the hydrophobic interior of the bilayer but somehow provide a microinterfacial environment that drives interfacial binding to the “polar” phospholipid headgroups (23). The nature of these interactions remains to be understood It is interesting to note that interfacial binding of cellulases to the “hydrophilic” surface of microcrystalline cellulose is driven by hydrophobic residues, including tryptophans on a cellulose-binding domain (24) Finally, the structure shown in Fig 1 provides a physical basis for the kinetic data that indicate that the interfacial recognition and catalytic sites are distinct (22).
The docking technique described in this study should be useful for determining the relative position of any macromolecule of virtually any size and of known three-dimensional structure with respect to any surface with known electrostatic properties, as long as there are no gross conformational changes in the structures of the components when they bind to each other However, useful membrane proximity data should also be obtainable for flexible membrane-bound peptides.
Acknowledgments
This work was supported by NIH grants HL36235 (M.H.G.), GM32681 (B.H.R.), and The Center for Environmental Health P30 ESO7033 (B.H.R.). D.M. is a Helen Hay Whitney Postdoctoral Fellow. We are grateful to J. M. Schurr, S. McLaughlin, and L. J. Slutsky for discussion of theory; to T. Lybrand and E. Adman (NIH University of Washington Center grant P30ES07033) for help with graphics; and to A. Beth for 15N spin labels.
Contributor Information
Ying Lin, Department of Chemistry and Department of Biochemistry, University of Washington, Box 351700, Seattle, WA 98195-1700, USA.
Robert Nielsen, Department of Chemistry, University of Washington, Box 351700, Seattle, WA 98195-1700, USA.
Diana Murray, Department of Physiology, State University of New York at Stony Brook, Health Science Center, Stony Brook, NY, 11794-8661, USA.
Wayne L. Hubbell, Jules Stein Eye Institute, Department of Chemistry and Biochemistry, University of California, Los Angeles, CA 90024-7008, USA
Colin Mailer, Department of Chemistry, University of Washington, Box 351700, Seattle, WA 98195-1700, USA.
Bruce H. Robinson, Department of Chemistry, University of Washington, Box 351700, Seattle, WA 98195-1700, USA.
Michael H. Gelb, Department of Chemistry and Department of Biochemistry, University of Washington, Box 351700, Seattle, WA 98195-1700, USA.
REFERENCES AND NOTES
- 1.Scott DL, Sigler PB. Adv Protein Chem. 1994;45:53. doi: 10.1016/s0065-3233(08)60638-5. [DOI] [PubMed] [Google Scholar]; Heinz DW, Ryan M, Bullock TL, Griffith OH. EMBO J. 1995;14:3855. doi: 10.1002/j.1460-2075.1995.tb00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Essen LO, Perisic O, Cheung R, Katan M, Williams RL. Nature. 1996;380:595. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]; van Tilbeurgh H, Sarda L, Verger R, Cambillau C. 1992;359:159. doi: 10.1038/359159a0. ibid. [DOI] [PubMed] [Google Scholar]; Dere-wenda ZS. Adv Protein Chem. 1994;45:1. [Google Scholar]
- 2.Thunnissen MMGM, et al. Nature. 1990;347:689. doi: 10.1038/347689a0. [DOI] [PubMed] [Google Scholar]; Scott DL, et al. Science. 1990;250:1541. doi: 10.1126/science.2274785. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dijkstra BW, Renetseder R, Kalk KH, Hol WG, Drenth J. J Mol Biol. 1983;168:163. doi: 10.1016/s0022-2836(83)80328-3. [DOI] [PubMed] [Google Scholar]; Achari A, et al. Cold Spring Habor Symp Quant Biol. 1987;52:441. doi: 10.1101/sqb.1987.052.01.051. [DOI] [PubMed] [Google Scholar]
- 3.Altenbach C, Marti T, Khorana HG, Hubbell WL. Science. 1990;248:1088. doi: 10.1126/science.2160734. [DOI] [PubMed] [Google Scholar]; Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. Proc Natl Acad Sci USA. 1994;91:1667. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.bvPLA2 mutants (I2C, N13C, K14C, S15C, R23C, F24C, T51C, T53C, K66C, I78C, F82C, K85C, and D92C) were prepared by a polymerase chain reaction (PCR)–based method (a list of PCR primers is available from the authors on request), and proteins were obtained as described [ Annand RR, et al. Biochemistry. 1996;35:4591. doi: 10.1021/bi9528412.Dudler T, et al. Biochim Biophys Acta. 1992;1165:201. doi: 10.1016/0005-2760(92)90188-2.]. Analysis with Ellman’s reagent revealed that the sulfhydryl group of the introduced cysteine was blocked, and electro-spray mass spectrometry revealed that this sulfhydryl group was disulfide-linked to free cysteine (from the refolding buffer). These surface-exposed disulfides can be selectively cleaved by treating protein [1 mg/ml in 50 mM tris (pH 8.5)] with 0.5 mole equivalents of dithiothreitol (DTT) (from a freshly made stock) for 30 min on ice. A small aliquot of the mixture was submitted to centrifugal gel filtration [DG P-6, Bio-Rad, in 500 mM tris (pH 7.5)] to remove DTT, and the sulfhydryl group content of the protein was estimated with Ellman’s reagent. This process was typically repeated two to four times until the sulfhydryl group content reached 0.2 to 0.5 protein equivalents. Protein was immediately treated with spin-labeling reagent (REANAL, Budapest; Scheme 1) [1 equivalent based on moles of sulfhydryl group (DTT + protein)] from a stock in CH3CN). After 40 min on ice, a small aliquot was submitted to gel filtration as above. If analysis of the void volume with Ellman’s reagent revealed the presence of sulfhydryl group, the protein was treated with an additional 0.5 equivalents of spin-labeling reagent. After a final round of centrifugal gel filtration, the eluant was dialyzed against 5 mM tris (pH 7.5) at 4°C. The sample was concentrated to approximately 10 mg/ml (optical density 280, A1% = 13 cm−1) in a Speed-Vac (Savant) and stored at 4°C for several months without loss of enzymatic activity. The specific enzymatic activities of all spin-labeled mutants except I78C-Sl are within a factor of 2 of that of wild-type enzyme when assayed fluorimetrically with anionic vesicles [ Bayburt T, et al. Anal Biochem. 1995;232:7. doi: 10.1006/abio.1995.9967.]. The specific activity of I78C-Sl was only 7% of the wild-type value.
- 5.EPR samples (3 μl) without membranes contained 0.25 to 0.3 mM spin-labeled bvPLA2 in 50 mM tris-HCl (pH 8.5) with or without 2.5 to 10 mM Crox. Samples with membranes contained 50 μM spin-labeled bvPLA2 in 50 mM tris-HCl (pH 8.5) with 0.5 mM CaCl2 and 22 to 27 mM DTPM as small unilamellar vesicles prepared by sonication [ Jain MK, Gelb MH. Methods Enzymol. 1991;197:112. doi: 10.1016/0076-6879(91)97138-o.]. EPR spectra were acquired with a resolution of 0.125 G per point and were averaged three times [ Mailer C, Danielson SJ, Robinson BH. Rev Sci Instrum. 1995;56:1917.Mailer C, Haas DA, Hustedt EJ, Gladden JG, Robinson BH. J Magn Reson. 1991;91:475.].
- 6.Ghomashchi F, Yu BZ, Jain MK, Gelb MH. Biochemistry. 1991;30:9559. doi: 10.1021/bi00104a001. [DOI] [PubMed] [Google Scholar]
- 7.Altenbach C, Flitsch SL, Khorana HG, Hubbell WL. 1989;28:7806. doi: 10.1021/bi00445a042. ibid. [DOI] [PubMed] [Google Scholar]; Haas DA, Mailer C, Robinson BH. Biophys J. 1993;64:594. doi: 10.1016/S0006-3495(93)81418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altenbach C, Froncisz W, Hyde JS, Hubbell WL. Biophys J. 1989;56:1183. doi: 10.1016/S0006-3495(89)82765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molin YN, Salikhov KM, Zamaraev KI. Spin Exchange: Principles and Applications in Chemistry and Biology. Springer-Verlag; New York: 1980. [Google Scholar]; Hyde JS, Shartz HM, Antholine WE. In: Spin Labeling II: Theory and Applications. Berliner LJ, editor. Academic Press; New York: 1979. pp. 71–113. [Google Scholar]
- 10.The following EPR data are given as mutant name, ε, and P2 (G2). The first pair of numbers is for +DTPM/ −Crox, the second is for +DTPM/+Crox, the third is for −DTPM/−Crox, and the fourth is for −DTPM/ +Crox. Errors in P2 are typically ±5% or less and a few are ± 10% or less. Numbers in parentheses were obtained with 10 mM Ni(ethylenediaminoediacetic acid) instead of Crox. I2C-Sl 0.81, 0.134, 0.75, 0.136, 1.05, 0.141, 0.53, 0.42; N13C-Sl 0.73 (0.85), 0.056 (0.097), 0.77 (0.78), 0.18 (0.32), 1.045 (1.045), 0.098 (0.098), 0.79 (0.78), 0.42 (0.384); K14C-Sl 0.97, 0.124, 0.92, 0.130,1.06, 0.1101, 0.78, 0.316; S15C-Sl 1.02 (1.02), 0.127 (0.127), 0.82 (0.67), 0.159 (0.227), 1.06(1.06), 0.115(0.115), 0.81 (0.86), 0.307 (0.42); R23C-sl 0.74, 0.082, 0.78, 0.228, 1.08, 0.081, 0.83, 0.54; F24C-sl 0.72, 0.077, 0.69, 0.19, 0.99, 0.082, 0.52, 0.51; T51C-Sl 0.94, 0.094, 0.99, 0.24, 1.32, 0.206, 1.06, 0.70; T53C-sl 0.92, 0.043, 0.86, 0.200, 0.92, 0.079, 0.97, 0.611; K66C-sl 0.92, 0.110, 0.92, 0.39, 1.17, 0.117, 0.86, 0.45; I78C-Sl 0.81, 0.157, 0.70, 0.162, 0.96, 0.092, 0.70, 0.72; F82C-Sl 0.93,0.091,0.87,0.14, 0.96,0.1003, 0.65, 0.28; K85C-Sl 0.86, 0.07, 1.01, 0.143, 0.67, 0.032,0.64, 0.15; D92C-sl 0.93,0.038,1.05,0.164, 1.05, 0.164, 1.04, 0.0875, 0.86, 0.38.
- 11.Changes in on the order of 10 to 20% occur upon membrane binding. Generally, relaxation is determined by the electron-nuclear coupling, [O2], and the motional rates of the molecules. The coupling is insensitive to structural alterations and [O2] is rather uniform, even through the outer surfaces of the membrane [ Altenbach C, Greenhaigh DA, Khorana HG, Hubbell WL. Proc Nat Acad Sci USA. 1994;91:1667. doi: 10.1073/pnas.91.5.1667.], and thus the major changes are expected to occur because of motional effects [ Robinson BH, Haas DA, Mailer C. Science. 1994;261:490. doi: 10.1126/science.8290958.]. These arguments suggest that structural rearrangements due to the membrane binding cause minimal changes in the reporter groups; the major change comes from altered local dynamics of the spin probe. More important, it can be assumed that χ (Eqs. 3 and 5) is independent of membrane binding, which depends primarily on the translational diffusion coefficient of Crox.
- 12.Lakshminarayanaiah N. Equations of Membrane Biophysics. Academic Press; Orlando, FL: 1984. [Google Scholar]
- 13.DTPM vesicles containing 3 mol% N-palmitoyl-4-amino-TEMPO [ Shin Y-K, Hubbell WL. Biophys J. 1992;61:1443. doi: 10.1016/S0006-3495(92)81950-1.] were added to the same buffer as that used in EPR studies with bvPLA2, except that 10 mM sodium citrate was used instead of Crox as a nonparamagnetic electrolyte. Saturation recovery and pulsed electron double resonance (EL-DOR) spectra of lipidated spin label were measured in the presence or absence of 1 mM relaxing agent [either 4-hydroxy-TEMPO or 4-ammonium-TEMPO, both labeled at N1 with 15N (gifts from A. Beth, Vanderbilt University)] at 51 °C. Following the theory [ Yin JJ, Hyde J. J Magn Res. 1987;74:82.], the Heisenberg exchange rate due to bimolecular collisions of 14N and 15N species was calculated. The ratio of exchange rates for the two relaxing agents of differing charge is equal to the ratio of local concentrations of these agents at the lipidated spin label location, and thus ψ(0) may be determined [ Shin Y-K, Hubbell W. Biophys J. 1992;61:1443. doi: 10.1016/S0006-3495(92)81950-1.Castle J, Hubbell W. Biochemistry. 1976;15:4818. doi: 10.1021/bi00667a011.].
- 14.Scott DL, Otwinowski Z, Gelb MH, Sigler PB. Science. 1990;250:1563. doi: 10.1126/science.2274788. [DOI] [PubMed] [Google Scholar]
- 15.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes. Cambridge Univ. Press; Cambridge: 1987. [Google Scholar]
- 16.Spin labels were modeled into bvPUA2 as follows. From x-ray crystal structure analysis of spin-labeled lysozyme, the spin label is observed to adopt two conformations for the Cα-Cβ, Cβ-Sγ, and Sγ-Sδ dihedral angles: −60, −60, −90 (major) or −180, −90, −90 (minor) (R. Langen, K.-J. Oh, H. Mchaourab, K. Hideg, W. L. Hubbell, unpublished data; angle conventions are as given by the Insight II program, Biosym Technologies). Using −60, −60, −90 angles, spin labels could be placed at positions 2, 14, 23, 53, 82, and 85 (−180, −90, −90 was used for 92), with no serious van der Waals clash with surrounding residues. For the other residues, it was obvious that neither of these two conformers is accommodated, and then the spin label was positioned into bvPUA2 by avoiding van der Waals conflict and by using reasonable dihedral angles including −90 for the S-S dihedral angle. Because there is some uncertainty in the placement of the spin labels, the regression fit of the modeled structure to the EPR results was repeated after one spin label at a time was omitted from the set. The changes in orientation and in distance from the membrane in all cases were within the errors given by the fit with all spin labels included. The spin labels at positions 14, 15, and 23 were removed as a set because they were deemed to have the most uncertainty in spin-labeling position. Again, the fit was within the stated errors. The good observed fit is due in part to the fact that 13 spin labels were used, and there are not just one or two residues that dictate the structure.
- 17.ψ(0) was allowed to vary during regression analysis as was zCrox from −2 to −3. In all cases, the root mean square deviation of experimental values of Φ from calculated Φ was less than 8%, with the optimum fit (zCrox = −2 and Ψ(0) = −80 mV) having a standard error <6%. The agreement between fitted and experimental values of ψ(0) = −77 ± 3 mV and zcrox = −2.3 (w. L. Hubbell, unpublished data) is gratifying, although the variation in the regression fit with zCrox varying from −2 to −3 is small (Y. Lin et al., data not shown). To account for the finite range of excursion of the nitroxide (as evidenced by the low-order parameters in the EPR spectra) and the finite size of Crox (~4 Å diameter), the potential ψ(r) was convolved with a Gaussian 6 Å wide. All of these changes, that is, zCrox, ψ(0) and a convolution over ψ(r), only affected the protein-membrane distance and did not alter the protein-membrane angular orientation. A third Euler angle is not needed because the membrane is of infinite extent. The Euler angles and protein-membrane distance have errors of less than 10° and 2 Å, respectively, as given by the standard error of propagation for nonlinear least squares algorithms. The data for I78C-Sl were not included in the fit because they are clearly anomalous (Table 1). Because this mutant has only 7% of the enzymatic activity of the wild type, EPR data are not useful. All fitting was done with MATLMB (Mathworks, Cambridge, MA).
- 18.Because the Crox-nitroxide spin exchange happens as fast as Crox diffuses to the nitroxide (9), the possibility that the presence of the membrane slows the rate of spin relaxation by slowing the rate of diffusion-limited Crox-nitroxide encounters was also considered, but the calculation (available from the authors on request) shows that diffusional effects only occur if the membrane is <3 Å from the spin label, and thus diffusional effects are not responsible for the deviation of Φ from unity measured when the spin label is tens of angstroms away from the membrane.
- 19.The electrostatic potentials for bvPLA2 bound to DTPM vesicles (ψE·M) as indicated in Fig. 1 and for the protein (ψE) and membrane (ψM) were calculated as previously described [ Ben-Tal N, Honig B, Peitzsch RM, Denisov G, McLaughlin S. Biophys J. 1996;71:561. doi: 10.1016/S0006-3495(96)79280-9.]. Each leaflet of the membrane bilayer consisted of 360 hexagonally packed DTPM lipids, and it was assumed that Ca2+ and one DTPM were bound in the enzyme’s active site. For each spin label, the potential values in the aqueous phase within 5 Å of the nitroxide nitrogen were used to calculate the average potential difference (ψE·M − ψE − ψM). The calculated potential differences (residue and average ± SD in millivolts) are: 2, −26 ± 15; 13, −7 ± 5; 14, −16 ± 10; 15, −6 ± 8; 23, −12 ± 6; 24, −9 ± 3; 51, −2 ± 3; 53, 0 ± 1; 66, 3 ± 1; 78, −9 ±3; 85, −6 ±2; and 92, −3 ± 1.
- 20.Honig B, Nicholls A. Science. 1995;268:1144. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 21.Thuren T, et al. Biochemistry. 1984;23:5129. [Google Scholar]
- 22.Jain MK, Yu BZ, Rogers J, Ranadive GN, Berg O. Biochemistry. 1991;30:7306. doi: 10.1021/bi00243a036. [DOI] [PubMed] [Google Scholar]; Yu BZ, et al. 1997;36:3870. doi: 10.1021/bi961541x. ibid. [DOI] [PubMed] [Google Scholar]
- 23.Ghomashchi F, et al. ibid. in press. [Google Scholar]
- 24.Din N, et al. Mol Microbiol. 1994;11:747. doi: 10.1111/j.1365-2958.1994.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 25.Kraulis P. J Appl Crystallogr. 1991;24:946. [Google Scholar]; Merritt EA, Bacon DJ. Methods Enzymol. 1997;277:505. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
Scheme 1.