Abstract
T cell receptor (TCR) allelic exclusion is believed to be primarily mediated by suppression of further recombination at the TCR locus after the expression of a functional TCR protein. Genetic allelic exclusion has been shown to be leaky for the β chain and, more commonly, for the α chain. Here, we demonstrate an additional mechanism by which T cells can maintain monoclonality. T cells from double TCR transgenic mice express only one or the other of the two available TCRs at the cell surface. This “functional allelic exclusion” is apparently due to control of the TCR assembly process because these T cells express RNA and protein for all four transgenic TCR proteins. Lack of cell surface expression of the second TCR may be controlled by a failure to assemble the TCR heterodimer.
The T cell receptor (TCR) genes are assembled by somatic recombination of gene segments during thymic development. Recombination is initiated before the CD4+CD8+ stage and normally occurs first at the TCR β locus. A productive rearrangement of one of the two available TCR β loci resulting in the synthesis of a β chain protein appears to be sufficient to suspend further recombination at the β locus (1). Suppression of recombination, or allelic exclusion, appears to be signaled by the pairing of the TCRβ protein with the invariant pre-Tα (2). Rearrangement of the TCR α locus commences after the β selection event (3, 4). Unlike the β locus, α chain recombination typically occurs on both chromosomes (1). Indeed, sequential recombination on the same chromosome can also occur (5). This correlates with the more rigorous positive selection step required for continued T cell development. Most T cells express a single TCR at the cell surface (6–9). Monoclonality of T cells is in part a result of the ordered process of gene rearrangement, especially for the β chain (1). Exclusion of α chains appears to be either at the genetic level owing to a defect in the message produced by the second chromosome (10) or, in some cases, because of phenotypic exclusion. In such cells, two in-frame α chain transcripts are produced, but only one is expressed at the cell surface. This has been suggested to be a passive effect due to competition for pairing of the α chains with the single available β chain (8) or an active process requiring signaling through CD45 (11). Such phenotypic exclusion of α chains appears to occur at the time of positive selection of the developing thymocyte (11, 12).
Despite these mechanisms, T cells bearing two TCR α chains have been detected (6–9), as have cells bearing two TCR β chains (13, 14). Indeed, a T cell clone expressing two different TCR, with each TCR being specific for a different MHC:peptide complex, has been isolated from a TCR transgenic mouse (15). As many as 30% of human T cells have been reported to express two α chains (6). Estimates for mouse lymphocytes expressing two α chains range from 2–21% (6, 8) to 60% in immature thymocytes (12). The frequency of cells expressing two β chains at the cell surface has been estimated at ≈1% for both mouse (14) and human (13).
The analysis of allelic exclusion in T cells from double TCR transgenic mice reported here reveals an additional level of complexity. Whereas allelic exclusion has primarily been thought to occur at the genetic level, we demonstrate that functional allelic exclusion of the T cell receptor can also occur at the level of cell surface expression of the heterodimeric protein. This exclusion affects cell surface expression of both the TCR α chain and also the β chain. Therefore, there appears to be multiple mechanisms for the maintenance of TCR clonality in T cells.
Materials and Methods
Fluorescence-Activated Cell Sorter (FACS) Analysis and Cell Sorting.
Single-cell suspensions were prepared by dissociation of tissues between glass slides. Cells (1 × 106) were incubated 30′ on ice with antibodies, washed, and then analyzed by FACS. TCR transgenic mouse (AND), myelin basic protein (MBP), and AND/MBP cells were stained with antibodies against CD4-Quantum Red (Sigma), Vβ3-R-phycoerythrin (PE), and Vβ8-FITC or Vα11-FITC (PharMingen). AND, D10, or AND/D10 cells were stained with CD4, Vβ3, and Vα2 (FITC, PharMingen). D10, MBP, or D10/MBP cells were stained with CD4, Vα2, and 19G (clonotypic for MBP TCR) followed by incubation with anti-mouse IgG (FITC, Sigma). Cells were prepared similarly for cell sorting.
Proliferation Assays.
Sorted T cells (1–3 × 104) were incubated with 1–2 × 105 irradiated splenocytes in round bottom 96-well plates with titrated peptide, pulsed with 1 μCi of [3H]thymidine per well after 48 h and harvested 24 h later. Controls with no added peptide were also preformed to ensure that the bound anti-TCR antibodies did not result in nonspecific T cell proliferation.
Reverse Transcription (RT)-PCR.
RNA was extracted using Trizol (GIBCO/BRL). cDNA was synthesized by using Ready-To-Go Beads (Amersham Pharmacia) and an oligo d(T)12–18 primer. cDNA added per PCR was normalized by comparing PCR amplification of the hypoxanthine phosphoribosyltransferase gene. DNA products of the PCR were visualized on ethidium bromide-stained 1.2% agarose gels.
Intracellular FACS Analysis.
Intracellular staining was done with the Cytofix/Cytoperm kit (PharMingen). Cells were incubated first with mAbs against cell surface proteins, then permeabilized and stained following the protocol. Control cells were handled identically. Anti-Vα2 and three different anti-Vβ8 antibodies gave substantial background (i.e., intracellular staining of T cells expressing only the AND TCR with F23.1-R-phycoerythrin (PE) resulted in a significant background signal). Increasing the number of washes, titration antibodies, etc. did not alleviate this problem.
Metabolic Labeling.
T cells were washed with methionine and cysteine-free DMEM labeling medium (GIBCO/BRL) and preincubated at 4 × 106 cells/ml for 60 min at 37°C. Cells were pelleted and resuspended at 1 × 107 cells/ml in fresh labeling medium plus 2 mCi of L-[35S]methionine in vitro cell labeling mix (Amersham Pharmacia). After incubation at 37°C for 30 min, cells were diluted, pelleted, and either lysed immediately or stored at −20°C until ready for use.
Immunoprecipitations.
Radiolabeled T cell pellets were extracted for 30 min, lysates were precleared with 2 μl of rabbit serum, 75 μl of Zysorbin (Zymed), and 25 μl of protein G Sepharose (Amersham Pharmacia) per ml of lysate overnight at 4°C before incubation with antibody (5 μg/precipitation) and protein G Sepharose (25 μl). Reducing Laemmli sample buffer was added; samples were boiled 5 min and then analyzed by SDS/PAGE (11%).
Results
TCR Expression in Mice Carrying Transgenes for Two Different αβ TCRs.
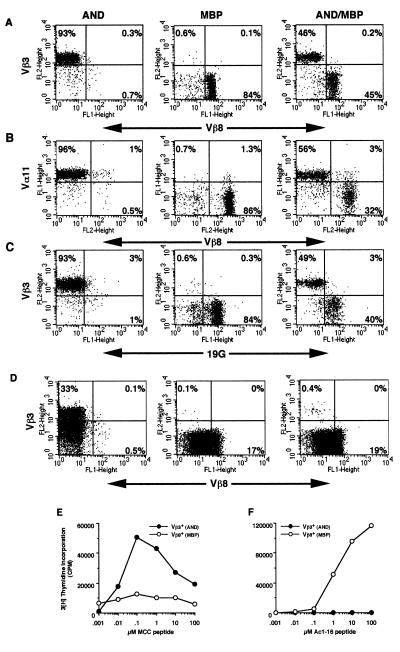
T cells in mice carrying transgenes encoding TCR α and β proteins predominantly express the transgene-encoded TCR proteins at the cell surface. Current data suggest that the presence of a functionally rearranged TCR β transgene will suppress recombination at the endogenous TCR β locus. Transgene-mediated allelic exclusion mimics the genetic allelic exclusion that occurs during the development of wild-type thymocytes. AND mice carry functionally rearranged genes for a Vα11, Vβ3 TCR (16). As a consequence of allelic exclusion, the vast majority of the CD4+ T cells from AND mice express only the transgenic TCR β chain at the cell surface (Fig. 1A, Left). Allelic exclusion of the endogenous TCR β locus also occurs in mice carrying the Vα4, Vβ8.2 TCR transgenes for the MBP TCR (Fig. 1A, Center) (17). Again, nearly all T cells from such mice express only the transgene-encoded Vβ8.2 TCR.
Figure 1.
Analysis of CD4+ T cells and thymocytes from AND, MBP, or AND/MBP double transgenic mice. FACS of lymphocytes with antibodies against (A) CD4, Vβ3, and Vβ8, (B) CD4, Vβ3, and Vα11, or (C) CD4, Vβ3, and 19G (clonotypic for the MBP TCR). Genotype of T cells is listed at the top (AND, AND transgenic mouse; MBP, MBP transgenic mouse; AND/MBP, double transgenic mouse). Plots are labeled with the specificity of the antibody. Only CD4+ T cells are shown. (D) Thymocytes were stained as in A. Quadrants in D separate TCRhi from TCRlo cells. The numbers are the percentage of cells within each quadrant. (E and F) Proliferation of CD4+ T cells from double TCR transgenic mice. Cells were sorted based on Vβ3 or Vβ8 expression. Sorted cells were incubated with (E) B10.BR (I-Ek) splenocytes + titrated MCC peptide or (F) B10.PL (I-Au) splenocytes + titrated Ac1–16 peptide, pulsed with 1 μCi of [3H]thymidine at 48 h and harvested at 72 h. Controls with splenocytes, but no added peptide, gave a low background cpm that was subtracted from each data point. Assays were done in triplicate and represent four different experiments.
Surprisingly, T cells from mice carrying the transgenes for both the AND TCR and the MBP TCR express only one of the two transgene-encoded αβ TCRs at the cell surface. As shown in Fig. 1A (Right), approximately half of the CD4+ lymphocytes from such a dual TCR transgenic mouse express only the AND Vβ3 protein at the cell surface, whereas the other half express only the MBP Vβ8 protein at the cell surface. Furthermore, only one of the two available TCR α chains in the double TCR transgenic T cells is expressed at the cell surface. As shown in Fig. 1B (Right), T cells that expressed the MBP-encoded β chain (Vβ8) did not express the AND-encoded Vα (Vα11) chain at cell surface. The Vα11-positive T cells in the double transgenic mice appeared to express only the AND-encoded TCRα chain, as the intensity of staining for Vα11 is equivalent to that seen for T cells from AND only (Fig. 1B, Left) transgenic mice. Finally, staining with the anti-MBP TCR clonotypic antibody, 19G, confirms that the MBP TCR α chain is paired with the MBP TCR β chain (Fig. 1C, Right). In summary, the AND, MBP double TCR transgenic mice have two populations of T cells: one population that expresses normal levels of only the AND αβ TCR and a second that expresses normal levels of only the MBP αβ TCR.
This unexpected “phenotypic” allelic exclusion is also apparent in thymocytes from these mice, as shown by FACS in Fig. 1D. Nearly all of the thymocytes from mice carrying just the AND TCR express Vβ3 at the cell surface, of which 33% are TCRhi (Fig. 1D, Left). Thymocytes from MBP TCR transgenic mice have a similar staining pattern to AND except, of course, that these cells are Vβ8+ with ≈17% being TCRhi (Fig. 1D, Center). Thymocytes from mice carrying both TCR transgenes predominantly (≈99%) express Vβ8 (MBP) at the cell surface, of which 19% are TCRhi (Fig. 1D, Right). A small, yet distinct, population expresses only Vβ3 at the cell surface (Fig. 1D, Right). No cells were detected that simultaneously express both TCRs. Cell surface expression of the AND transgene-encoded Vα11 protein was also not seen in the Vβ8-expressing population (data not shown). Cell surface exclusion of TCR expression, therefore, is also apparent in thymocytes; however, the ratio of AND TCR to MBP TCR expressing lymphocytes is substantially different in thymocytes as compared with lymphocytes. It is important to note that although the overall percentage of AND TCR+ thymocytes is low in these double transgenic mice, the actual number of mature, AND TCRhi thymocytes is greater than 1 × 106.
Next, T cells from double transgenic mice were sorted by FACS for either Vβ3 (AND) or Vβ8 (MBP) expression. These two sorted populations of cells were incubated with either irradiated B10.BR (I-Ek) splenocytes plus the MCC peptide or with irradiated B10.PL (I-Au) splenocytes plus the MBP Ac1–16 peptide. As predicted by the cell surface staining, the Vβ3+, AND T cells responded only to the B10.BR, MCC peptide-coated splenocytes (Fig. 1 E and F). The Vβ8+, MBP T cells, however, proliferated only to the B10.PL, MBP Ac1–16-coated splenocytes (Fig. 1 E and F). The low background response of the Vβ8+ T cells to the B10.PL, MBP Ac1–16-coated splenocytes was not peptide dependent and may be due to the bound Vβ8 antibody. These data demonstrate that the observed exclusion of cell surface expression of one of the transgenic TCR results in functionally distinct subsets of T cells.
Other TCR Transgene Combinations also Reveal Phenotypic Allelic Exclusion.
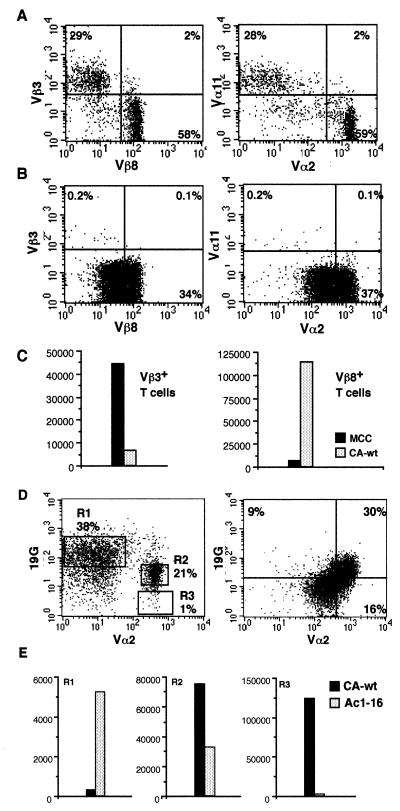
We next examined T cells from mice transgenic for the D10 (18) and AND TCRs. FACS with antibodies against Vβ3 (AND) and Vβ8 (D10) revealed that very few CD4+ T cells from AND/D10 mice simultaneously express both TCRβ chains (Fig. 2, Left). A similar staining pattern was found when antibodies against the different TCR Vα proteins were used (Fig. 2, Right). The clonotypic anti-D10 TCR antibody, 3D3, was used to confirm that the Vβ8+ cells were also expressing the D10α chain (not shown). Thymocytes from AND/D10 mice predominantly express only one of the two TCRs at the cell surface (Fig. 2B).
Figure 2.
Phenotypic and functional allelic exclusion in double TCR transgenic mice. (A) CD4+ Lymphocytes and (B) thymocytes from mice transgenic for both the D10 and AND TCR stained with antibodies against CD4, Vβ3, and Vβ8 (Left) or CD4, Vα2, and Vα11 (Right). (C) Lymphocytes were sorted based on Vβ expression and then stimulated with B10.BR splenocytes plus either 1 μg/ml MCC peptide recognized by the AND TCR or 1 μg/ml CA-wt peptide recognized by the D10 TCR. (D, Left) CD4+ cells from double transgenic D10/MBP mice stained with antibodies against CD4, Vα2 (to detect the D10 TCR), and 19G (clonotypic for the MBP TCR). T cells express only the MBP TCR (R1), only the D10 TCR (R3), or the D10 TCR plus reduced levels of the MBP TCR (R2). Most thymocytes from D10/MBP mice (D, Right) express both the D10 TCR and lower level of the MBP TCR. (E) T cells were sorted as shown in the left panel of D and then incubated with B10.BR splenocytes plus 1 μg/ml CA-wt peptide or B10.PL splenocytes plus 1 μg/ml Ac1–16 peptide. Assays were done in triplicate and represent three different experiments.
Functional exclusion was examined by sorting T cells from AND/D10 mice into two populations using antibodies against Vβ3 and Vβ8, followed by incubation with irradiated B10.BR (I-Ak/Ek) splenocytes loaded with either the MCC peptide or the CA-wild-type (wt) peptide that the D10 TCR recognizes. The sorted Vβ3+ (AND) T cells responded strongly to B10.BR splenocytes coated with MCC peptide, but not with CA-wt peptide. The Vβ8+ (D10) T cells responded weakly to B10.BR splenocytes with MCC peptide and strongly to splenocytes with CA-wt (Fig. 2C).
We also examined mice transgenic for both the D10 and MBP TCRs. Both of these TCRs use the Vβ8.2 gene segment to encode their TCRβ chain; therefore, we resorted to using the anti-Vα2 antibody to detect the D10 TCR and the clonotypic antibody, 19G, to detect the MBP TCR. A large percentage of CD4+ lymphocytes in these mice expressed the MBP TCR but not the D10 TCR α chain (Fig. 2D, R1). A much smaller population of T cells expressed the D10α chain (Vα2) but did not stain positive with the MBP TCR-specific 19G antibody (Fig. 2D, R3). Unlike the other double transgenic mice we examined, D10/MBP mice also had a significant population of T cells expressing both TCR at the cell surface (Fig. 2D, R2). The level of MBP TCR expression is, however, markedly reduced as compared with the MBP TCR-only T cells. These cells, therefore, have down-regulated the MBP TCR while maintaining normal levels of D10 TCR cell surface expression. Approximately one-third of the thymocytes from the D10/MBP mice appear to express high levels of both the D10 and MBP TCRs at the cell (Fig. 2E, Left). Lastly, we sorted the T cells from the MBP/D10 mice into the three populations shown in Fig. 2D. Proliferation assays with each of these populations of T cells demonstrated that the Vα2 (D10) cells responded only to I-Ak APCs + CA-wt peptide, the Vα2+19Glo cells respond to both CA-wt and Ac1–16 when presented by I-Ak and I-Au APCs, respectively, and the Vα2-19G+ cells respond only to Ac1–16 + I-Au APCs (Fig. 2E, Left). The fairly weak response of the Vα2−19G+ cells is presumably due to the interference of the clonotypic antibody because cells collected based on lack of Vα2 expression responded strongly to I-Au + Ac1–16 (≈40,000 cpm, data not shown).
Exclusion Is Not Mediated at the Level of RNA Transcription.
The phenotypic allelic exclusion observed in T cells from double TCR transgenic mice may occur as a result of several different mechanisms. One of the most likely possibilities is the loss of RNA transcription of the nonsurface-expressed TCR. This could possibly be caused by down-regulation of transcription or a deletion or mutation of the nonsurface-expressed TCR transgene. Such deletions have been observed in T cells derived from mice carrying the transgenes for a self-reactive TCR (19). Therefore, we sorted T cells from double transgenic mice as described above, extracted total RNA, and assessed message transcription by RT-PCR. Some of the T cells collected for these experiments were also used in T cell proliferation assays to ensure that the exclusion was complete (data not shown).
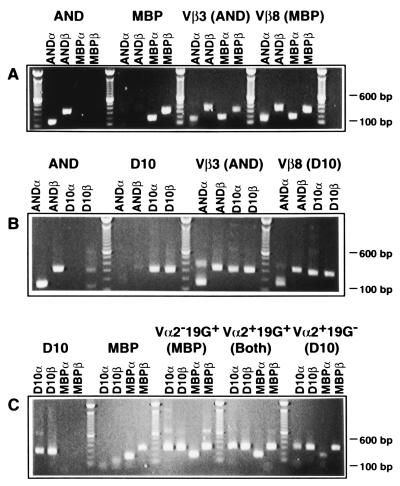
RT-PCR was done with primers for the Vα-Jα or Vβ-Jβ joins for each of the transgenic TCR. Using antisense primers specific for the Jα or Jβ gene segment encoded by the transgenes greatly reduced the possibility of amplification of endogenous TCR α or β chains. RNA from the AND TCR α and β chains can be detected by RT-PCR, as shown in Fig. 3A. Amplification of the AND-derived cDNA with primers specific for the MBP TCR, however, yields no visible product. Amplification of cDNA derived from an MBP TCR single transgenic mouse yields PCR products only for the MBP-specific primers and not the AND-specific primers. The right two panels of Fig. 3A show the results of PCR done with the same primers on sorted T cells from double TCR transgenic mice. The sorted CD4+Vβ3+ (AND) cells also express the mRNA for the MBP TCR, and the sorted CD4+Vβ8+ (MBP) cells also express mRNA for the AND TCR. The quantity of cDNA added to each PCR reaction was normalized based on amplification of hypoxanthine phosphoribosyltransferase (data not shown); however, subtle variations in mRNA levels would not necessarily be detected by this RT-PCR analysis. It is also possible that RNAs for both chains of the nonsurface-expressed TCR are not expressed in the same cells (i.e., half the T cells express the α chain and the other half express the β chain). Analysis of protein expression shown below demonstrates that this was not occurring.
Figure 3.
RNA expression in CD4+ T cells sorted based on cell surface TCR expression. PCR of cDNA from single TCR transgenic mice and from sorted lymphocytes from doubly transgenic mice. (A) AND mice express Vα11 and Vβ3 mRNA, and MBP mice express Vα4 and Vβ8.2 mRNA, whereas T cells from AND/MBP mice sorted for cell surface expression of Vβ3 (AND) or Vβ8 (MBP) express mRNA from both transgenic TCR. (B) Both sorted Vβ3+ (AND) and Vβ8.2+ (MBP) T cells from double transgenic AND/D10 mice express mRNA from both TCR transgenes. Only Vα11 and Vβ3 cDNA are amplified from AND mice, and only Vα2 and Vβ8 cDNA from D10 mice are amplified. (C) All four TCR transcripts are amplified from D10/MBP T cells sorted as shown in Fig. 2D. Different Jβ usage in D10 and MBP allowed for each transcript to be separately amplified (as demonstrated in A and B), although both TCRs use the same Vβ8.2 gene segment. RNA was extracted and reverse transcribed, and the cDNA for the TCR α and β chains was amplified by PCR using primers specific for the Vα-Jα and Vβ-Jβ joins. The AND α chain was amplified using primers specific for Vα11 and Cα. Potential contamination of the cDNA samples with genomic DNA was evaluated by the use of primers specific for the second intron of the MHC class II Eb gene (42). Using identical conditions for PCR to those used to amplify the cDNA, no PCR product could be detected on ethidium bromide-stained agarose gels (data not shown).
The same RT-PCR analysis was completed for the other two combinations of TCR transgenic mice with similar results. Both the Vβ3+ and Vβ8+ T cells from the AND/D10 double TCR transgenic mice express message for both transgenic TCR (Fig. 3B). The T cells from MBP/D10 mice were sorted into the three described populations (Fig. 3A). All three of these populations expressed the message of both transgenic TCR (Fig. 3C). Therefore, exclusion of cell surface expression of the transgenic TCR seen in these various mice is not due to a lack of mRNA transcription.
Intracellular TCR Protein Expression Detected by FACS.
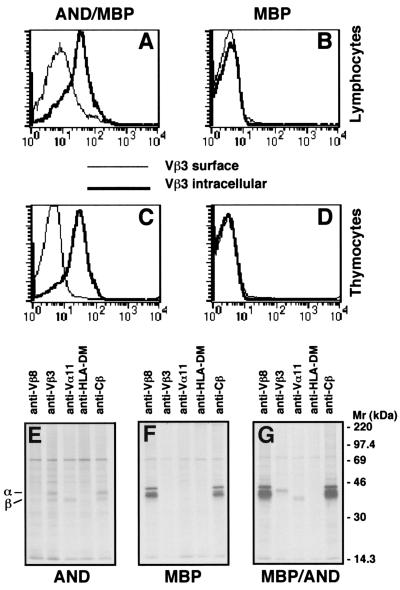
Intracellular staining of AND/MBP double transgenic T cells was next used to determine whether intracellular protein was present for the nonsurface-expressed TCR. For all staining of double transgenic T cells, T cells from single transgenic mice (AND or MBP only) were simultaneously stained to control for background. Unfortunately, various Vβ8-specific antibodies (F23.1, F23.2, and MR5–2) as well as the anti-Vα2 antibody (B20.1) and the MBP TCR clonotypic antibody (19G) all proved to be nonspecific when used for intracellular staining, in our hands. The Vβ3 antibody (KJ25), however, gave clear results as shown in Fig. 4. Lymphocytes from AND/MBP mice were stained with anti-Vβ8 and anti-Vβ3 or with anti-Vβ8 antibodies followed by permeabilization and then incubation with the anti-Vβ3 antibody. The thin line in Fig. 4A shows lack of Vβ3 cell surface staining on Vβ8+ T cells from AND/MBP mice. The overlaid thick line shows bright staining of the same cells that were permeabilized before incubation with the anti-Vβ3 antibody. Importantly, specificity of staining was confirmed by carrying out the same procedure on mice transgenic only for the MBP TCR (Fig. 4B). Similar results are obtained with thymocytes from AND/MBP transgenic mice (Fig. 4C). Again, specificity was confirmed by using thymocytes from MBP TCR transgenic mice (Fig. 4D). This staining clearly indicates that the Vβ8+ T cells and thymocytes from AND/MBP mice also express copious amounts of intracellular Vβ3 TCR protein. Importantly, this analysis also demonstrates that nearly all of the Vβ8+ T cells and thymocytes in AND/MBP mice express intracellular Vβ3. These data demonstrate that lack of cell surface expression of the second TCR is not due to lack of protein expression or, as considered above, expression of only the TCR α chain or the TCR β chain within different T cells.
Figure 4.
Intracellular staining of Vβ3 in Vβ8+ T cells from AND/MBP mice. FACS analysis of lymphocytes and thymocytes harvested from either double transgenic AND/MBP mice or single transgenic MBP mice that were stained with antibodies against CD4, Vβ3, or Vβ8. The histograms show the level of cell surface Vβ3 (thin line) and intracellular Vβ3 staining (thick line) on electronically gated CD4+, Vβ8+ T cells or on total thymocytes. Vβ3 staining of Vβ8+ (A) T cells and (C) thymocytes from AND/MBP mice can only be seen if cells are permeabilized before the addition of the Vβ3 antibody. The specificity of the intracellular staining of the AND TCRβ chain is confirmed by carrying out the same staining procedure on (B) T cells and (D) thymocytes from MBP TCR-only transgenic mice. Sorted T cells from AND (E), MBP (F), and Vβ8+ T cells from AND/MBP (G) transgenic mice were pulse labeled for 30 min with [35S]methionine. The TCR α and β proteins were immunoprecipitated from Triton X-100 solubilized lysates with anti-Vβ8 (mAb F23.2), anti-Vβ3 (mAb KJ25), anti-Vα11 (mAb RR8–1), anti-Cβ (mAb H57), or control anti-HLA-DM (mAb MaP.DMB/c) and analyzed by SDS/PAGE (11%). The positions of the individual TCR β and α chains are indicated.
Posttranslational TCR Functional Allelic Exclusion.
The expression of mRNA from all four TCR chains and the intracellular detection of Vβ3 in Vβ8+ cells in these doubly transgenic T cells suggested that protein from all four chains of these two TCRs is present. To confirm this, we metabolically labeled sorted Vβ8+ (MBP)-expressing T cells from the double transgenic AND/MBP mice and lymphocytes from single transgenic AND TCR or MBP TCR mice. To increase the total cell number, these T cells were first cultured by adding T cell-depleted, irradiated splenocytes bearing the appropriate MHC and antigenic peptide. After 6 days, T cells were restimulated. Before restimulation, the T cells were rechecked for TCR expression patterns by FACS (not shown). The activated T cells were depleted of the added APCs and then pulsed with [35S]methionine for 30 min and lysed in 1% Triton X-100; the TCR proteins were immunoprecipitated using antibodies against Vα11 (RR8–1, ref. 20) and Vβ3 (KJ25, ref. 21) for the AND TCR or Vβ8 (F23.2, ref. 22) for the MBP TCR. Anti-Cβ (H57–597, ref. 23) was used as a positive control and anti-HLA-DM (MaP.DMB/C, ref. 24) as a negative control. The immunoprecipitated proteins were then analyzed by SDS/PAGE.
Both the TCR α and β chain proteins can be immunoprecipitated with anti-Vβ3 from these AND T cells (Fig. 4E). Only the α chain, however, is precipitated by anti-Vα11. The anti-Cβ antibody brings down both the TCR α and β chains. As shown in Fig. 4F, anti-Vβ8 immunoprecipitates both the α and β chain proteins from MBP T cell lysates. An antibody is not available for the MBP TCR α chain, however, both the α and β chains were detected by immunoprecipitation with the anti-clonotype antibody, 19G (not shown). Next, the TCR proteins from sorted Vβ8+ T cells from the AND/MBP double transgenic mice were immunoprecipitated and analyzed by SDS/PAGE (Fig. 4G). As expected, both the α and β chains of the cell surface-expressed MBP TCR can be immunoprecipitated with an antibody against Vβ8. Remarkably, expression of the nonsurface-expressed AND TCR is also clearly detected by immunoprecipitation with antibodies against Vβ3 and Vα11. Intriguingly, immunoprecipitation of the AND TCR with anti-Vβ3 from the Vβ8+ double transgenic T cells resulted only in detection of the AND β chain (Fig. 4G). This is in contrast to what was seen in single AND TCR transgenic T cells in which the anti-Vβ3 antibody precipitated both the β and the α chains (Fig. 4E). Similar results were obtained using freshly isolated thymocytes from these same mice (data not shown).
These data confirm that the proteins for both the MBP and the AND transgenic TCR α and β chains are being synthesized; however, only one of the two available TCRs is expressed at the cell surface. Additionally, these data suggest that the mechanism for preventing cell surface expression of one of the two TCRs involves disruption of the assembly of the αβ heterodimer. Regardless of the mechanism, it is clear that allelic exclusion of TCRs can occur at the protein level.
Discussion
In this report, we demonstrate that allelic exclusion of the TCR is mediated not only at the genetic level but also at the level of the protein. Our data demonstrate that T cells from doubly transgenic mice do not simultaneously express both available TCRs at the cell surface. These transgenic T cells, however, transcribe the mRNA for all four of the TCR chains encoded by the transgenes, suggesting that the proteins are being synthesized as well. Indeed, for one combination we directly demonstrate the presence of all four proteins (two TCR α and two TCR β) by intracellular FACS and also by metabolic labeling of the T cells followed by immunoprecipitation of the TCR complexes. Perhaps most remarkably, either of the two TCR transgenes has the ability to exclude cell surface expression of the other. Therefore, in these mice approximately half of the peripheral T cells express one of the TCRs at the cell surface, whereas the second half express the other TCR at the cell surface.
Expression of only one of the two TCRs at the cell surface may be explained by several mundane explanations. For example, the transgenes encoding the cell surface-expressed TCR may be transcribed earlier in development or at a much higher level. Or perhaps transport of the α or β chain of the nonsurface-expressed TCR out of the endoplasmic reticulum or to the cell surface is somehow less efficient than the cell surface-expressed TCR. These explanations, however, are not likely because any such differences between transcription, expression, or transport of the two TCRs should result in all of the T cells in double transgenic mice expressing the same TCR at the cell surface. This is clearly not the case in any of the mice we have characterized. It is also important to note that genomic or genomic “equivalent” genes were used to construct all three of these transgenic mice (16–18). Therefore, transcription of these TCR transgenes is being controlled by endogenous promoters and enhancers.
The only clues to the mechanism of this functional TCR exclusion come from our immunoprecipitation experiments of metabolically labeled T cells and thymocytes (Fig. 4 and data not shown). Immunoprecipitation of the TCR proteins from AND-only transgenic T cells (Fig. 4E) and thymocytes (not shown) with anti-Vβ3 results in precipitation of both the AND TCR β chain and the associated AND TCR α chain. Intriguingly, however, immunoprecipitation of the intracellular AND TCR proteins from double TCR transgenic T cells that express the MBP TCR at the cell surface brought down only the AND β chain and not the AND α chain (Fig. 4G). This implies that in these T cells, the AND heterodimer never assembles. These data suggest some type of competition between the assembling heterodimers. The TCR assembly process appears to be regulated and will not occur in the absence of certain proteins (25). Perhaps TCR assembly in the endoplasmic reticulum requires multimerization of many αβ heterodimers plus the components of CD3. Supermolecular structures of soluble TCR:peptide:MHC complexes have been observed in vitro as a result of oligomerization (26). In the double TCR transgenic T cells, it is possible that the first αβ heterodimers to multimerize have an advantage for assembly with limiting CD3 components. This, in turn, would prevent the second αβ heterodimer from assembling. Presumably, these unassembled polypeptides would then be susceptible to rapid degradation (27, 28).
Although allelic exclusion is mediated primarily at a genetic level, many exceptions have been noted. For example, T cell clones have been identified that have both α chain loci productively rearranged but only express one of the α chains at the cell surface (29, 30). More impressive was the use of intracellular staining to determine that a high percentage of T cells from B6 mice express two different α chains, one at the cell surface and a second only in the cytoplasm (8). Only a small percentage of peripheral T cells from these mice, however, express two TCR α chains at the cell surface. Competition between the two α chains for binding the one available β chain during positive selection (12) is suggested to explain the discrepancy. Because in our system both TCR β chains are available to associate with the respective α chains, a mechanism other than simple competition must be in effect. Indeed, work by Boyd et al. (11) demonstrated a CD45-dependent signal during intrathymic positive selection causes down-regulation of a second, nonselectable, Vα chain.
T cells with two productively rearranged β chains that express only one of the β chains at the cell surface have been observed in both transgenic and wild-type mice. In a panel of 14 influenza hemagglutinin CD4+ T cell clones, nine expressed cDNA for two different β chains (31). Of these nine clones, two clones had two β chain transcripts that were found to be in-frame. FACS analysis, however, demonstrated that only one of the two TCR was expressed at the cell surface. Similar results were found in T cells from mice carrying both a T cell receptor-β minilocus transgene and a completely rearranged Vβ2 transgene (32). Although the minilocus was productively rearranged, only the completely rearranged Vβ2 transgene was expressed at the cell surface. T cells expressing nontransgene-encoded TCR at the cell surface are easily detected in αβ TCR transgenic mice bred onto deleting MHC haplotypes (19, 33, 34).
Reports have indicated that T cells carrying two productive β chains will express both of the β chains at the cell surface. For example, mice double transgenic for rearranged Vβ2 and Vβ8.2 transgenes express both TCR at the cell surface (35). FACS analysis, however, clearly demonstrated that the majority of the T cells have greatly reduced cell surface levels of one or the other of β chains. The same is true in a report in which double TCR transgenic, Rag1−/− mice are bred (36). The T cells in these mice have clearly down-regulated the cell surface expression of one of the two TCRs. Dual T cell β chain expression has been detected in human lymphocytes (13, 14). Both these reports, however, also demonstrate that the surface expression of the two β chains is not equal. In all three of these reports, it is suggested that the variation of surface β expression is due to competition for pairing with the available α chain.
It is clear, however, that T cells can be identified that have stable expression of equal amounts of two TCR α chains or two TCR β chains at the cell surface (6, 7, 15, 37). Dual expressing clones have even been derived from the D10/MBP double transgenic mice described in this report (38). It should be noted that similar attempts to derive double TCR-expressing T cell clones from AND/D10 mice were not successful (A. Barlow, D.B.S., and C.A.J., unpublished data). Therefore, the question remains as to which of these cell types represents the general rule and which is the exception to the rule. The frequency of normal lymphocytes that express two α or two β chains that have functionally excluded one of the TCR is difficult to determine. A recent analysis of lymphocytes from normal B6 mice suggests that 18–27% of peripheral mouse T cells express two α chains in their cytoplasm (8). However, only 2–4% of T cells from the same mice express two TCR at the cell surface, strongly suggesting posttranslational allelic exclusion (8). Therefore, such cells may not be exceedingly rare.
More importantly, the biological significance of both types of these dual TCR T cells remains to be explored. For example, a TCR “hidden” in the cytoplasm of a thymocyte may allow for a self-reactive TCR to escape intrathymic negative selection. Cell surface expression of the internalized TCR in the periphery may then potentially lead to autoimmunity. To directly test if the presence of a second TCR α chain enhanced the susceptibility of nonobese diabetic (NOD) mice to autoimmune diabetes, NOD mice have been constructed that were heterozygous for a mutation in the TCR Cα gene. An initial report suggested that such mice were protected from disease (39), but upon further backcrossing the differences in the onset of diabetes no longer proved to be significant (ref. 40; S. Wong and C.A.J., unpublished results). However, a recent report using TCR transgenic mice demonstrates that T cells can escape negative selection due to expression of an endogenous α chain (41). These T cells are then capable of infiltrating pancreatic tissue.
Our data suggest that that clonality of TCR expression is not only accomplished at the genetic level but also, to some degree, as a result of a posttranslational mechanism. Most previous work concerning this issue has focused on dual TCRα chain-expressing T cells where lack of surface expression of one of the two α chains is thought to be due to competition for the one available TCRβ chain. Our work clearly demonstrates that posttranslation allelic exclusion functions even when two complete TCRs are expressed and that either of the two TCRs can dominate. Preliminary data suggest that control of this process is likely to be during the assembly of the αβ heterodimer.
Acknowledgments
We thank M. Tector for discussions and critical reading of the manuscript and O. Vandal, D. Guo, and C. Annicelli for technical assistance. We are especially grateful for the thorough review and critical comments from Dr. Mark Davis (Stanford University).
Abbreviations
- TCR
T cell receptor
- FACS
fluorescence-activated cell sorter
- AND
TCR transgenic mouse
- MBP
myelin basic protein
- RT
reverse transcription
- MCC
moth cytochrome c peptide
- CA
chicken conalbumin
- wt
wild type
References
- 1.Malissen M, Trucy J, Jouvin-Marche E, Cazenave P A, Scollay R, Malissen B. Immunol Today. 1992;13:315–322. doi: 10.1016/0167-5699(92)90044-8. [DOI] [PubMed] [Google Scholar]
- 2.Von Boehmer H. Ann NY Acad Sci. 1995;766:52–61. doi: 10.1111/j.1749-6632.1995.tb26648.x. [DOI] [PubMed] [Google Scholar]
- 3.Mallick C A, Dudley E C, Viney J L, Owen M J, Hayday A C. Cell. 1993;73:513–519. doi: 10.1016/0092-8674(93)90138-g. [DOI] [PubMed] [Google Scholar]
- 4.Dudley E C, Girardi M, Owen M J, Hayday A C. Curr Biol. 1995;5:659–669. doi: 10.1016/s0960-9822(95)00131-x. [DOI] [PubMed] [Google Scholar]
- 5.Petrie H T, Livak F, Burtrum D, Mazel S. J Exp Med. 1995;182:121–127. doi: 10.1084/jem.182.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Science. 1993;262:422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 7.Heath W R, Carbone F R, Bertolino P, Kelly J, Cose S, Miller J F. Eur J Immunol. 1995;25:1617–1623. doi: 10.1002/eji.1830250622. [DOI] [PubMed] [Google Scholar]
- 8.Alam S M, Gascoigne N R. J Immunol. 1998;160:3883–3890. [PubMed] [Google Scholar]
- 9.Alam S M, Travers P J, Wung J L, Nasholds W, Redpath S, Jameson S C, Gascoigne N R. Nature (London) 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 10.Casanova J L, Romero P, Widmann C, Kourilsky P, Maryanski J L. J Exp Med. 1991;174:1371–1383. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd R, Kozieradzki I, Chidgey A, Mittrucker H W, Bouchard D, Timms E, Kishihara K, Ong C J, Chui D, Marth J D, et al. J Immunol. 1998;161:1718–1727. [PubMed] [Google Scholar]
- 12.Alam S M, Crispe I N, Gascoigne N R. Immunity. 1995;3:449–458. doi: 10.1016/1074-7613(95)90174-4. [DOI] [PubMed] [Google Scholar]
- 13.Davodeau F, Peyrat M A, Romagne F, Necker A, Hallet M M, Vie H, Bonneville M. J Exp Med. 1995;181:1391–1398. doi: 10.1084/jem.181.4.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padovan E, Giachino C, Cella M, Valitutti S, Acuto O, Lanzavecchia A. J Exp Med. 1995;181:1587–1591. doi: 10.1084/jem.181.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardardottir F, Baron J L, Janeway C A., Jr Proc Natl Acad Sci USA. 1995;92:354–358. doi: 10.1073/pnas.92.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaye J, Hsu M L, Sauron M E, Jameson S C, Gascoigne N R, Hedrick S M. Nature (London) 1989;341:746–749. doi: 10.1038/341746a0. [DOI] [PubMed] [Google Scholar]
- 17.Lafaille J J, Nagashima K, Katsuki M, Tonegawa S. Cell. 1994;78:399–408. doi: 10.1016/0092-8674(94)90419-7. [DOI] [PubMed] [Google Scholar]
- 18.Sant'Angelo D B, Waterbury G, Preston-Hurlburt P, Yoon S T, Medzhitov R, Hong S C, Janeway C A., Jr Immunity. 1996;4:367–376. doi: 10.1016/s1074-7613(00)80250-2. [DOI] [PubMed] [Google Scholar]
- 19.Bluthmann H, Kisielow P, Uematsu Y, Malissen M, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. Nature (London) 1988;334:156–159. doi: 10.1038/334156a0. [DOI] [PubMed] [Google Scholar]
- 20.Jameson S C, Nakajima P B, Brooks J L, Heath W, Kanagawa O, Gascoigne N R. J Immunol. 1991;147:3185–3193. [PubMed] [Google Scholar]
- 21.Pullen A M, Marrack P, Kappler J W. Nature (London) 1988;335:796–801. doi: 10.1038/335796a0. [DOI] [PubMed] [Google Scholar]
- 22.Staerz U D, Rammensee H G, Benedetto J D, Bevan M J. J Immunol. 1985;134:3994–4000. [PubMed] [Google Scholar]
- 23.Kubo R T, Born W, Kappler J W, Marrack P, Pigeon M. J Immunol. 1989;142:2736–2742. [PubMed] [Google Scholar]
- 24.Robbins N F, Hammond C, Denzin L K, Pan M, Cresswell P. Hum Immunol. 1996;45:13–23. doi: 10.1016/0198-8859(95)00152-2. [DOI] [PubMed] [Google Scholar]
- 25.Klausner R D, Lippincott-Schwartz J, Bonifacino J S. Annu Rev Cell Biol. 1990;6:403–431. doi: 10.1146/annurev.cb.06.110190.002155. [DOI] [PubMed] [Google Scholar]
- 26.Reich Z, Boniface J J, Lyons D S, Borochov N, Wachtel E J, Davis M M. Nature (London) 1997;387:617–620. doi: 10.1038/42500. [DOI] [PubMed] [Google Scholar]
- 27.Kearse K P, Roberts J L, Munitz T I, Wiest D L, Nakayama T, Singer A. EMBO J. 1994;13:4504–4514. doi: 10.1002/j.1460-2075.1994.tb06772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huppa J B, Ploegh H L. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- 29.Kuida K, Furutani-Seiki M, Saito T, Kishimoto H, Sano K, Tada T. Int Immunol. 1991;3:75–82. doi: 10.1093/intimm/3.1.75. [DOI] [PubMed] [Google Scholar]
- 30.Couez D, Malissen M, Buferne M, Schmitt-Verhulst A M, Malissen B. Int Immunol. 1991;3:719–729. doi: 10.1093/intimm/3.7.719. [DOI] [PubMed] [Google Scholar]
- 31.Smith C A, Graham C M, Thomas D B. Immunology. 1994;81:502–506. [PMC free article] [PubMed] [Google Scholar]
- 32.van Meerwijk J P, Iglesias A, Hansen-Hagge T, Bluethmann H, Steinmetz M. J Immunol. 1991;147:3224–3228. [PubMed] [Google Scholar]
- 33.Sha W C, Nelson C A, Newberry R D, Kranz D M, Russell J H, Loh D Y. Nature (London) 1988;336:73–76. doi: 10.1038/336073a0. [DOI] [PubMed] [Google Scholar]
- 34.Kisielow P, Bluthmann H, Staerz U D, Steinmetz M, von Boehmer H. Nature (London) 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 35.van Meerwijk J P, Romagnoli P, Iglesias A, Bluethmann H, Steinmetz M. J Exp Med. 1991;174:815–819. doi: 10.1084/jem.174.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zal T, Weiss S, Mellor A, Stockinger B. Proc Natl Acad Sci USA. 1996;93:9102–9107. doi: 10.1073/pnas.93.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heath W R, Miller J F. J Exp Med. 1993;178:1807–1811. doi: 10.1084/jem.178.5.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dittel B N, Stefanova I, Germain R N, Janeway C A., Jr Immunity. 1999;11:289–298. doi: 10.1016/s1074-7613(00)80104-1. [DOI] [PubMed] [Google Scholar]
- 39.Elliott J I, Altmann D M. J Exp Med. 1995;182:953–959. doi: 10.1084/jem.182.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elliott J I, Altmann D M. Eur J Immunol. 1996;26:953–956. doi: 10.1002/eji.1830260436. [DOI] [PubMed] [Google Scholar]
- 41.Sarukhan A, Garcia C, Lanoue A, von Boehmer H. Immunity. 1998;8:563–570. doi: 10.1016/s1074-7613(00)80561-0. [DOI] [PubMed] [Google Scholar]
- 42.Sant'Angelo D B, Lafuse W P, Passmore H C. Genomics. 1992;13:1334–1336. doi: 10.1016/0888-7543(92)90061-v. [DOI] [PubMed] [Google Scholar]