Summary
ALC1, a novel PARP1-stimulated chromatin-remodelling enzyme promotes DNA repair.
Post-translational modifications play key roles in orchestrating chromatin plasticity. Although various chromatin-remodelling enzymes have been described that respond to specific histone modifications, little is known about the role of poly(ADP-ribose) in chromatin remodelling. Here, we identify a novel chromatin-remodelling enzyme, ALC1 (Amplified in Liver Cancer 1), that is specifically regulated by poly(ADP-ribosyl) ation. ALC1 binds poly(ADP-ribose) via a C-terminal Macro domain and catalyzes PARP1-stimulated nucleosome sliding, conferred by an N-terminal ISWI-related helicase core. Our results define ALC1 as a novel DNA damage-response protein, whose role in this process is sustained by its association with known DNA repair factors and its rapid poly(ADP-ribose)-dependent recruitment to DNA damage sites. Furthermore, we show that depletion or overexpression of ALC1 results in sensitivity to DNA-damaging agents. Collectively, these results provide new insights into the mechanisms by which poly(ADP-ribose) regulates DNA repair.
The restricted accessibility of DNA within chromatin presents a barrier to DNA manipulations that require direct protein-DNA interactions (1-3). Processes such as transcription, repair and replication that require efficient DNA recognition are therefore dependent on the appropriate modulation of chromatin structure. Chromatin relaxation is a critical event that occurs during DNA repair and is associated with post-translational poly(ADP-ribose) (PAR) modification (4). PAR is synthesized in a reaction that utilizes NAD+ as a substrate by the PARP family of enzymes, of which PARP1 (and to a lesser extent PARP2) respond to DNA strand breaks (5-7). As a consequence of poly(ADP-ribosyl)ation chromatin has been shown to adopt a more relaxed structure than its unmodified counterpart (8-11), and this is thought to facilitate DNA repair events both by nucleosome displacement and by the recruitment of DNA repair factors (4, 12-14). However, the molecular mechanism by which PAR modulates chromatin during DNA repair is largely unknown.
One potential mechanism for PAR function is to bind and recruit chromatin-modifying components. In a search for DNA repair associated chromatin factors we investigated the function of human ALC1 (Amplified in Liver Cancer; also know as CHD1L). ALC1 contains a putative helicase domain at its N-terminus (Fig. 1A) that is related to the helicase domains found in the Snf2 family of ATP-dependent chromatin remodelling complexes (such as Snf2, ISWI and CHD1) (15). ATPases of this family are modular in nature and often combine a helicase domain with motifs that mediate selective recognition of protein modifications. Interestingly, unlike CHD1, ALC1 does not contain a Chromo domain, which can recognize methylated histone tails. Instead, the modular structure of ALC1 reveals the presence of a Macro domain, an ADPribose/PAR binding element (16). Based on these observations, we speculated that ALC1 might possess a PAR-dependent chromatin remodelling activity that could be utilized to facilitate DNA repair reactions within a chromatin context.
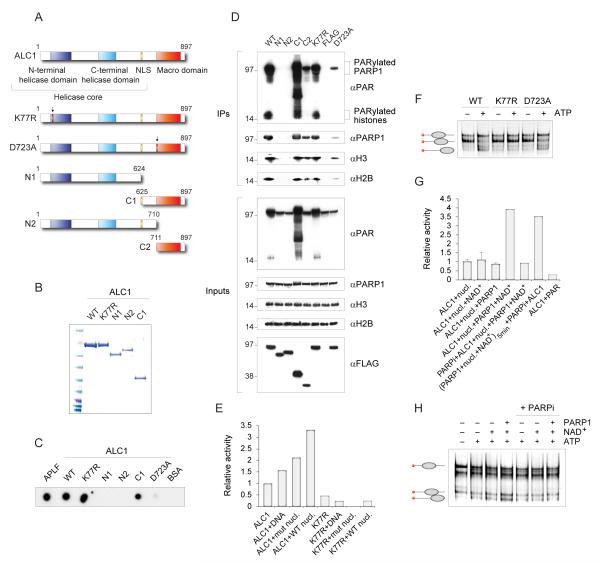
Figure 1.
ALC1 is a chromatin remodelling enzyme regulated by PAR
(A) Schematic representation of the wild-type and mutant ALC1 proteins.
(B) Purified FLAG- tagged ALC1 proteins stained by Coomassie following SDS gel electrophoresis.
(C) In vitro analysis of PAR binding by the wild-type and mutant ALC1 proteins. Proteins were dot-blotted onto a nitrocellulose membrane and incubated with 32P-labelled PAR. APLF was used as a positive control.
(D) Interactions with poly(ADP-ribose), PARP1 and core histones are Macro domaindependent. PAR, PARP1 and histones were immunoprecipitated from cells expressing wild-type or mutant FLAG-tagged ALC1 proteins.
(E) Quantification of ATPase activities measured by a fluorescence-based assay. ATPase activity of ALC1 (2.3 pmol) is stimulated by the wild-type nucleosomes, but not by the mutant nucleosomes assembled with H4(16-19)A. K77R mutation in the Walker A motif completely abolishes ATPase activity of ALC1. The reactions contained 0.1 mM ATP and 20 nM DNA and nucleosomes.
(F) ALC1 repositions nucleosomes in an ATP-dependent manner. 1 pmol of histone octamer assembled on the Cy5-labelled 54A18 DNA was incubated with 2.6 pmol of the wild-type or mutant ALC1 proteins in the absence or presence of 1 mM ATP. Nucleosome-sliding activity of ALC1 is abolished by the Walker A K77R mutation, but not by the Macro domain D723A mutation.
(G) Stimulation of ALC1 ATPase activity by PARP1 and NAD+. ATP hydrolysis by recombinant ALC1 (2.3 pmol) was monitored in the presence of nucleosomes. The reactions contained 0.1 mM ATP and 20 nM DNA and nucleosomes. NAD+ (0.5mM), PARP1 (0.16 pmol), PARP inhibitor (1 μM) and purified PAR (15 nM) were added where indicated. Where indicated, PARP1, NAD+ and nucleosomes were preincubated for 5 min before the sequential addition of PARP inhibitor and ALC1. Rates of ATP hydrolysis are expressed relative to the activity of ALC1 in the presence of nucleosomes.
(H) Stimulation of ALC1 nucleosome repositioning by PARP1 and NAD+. Nucleosome repositioning assays as in F were performed in the presence of PARP1 (0.16 pmol), NAD+ (0.5mM) and PARP inhibitor (1 μM) as indicated. ATP-dependent redistribution of nucleosomes to distal positions on the fragment is stimulated by PARP in the presence of NAD.
To assess whether ALC1 might possess chromatin remodelling activity regulated by poly(ADP-ribosyl)ation, we first determined whether ALC1 binds PAR in vitro. We have constructed a number of ALC1 domain deletions, as well as the putative ATPase dead mutant K77R, and the Macro domain mutant whose ability to bind PAR is predicted to be affected by the mutation of the conserved Asp residue (D723A, Fig. 1A; (16, 17)). FLAG-tagged wild-type (WT) ALC1 and the various mutated ALC1 derivatives were expressed and purified by affinity chromatography from human 293T cells (Fig. 1B). Recombinant proteins were subsequently dotted onto a nitrocellulose membrane, and their ability to bind 32P-radiolabelled PAR was measured. APLF, which was previously shown to bind PAR via an unrelated PAR-binding Zn-finger, was used as a positive control (18). Notably, PAR-binding was detected with the Macro domain-containing ALC1 proteins (WT, K77R, C1), but not with the N-terminal fragments N1 and N2 that contain the helicase domain alone (Fig. 1C). Moreover, the ability to bind PAR was abrogated by the D723A mutation within the ALC1 Macro domain. We also demonstrated PAR-binding in cells by immunoprecipitation of the FLAG-tagged ALC1 proteins from transiently transfected 293T cells. As shown in Fig. 1D, endogenous PAR was immunoprecipitated from extracts of cells expressing the Macro domain proteins, but not from extracts of cells expressing the D723A mutant and the helicase domain fragments N1 and N2. Most of the immunoprecipitated PAR appeared to be associated with PARP1 and histones, as we observed signals on the anti PAR blot whose molecular weights corresponded to these proteins (Fig. 1D). Interestingly, we also detected increased levels of endogenous PAR in the extracts of cells that expressed the Macro domain proteins (WT, K77R and, most notably, C1; Fig. 1D inputs). Possibly, overexpression of these PAR-binding proteins restricted accessibility of PAR to be degraded by the catabolic enzyme PARG, giving rise to the observed effect. The ALC1 immunoprecipitates also revealed interactions of the Macro domain proteins with PARP1 and core nucleosome components (Fig. 1D). The interaction with PARP1 was severely reduced in D723A mutant. Collectively, these results demonstrated Macro domain-dependent associations of ALC1 with PAR, PARP1 and histones.
To examine the potential role of ALC1 in modulating chromatin structure, we tested it for nucleosome-stimulated ATPase activity. ALC1 displayed only weak ATPase activity on its own, but the activity was stimulated modestly by the addition of DNA, and more markedly by the addition of nucleosomes (Fig. 1E). This enhanced activity was, however, less pronounced with mutant nucleosomes containing histone H4 in which residues 16-19 have been replaced by alanine (H4(16-19)A), demonstrating that the ALC1 ATPase activity required the histone H4 N-terminal tail (Fig. 1E). Importantly, a single amino acid change in the ALC1 Walker A motif (K77R) abolished its ATPase activity (Fig. 1E).
To establish ALC1 as a bona fide chromatin-remodelling enzyme, we tested its ability to reposition nucleosomes. Native-gel analyses showed that wild-type ALC1, but not the ALC1 ATPase dead mutant K77R, catalyzed nucleosome sliding in an ATP-dependent manner (Fig. 1F). In contrast, this activity was not compromised by mutation of the Macro domain (D723A) (Fig. 1F). We also demonstrated that the ability of ALC1 to reposition nucleosomes was histone H4 tail-dependent, as it was not observed with the mutant nucleosome H4(16-19)A (fig. S1D). The dependence on this epitope within the H4 tail has been observed with other remodelling enzymes (19, 20) and provides strong evidence that nucleosomes are the relevant substrate for this enzyme. Our results also show that the directionality with which ALC1 repositions mononucleosomes generally involves movement from central locations to positions towards the ends of the DNA fragments (fig. S1E). This aligns the action of ALC1 with the generation of chromatin accessibility rather than the reorganisation or spacing of nucleosomes, which is carried out more efficiently by enzymes that reposition mononucleosomes with the opposite directionality.
Given that ALC1 interacts with PAR and PARP1, we next investigated the effect of PARP1 action on the activities of ALC1. Strikingly, PARP1 stimulated the ATPase activity of recombinant ALC1 approximately 3-4 fold, depending on the assay conditions (with nucleosomes or with DNA, Fig. 1G and fig. S1G). Stimulation was not observed in the absence of NAD+ or DNA, and was abolished by the presence of a PARP inhibitor, indicating that poly(ADP-ribosyl)ation was required for stimulation of ALC1 activity. Interestingly, pre-incubation of PARP1 with nucleosomes and NAD+, followed by the addition of PARP inhibitor, was sufficient for the stimulation of the ALC1 activity (Fig. 1G), suggesting that the stimulation is not dependent on the PARylation of ALC1 itself (fig. S1B). Moreover, addition of purified PAR did not stimulate ALC1 (Fig. 1G), indicating that PAR binding alone is insufficient to stimulate ATPase activity. Based on these results we propose that stimulation of ALC1 requires PARylated PARP1. PARP1 also stimulated the nucleosome repositioning activity of ALC1 in the presence of NAD+ (Fig. 1H), thereby defining ALC1 as a PARP1-stimulated chromatin-repositioning enzyme.
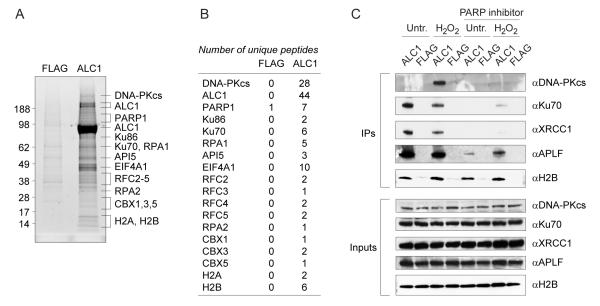
To investigate the cellular functions of ALC1, we analyzed ALC1-associated immunocomplexes by mass spectrometry (MS). We generated stable Flp-In cell lines by integration of FLAG-tagged constructs into a single genomic FRT site expressed from the CMV promoter. The resulting Flp-In-FLAG and Flp-In-ALC1 cells were then used to prepare chromatin extracts, which were subjected to anti-FLAG immunoprecipitation. Comparative MS analysis revealed the presence of proteins implicated in DNA repair, but also in other cellular processes.However, we were most intrigued by the identification of several known DNA repair factors in the ALC1 immunoprecipitates, including DNA-PKcs, Ku and PARP1 (Fig. 2, A and B) (21). These interactions, as well as those with the DNA damage responsive XRCC1 and APLF proteins, were confirmed by Western blot analyses following the immunoprecipitation of ALC1 from transiently transfected cells (Fig. 2C). Moreover, these interactions were largely abrogated by inhibiting PAR synthesis with the PARP inhibitor, indicating that the association of ALC1 with these DNA repair proteins is dependent on PAR modifications.
Figure 2.
ALC1 associates with DNA repair factors in vivo.
(A) Identification of ALC1 interacting partners by mass spectrometry. Chromatin extracts of stable FLAG control and FLAG-tagged ALC1 expressing cells were used for anti-FLAG immunoprecipitation.
(B) Numbers of unique peptides identified by mass spectrometry are indicated in the table.
(C) ALC1 immunoprecipitates from transiently transfected 293T cells contain known DNA repair factors. Interactions are largely dependent on active PAR synthesis. To minimize nucleic-acid-mediated interactions, extracts were treated with benzonase before anti-FLAG immunoprecipitation.
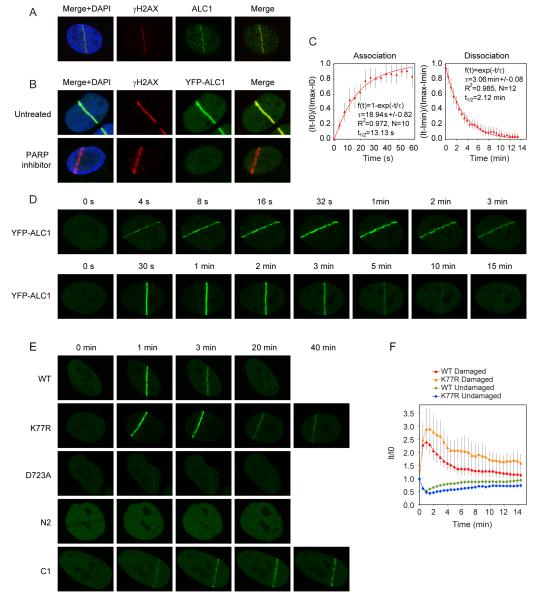
The stimulation of ALC1 nucleosome-repositioning activity by PARP1 and the PAR-dependent interactions between ALC1 and several known DNA repair factors indicates a possible role for ALC1 in the DNA damage response. To examine this we analyzed the association of ALC1 with chromatin before and after treatment of cells with H2O2, which yields single-strand DNA breaks that activate PARP1. Fig. S2A shows the reversible mobilization of ALC1 from the soluble to chromatin-bound fraction after H2O2 treatment, which was largely dependent on active PAR synthesis. Furthermore, we observed the co-localization of ALC1 with sites of active PAR synthesis in mouse 3T3 cells following exposure to H2O2 (fig. S2B). To further investigate the role of ALC1 in the DNA damage response, we analyzed the recruitment of ALC1 to sites of DNA damage induced by laser micro-irradiation. Strikingly, this revealed the efficient localization of endogenous ALC1 to sites of laser-induced DNA breaks, as marked by γH2AX staining (Fig. 3A). Recruitment of YFP-ALC1 to laser-induced DNA breaks was also observed in transiently-transfected U2OS cells (Fig. 3B). Importantly, these recruitment events were entirely dependent on PAR synthesis, as ALC1 recruitment was not detected in cells treated with the PARP inhibitor (Fig. 3B, S2D). Furthermore, ALC1 was also observed to co-localize with PARP1, XRCC1 and APLF at sites of laser micro-irradiation (fig.S3A), consistent with our interaction data (fig. S3A).
Figure 3.
Recruitment of ALC1 to sites of laser-induced DNA damage.
(A) Endogenous ALC1 localizes to sites of laser-induced DNA damage. 1 minute after laser damage, U2OS cells were fixed and stained against γH2AX and ALC1.
(B) Recruitment of ALC1 to laser-induced DNA breaks requires active PAR synthesis. U2OS cells were transiently transfected with YFP-ALC1 and treated with PARP inhibitor where indicated. 1 minute after laser damage cells were fixed and stained against γH2AX.
(C) Kinetics of ALC1 association and dissociation from DNA breaks. Cells transiently expressing YFP-ALC1 were laser micro-irradiated and YFP fluorescence intensities at sites of laser damage were recorded over time. Mathematical modelling of ALC1 association and dissociation at sites of laser damage was carried out as described in Material and Methods. τ: time constant, N: number of cells. Error bars represent standard deviations.
(D) Rapid mobilization and transient association of ALC1 with sites of DNA damage. Shown are representative images of YFP-ALC1 recruitment and dissociation from sites of laser-induced DNA damage.
(E) Recruitment of ALC1 to DNA damage sites is Macro domain-dependent, but its efficient dissociation requires active helicase domain. The helicase core fragment of ALC1 (N2 mutant) is not recruited to DNA damage sites and the recruitment of the D723A mutant is dramatically reduced. In contrast, the ATPase dead mutant (K77R) and the Macro domain fragment (C1) show prolonged retention at sites of laser-induced damage.
(F) Comparative kinetics of the wild-type and K77R mutant ALC1 recruitment to laser-induced DNA breaks in cells transiently expressing YFP-ALC1 constructs. YFP fluorescence intensities were recorded over time in the laser-damaged region (Damaged) or within an undamaged area in the same nucleus (Undamaged) and normalized against the corresponding fluorescence intensity at the time of laser micro-irradiation. Plotted data are averaged values of a minimum of 10 cells from at least two independent experiments. Error bars represent standard deviations.
Interestingly, the kinetics of ALC1 recruitment to DNA damage sites closely mirrored the dynamics of PAR, which is known to be a short-lived modification. Analysis of ALC1 recruitment kinetics using live cell imaging revealed an almost instant mobilization of ALC1 to DNA damage sites, and a relatively transient retention of ALC1 at such regions (Fig. 3, C and D; association t1/2=13.13 s; dissociation t1/2=2.12 min). Moreover, the ALC1 K77R mutant, which is defective for nucleosome sliding in vitro, exhibited prolonged retention at DNA damage sites compared to WT ALC1 (Fig. 3, E and F). This was even more pronounced for the Macro domain fragment of ALC1 (C1 mutant), which persisted at DNA damage sites beyond 40 min (Fig. 3E). In contrast, the helicase core fragment (N2 mutant) was not mobilized to DNA damage sites and the recruitment of the D723A mutants was severely impaired (Fig. 3E). Collectively, these results show that ALC1 recruitment requires Macro domain-mediated recognition of PAR at sites of DNA damage. Conversely, the timely disengagement of ALC1 from DNA damage sites required the ATPase activity of its helicase core, despite this region being dispensable for initial ALC1 recruitment. Interestingly, expression of the Macro domain fragment (C1) also led to prolonged accumulation of the single-strand-break repair factor XRCC1 at sites of DNA damage (fig. S2E), potentially indicating inefficient DNA-break processing and delayed repair kinetics in the absence of ALC1-mediated chromatin remodelling.
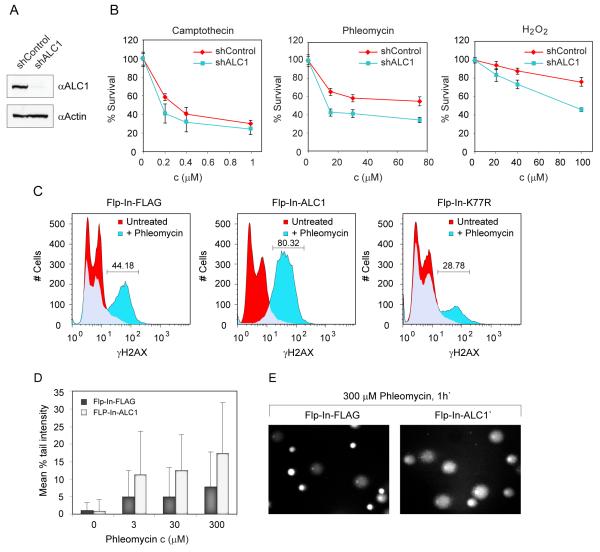
We further explored ALC1 function by assessing the sensitivity of ALC1-deficient cells to various DNA-damaging agents. To do this, we constructed a U2OS cell line in which ALC1 expression was stably reduced by an integrated ALC1 shRNA construct (Fig. 4A). Analyses of these cells revealed them to be sensitive towards H2O2 and the radiomimetic drug phleomycin, but not to other DNA-damaging agents such as camptothecin, mitomycin and hydroxyurea (Fig. 4B and data not shown). DNA damage sensitivity conferred by ALC1 depletion was further confirmed with three different ALC1 shRNA-expressing stable cell lines (data not shown).
Figure 4.
Dysregulation of ALC1 expression confers sensitivity to DNA damage.
(A) Knockdown efficiency of ALC1 in U2OS stable sh cell lines.
(B) Sensitivity of ALC1 deficient cells to DNA damaging agents measured by MTS assay. Plotted data are averaged values from 3 experiments.
(C) Over-expression of ALC1 sensitises cells to H2AX phosphorylation. Stable control (Flp-In-FLAG) and FLAG-tagged ALC1 over-expressing cells (Flp-In-ALC1) were either untreated or exposed to 300 μM phleomycin for 1 h, fixed, stained with FITC-γH2AX antibody and analyzed by FACS. Increase in H2AX phosphorylation is abolished by K77R mutation.
(D) Assessment of DNA damage in ALC1 over-expressing cells by Comet assay. Stable control (Flp-In-FLAG) and FLAG-tagged ALC1 over-expressing cells (Flp-In-ALC1) were untreated or exposed to the indicated concentrations of phleomycin for 1h, and immediately processed. Increase in DNA breaks is observed in cells overexpressing ALC1 following phleomycin treatment. The difference between untreated FLAG and ALC1 Flp-In cell lines is not statistically significant (P value 0.3941). The differences between phleomycin treated FLAG and ALC1 Flp-In cell lines are statistically significant (P values less than 0.0001).
(E) Representative images of control and ALC1 over-expressing cells treated with 0.5 mg/ml phleomycin for 1h and analyzed by Comet assay.
ALC1 was recently identified as a target oncogene within the 1q21 amplicon, which is the most frequent genetic alteration in human hepatocellular carcinoma (HCC) and occurs in 58%-78% of primary HCC cases (22). Furthermore, while the precise molecular impact of ALC1 over-expression is unclear, it was shown that ALC1 over-expressing cells display increased colony formation in soft agar and tumorigenicity in nude mice. Our findings raised the possibility that such effects might result from alterations to the DNA damage response. To analyze the possible consequences of ALC1 over-expression, we exposed stable ALC1 over-expressing and control cells to phleomycin and monitored induction of H2AX phosphorylation (a DNA damage marker) by quantitative FACS analysis. Although no measurable differences in γH2AX profiles were observed between untreated control and ALC1 over-expressing cells, the γH2AX profiles showed a marked distinction following phleomycin treatment. Specifically, phleomycin induced H2AX phosphorylation in 44-50% of control cells, whereas 80-93% of ALC1 over-expressing cells exhibited H2AX phosphorylation (Fig. 4C and fig S4B). Interestingly, the enhanced H2AX phosphorylation was compromised in cells over-expressing the ALC1 Macro domain fragment C1 (fig. S4B), suggesting that the phenotype required ALC1 chromatin remodelling activity. In accord with these findings, enhanced H2AX phosphorylation was abrogated by the K77R ALC1 mutation (Fig. 4C). Interestingly, γH2AX levels were not affected by the treatment of ALC1 overexpressing cells with PARP inhibitor (Fig. S4C), suggesting that enhanced H2AX phosphorylation is not dependent on the recruitment of ALC1 to the sites of DNA damage, but rather that it arises as a consequence of dysregulated ALC1 expression. Furthermore, the differential phenotype was specific to phleomycin and could not be observed with IR or H2O2 (data not shown), possibly reflecting a difference in the accessibility of chromatin-embedded DNA to these DNA damaging agents. Indeed, these results are in agreement with the observation that the structurally related DNA damaging agent bleomycin induces DNA breaks in the linker region, but not in core DNA and therefore acts preferentially on relaxed chromatin (23). To confirm that the stimulation of H2AX phosphorylation indeed reflected an increase in the levels of DNA damage induced in ALC1 over-expressing cells we analyzed DNA damage directly using an alkaline Comet assay. We found that phleomycin exposure consistently produced longer tails (i.e. more damage) in ALC1 over-expressing cells compared to control cells (Fig. 4, D and E). We thus conclude that ALC1 over-expression leads to increased susceptibility to phleomycin-induced DNA damage.
Modulation of chromatin is principally achieved by two activities: post-translational modifications of histones (24, 25) and ATP-dependent chromatin remodelling (26, 27). Consequently, DNA helicase and histone-modification-binding domains frequently synergize to reorganize chromatin into productive configurations, as exemplified by the modularity within the members of the Snf2 enzyme family (15, 28). While chromatin-remodelling factors that respond to acetylated and methylated nucleosomes are well-documented (12, 29-33), PAR-stimulated chromatin remodelers have not been previously identified. Interestingly, a recent report demonstrates negative regulation of Drosophila ISWI by PAR, suggesting a role of PAR in modulating the activity of chromatin remodelling enzymes (34). However, despite the firmly established connection between PAR and the DNA damage response, the molecular mechanism of how PAR regulates DNA repair within the context of chromatin has not been resolved. Here we provide molecular evidence that defines the role of PAR in active chromatin remodelling. We identify a novel nucleosome-repositioning activity that is specifically targeted to sites of DNA damage through interaction with PAR and is required for DNA repair. Intriguingly, we show that PAR not only acts to recruit ALC1 to DNA damage sites, but that it also stimulates its enzymatic activity, thus providing an additional level of regulation. Furthermore, our results demonstrate interdependence between the chromatin remodelling and DNA repair functions of ALC1, as separation of the constituent domains leads to defective recruitment or delayed repair kinetics. Finally, the requirement for appropriate ALC1 regulation is highlighted by its frequent over-expression in human liver cancers (22). Our findings demonstrating that such over-expression sensitizes cells to DNA damage provide a key insight into the molecular consequence of ALC1 deregulation, emphasizing the importance of chromatin reorganization as a significant element in genome stability and cancer.
Materials and methods
Cell lines, cell culture, inhibitors
U2OS and HEK293 cell lines (Cancer Research UK Cell Services) were maintained as adherent monolayers or in cell suspension cultures in DMEM media containing 10% FBS and 1% Pen/Strep at 37°C in a humidified atmosphere of 5% carbon dioxide. Stable HEK293-Flp-In cell lines (Invitrogen) were created by transfection of pDEST-Flag/FRT/TO-ALC1 according to the Flp-In cell lines manual and selected in media containing 150 μg/ml hygromycin B (Invitrogen). Stable U2OS shALC1 and sh Control cell lines were generated by transfection of pSuperior containing hairpin targeting the untranslated region of ALC1 mRNA (sense target sequence TTGCTAGTTGCATAATAA) or an empty vector. The cells were selected in media containing 2 μg/ml Puromycin (Sigma). PARP-1/2 inhibitor (KU-0058948, kindly provided by G.C. Smith, KuDOS Pharmaceuticals Ltd) was added to the culture medium at a final concentration of 10 μM, 1 h prior to subsequent cell treatment.
Plasmids and Proteins
ALC1 was amplified from human HeLa cDNA library. N1, N2, C1 and C2 ALC1 variants were subcloned from ALC1 pDONR221 entry vector, whereas K77R and D723A mutations were introduced using the QuickChange II site-directed mutagenesis kit (Stratagene). For transient transfection the ALC1 constructs were cloned by Gateway LR reaction to pDEST-Flag and pDEST-YFP/FRT/TO, while pDEST-Flag/FRT/TO was used for the stable transfections. The pDEST-YFP/FRT/TO was created by changing the FLAG sequence in pDEST-Flag/FRT/TO to YFP from pYFP-C1 (Clontech).
Human embryonic kidney 293T cells were transiently transfected using Lipofectamine and Plus reagent (Invitrogen), according to the manufacturer’s specifications, with FLAG-tagged wild-type and mutant ALC1 constructs. Approximately 30 h after transfection cells were solubilized in lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% Triton X-100, 1 mM DTT, 1 mM EDTA) supplemented with 50 U/μL benzonase (Novagen) and protein inhibitors (Sigma). Whole cell extracts were obtained by centrifugation and incubated with anti-FLAG M2 agarose beads (Sigma) for 2 hours at 4 °C. Beads were successively washed in lysis buffer and wash buffer (50 mM Tris-HCl pH 8.0, 2 M NaCl, 1% Triton X-100, 1 mM DTT, 1 mM EDTA). Proteins were eluted with 3×FLAG peptide as specified by the manufacturer.
For ATPase and chromatin remodeling assays ALC1 proteins were subcloned into pFastBac HTa (Invitrogen). FLAG-tagged wild-type ALC1, K77R and D723A proteins were expressed in Sf9 insect cells after bacmid transfection and virus amplification according to the manufacturer’s instructions (BAC-TO-BAC Baculovirus Expression System, Invitrogen). Recombinant proteins were purified as described previously for Acf1 (1). Yeast Chd1 was purified as previously described (2). APLF was purified as described (3).
Antibodies
Rabbit anti-ALC1 polyclonal and mouse anti-ALC1 (CHD1L) monoclonal antibodies, as well as rabbit anti-PARP1, H2B, H4, DNA-PKcs, XRCC1 and mouse actin antibodies were from Abcam. Rabbit anti-XRCC1 anti PARP1 antibodies used in immunufluorescence were from Bethyl Laboratories and Cell Signalling, respectively. Mouse monoclonal and rabbit polyclonal anti-PAR antibodies were from Trevigen. Rabbit anti-Ku70 was from Serotec. Rabbit anti–γH2AX antibody was from Cell Signaling. Anti–γH2AX, anti-HP1α, ubH2A and anti-H3K9ac antibodies were from Upstate Biotechnology. Anti phospho-SMC1 was from Bethyl, and anti-H3 was from Abcam. Anti GFP antibody was from Abcam. Mouse anti-Tubulin antibody and the mouse anti-Flag antibody conjugated with peroxidase was from Sigma-Aldrich. The secondary antibodies for immunofluorescence coupled to Alexa 488 or Alexa 594 or 647 were from Molecular Probes. Rabbit anti-APLF polyclonal antibody was raised against purified recombinant APLF protein prepared from E. coli (3).
PAR-binding assay
Proteins (2 pmol) were dot-botted onto a nitrocellulose membrane, which was subsequently blocked with TBS-T buffer (Tris-HCl pH 7.5, 150 mM NaCl, 0.05% Tween) supplemented with 5% milk. 200 ng of automodified PARP1 was used to produce radioactively labeled PAR as described (3). PAR polymer was then detached from PARP1 by DNAse I and proteinase K digestion. Following phenol–chloroform extraction, the water-soluble PAR polymer was diluted in 10 ml with TBS-T and incubated with the nitrocellulose membrane. The membrane was extensively washed with TBS-T, and TBS-T containing 1 M NaCl, air-dried and subjected to autoradiography.
Modification by PARP1 in vitro
Recombinant proteins were modified using a PARP activity assay kit (Trevigen). Typical 10 μl reaction contained 30 ng of purified PARP1, 1 μM substrate protein, and 100 μM of NAD+ spiked with [32P]-labeled NAD+ (Amersham Biosciences). Reactions were incubated for 10 min at room temperature. Modified proteins were analyzed by SDS-PAGE and visualized by autoradiography.
Octamer and Nucleosome assembly
Octamers were assembled from recombinant Xenopus laevis histones expressed and purified as previously described (4). Nucleosomes were assembled on a DNA fragment containing the MMTV nucleosome A sequence by mixing equimolar amounts of histone octamer and DNA in high salt concentrations and performing stepwise dialysis into low salt concentrations (5). DNA was generated by preparative PCR using Cy5 labeled primers (MWG). The diagram represents the length of the DNA extensions on either side of the MMTV nucleosome A (6) or 601.3 (W) (7) positioning sequence (8).
ATPase Assay
ATPase activity was measured in an assay where the amount of inorganic phosphate (Pi) produced by ATP hydrolysis is in proportion to the fluorescence intensity of a phosphate binding protein (PBP) (8, 9) labelled with a coumarin-based fluorescent dye, 7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl) coumarin (MDCC). On binding Pi, the fluorescence of the labelled protein (MDCC-PBP) increases and its emission wavelength shifts. The increase in fluorescence was measured in solution in real-time on a Cary Eclipse fluorometer (Varian, Australia), as described previously (8). MDCC-Phosphate Sensor was diluted using 15 mM Pipes (pH 6.8) to 50 μM, and mixed with nucleosome and enzyme (2.3 pmol) in a final buffer concentration of 50 mM NaCl, 50 mM Tris HCl pH 7.5 and 3 mM MgCl2 to a final concentration of 1.5 μM. Reactions were started by addition of 0.5 mM ATP which had previously been treated to remove inorganic phosphate contamination (9) The rate of ATP hydrolysis calculated from the initial gradient.
Nucleosome repositioning
Nucleosome repositioning was carried out in 10 μl reactions containing 50 mM Tris HCl pH 8.0, 35 mM NaCl, 3 mM MgCl2, 1 mM ATP, 1 pmol of nucleosome and various concentrations of enzymes. After 20 min incubation at 30 °C reactions were stopped by addition of competitor DNA (0.1 μg/μl final), salt (200 mM NaCl final) and sucrose to 5 % (w/v), and placed on ice. The reactions were run on a 5% native polyacrylamide gel at 4° C in 0.2x Tris borate-EDTA, for 3.5 h at 300 V with running buffer recirculation. Gels were scanned in a Fuji Phosphoimager FLA-5100 and the bands were analysed with Aida software (Fujifilm).
Immunoprecipitation
293T cells were transiently transfected using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s specifications, with FLAG-tagged wild-type and mutant ALC1 constructs. 30 h after transfection cells were solubilised in lysis buffer (50 mM Tris-HCl pH 8.0, 200 mM NaCl, 1% Triton X-100, 1 mM DTT, 1 mM EDTA) supplemented with 50 U/μL benzonase (Novagen) and protein inhibitors (Sigma). Whole cell extracts were clarified by centrifugation and incubated with anti-FLAG M2 agarose (Sigma) for 30 min at 4 °C. Following repeated washes with lysis buffer, the immunoprecipitates were boiled in SDS-PAGE loading buffer and analysed by immunoblotting.
Immunoblotting
Proteins were resolved on 6% SDS-PAGE and transferred onto nitrocellulose membrane (Whatman Protran). The membrane was blocked in Tris Buffered Saline-0.1% Tween-5% non-fat dry milk. Proteins were detected with the approriate primary antibodies and secondary antibodies coupled to horse-radish peroxidase (from Pierce and DAKO). Revelation was performed with ECL Western Blotting detection reagent (GE Healthcare)
Immunofluorescence
3T3 cells were grown on glass coverslips in 24 well plate. Where indicated, they were treated with hydrogen peroxide for 10 min and fixed with methanol / acetone (1/1, v/v) for 10 min at 4 °C. Cells were then washed three times with PBS, permeabilized with 0.1% Triton X-100 in PBS and blocked with 2% BSA in PBS. Following incubation with monoclonal anti-PAR antibody and Alexa Fluor 546 goat anti-mouse IgG secondary antibody (Invitrogen), samples were analysed using a Deltavision system.
For immunofluorescence on laser micro-irradiated cells, U2OS cells were fixed in 2% paraformaldehyde and permeablized with 0.2% Triton-X-100 in PBS, except for XRCC1 immunodetection where permeablization was carried out prior to fixation in extraction buffer (20 mM Hepes pH 7.5, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 0.5% Triton-X-100). Appropriate primary antibodies and secondary antibodies coupled to AlexaFluor 488, 594 or 647 (Molecular Probes) were used for immunodetection. Confocal images were captured on FluoView 1000 microscope using 60× oil objective. To avoid bleed-through artefacts, signals from different fluorescent channels were acquired sequentially using a multi-tracking mode.
Purification of ALC1 associated immunocomplexes for mass spectrometry analysis
Purification of ALC1 associated immunocomplexes was performed essentially as described (10). Stable Flp-In FLAG and ALC1 cells were grown in roller bottles (6 litres), pelleted, washed in PBS and lysed for 10 min at 4 °C in sucrose buffer (10 mM HEPES pH 7.9, 0.34 M sucrose, 3 mM CaCl2, 2 mM magnesium acetate, 0.1 mM EDTA, protease inhibitors) containing 0.5 % NP40. Nuclei were then pelleted by centrifugation at 3900 g for 20 min. Residual cytoplasmic contamination was removed by washing with sucrose buffer and subsequent centrifugation step at 3900 g for 20 min. Nuclei were resuspended in nucleoplasmic extraction buffer (20 mM HEPES pH 7.9, 3 mM EDTA, 10% glycerol, 150 mM potassium acetate, 1.5 mM MgCl2, 1 mM DTT, protease inhibitors), homogenised and rotated for 20 min at 4 °C. Chromatin enriched fraction was pelleted by centrifugation at 13,000 rpm for 30 min. The pellet was resuspended in digestion buffer (150 mM HEPES pH 7.9, 1.5 mM MgCl2, 150 mM potassium acetate, protease inhibitors), homogenised and incubated with benzonase (25 units/ul stock) for 1 h at room temperature. The digested chromatin was cleared by centrifugation at 38,000 g for 30 min. Soluble chromatin extract was recovered and used for anti-FLAG immunoprecipitation with anti-FLAG M2 agarose. Immunoprecipirated proteins were eluted by 3×FLAG peptide and resolved by SDS protein gel electrophoresis. Protein bands were visualized by staining with SYPRO Rubi, excised and identified by mass spectrometry.
Mass spectrometric analyses and protein identification
Polyacrylamide gel slices (1-2 mm) were excised using a scalpel and processed for mass spectrometry using the Janus automated liquid handling system (PerkinElmer, U.K.). Briefly, the excised protein gel pieces were placed in individual wells of a 96-well microtitre plate and destained with 50% v/v acetonitrile and 50 mM ammonium bicarbonate, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide. After alkylation, proteins in gel pieces were digested with 6 ng/uL trypsin overnight at 37 °C. The resulting peptides were extracted in 1% v/v formic acid, 2% v/v acetonitrile and subsequently adjusted to 10% v/v formic acid immediately prior to analysis. The digests were analysed by nano-scale capillary LC-ESIMS/MS using a Waters nanoACQUITY UPLC to deliver a flow of 300 nL/min. A Waters μ-Precolumn, C18 Symmetry 5 mm, 180 mm × 20 mm (Waters, U.K.) guard column trapped the peptides prior to separation on a C18 BEH130 1.7 mm, 75 μm × 100 mm nanoAcquity UPLC column. Peptides were flushed from the guard column onto the analytical column at 300 nL/min and eluted with a gradient of acetonitrile. The column outlet was directly coupled to a Triversa nanomate microfluidic chip interface (Advion, U.K.). Mass spectrometric information was obtained using an orthogonal acceleration Quadrupole-Time of Flight mass spectrometer (SYNAPT HDMS, Waters, U.K.). Data dependent analysis was carried out where automatic MS/MS was acquired on the 8 most intense, multiply charged precursor ions in the m/z range 400–1500. MS/MS data were acquired over the m/z range 50–1995. LC/MS/MS data were processed using ProteinLynx Global Server 2.2.5 (Waters, U.K.) to generate pkl files. The processed data was then searched against a concatenated, non-redundant protein database (UniProt 13.6) using the Mascot search engine programme V2.2 (Matrix Science, U.K.). Oxidation (Met), phosphorylation (STY) and carbamidomethylation (Cys) were included as variable modifications and a maximum of one missed tryptic cleavage was allowed. Precursor and fragment ion tolerances were set to 0.25 Da and 0.1 Da respectively.
Biochemical fractionation
U2OS cells were harvested by scraping and washed in cold phosphate-buffered saline (PBS). After centrifugation, the cell pellets were resuspended in extraction buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, protease and phoshatase inhibitors) supplemented with 0.1% Triton-X-100, and incubated on ice for 10 min. The cell lysates were centrifuged at maximum speed for 5 min at 4°C and the pellets subjected to a second extraction step in extraction buffer adjusted to 400 mM NaCl. The 150 mM and 400 mM supernatants and the final pellets were boiled in SDS-PAGE loading buffer (0.8% SDS, 4% glycerol, 280 mM β-mercaptoethanol, 25 mM Tris-Hcl pH 6.8, 0.005% bromophenol blue) before analysis by immunoblotting.
Laser micro-irradiation and live-cell imaging analysis
For generation of localized damage in cellular DNA by exposure to a UV-A laser beam (11, 12), cells were plated on glass-bottom dishes (Willco Wells) and transfected using Lipofectamine 2000 (Invitrogen) 48 h prior to laser-damage. Cells were sensitized with 10 μM 5-bromo-2′-deoxyuridine (BrdU, Sigma Aldrich) in phenol red-free medium (Invitrogen) for 24 h at 37°C. Laser micro-irradiation was carried out on a FluoView 1000 confocal microscope (Olympus) equipped with a 37°C heating stage (Ibidi) and a 405 nm laser diode (6 mW, SIM scanner) focused through a 60x UPlanSApo/1.35 oil objective to yield a spot size of 0.5-1 μm. Time of cell exposure to the laser beam was around 250 ms (fast scanning mode). Laser settings (0.40 mW output, 50 scans) were chosen as to generate a detectable damage response restricted to the laser path in a presensitization-dependent manner without noticeable cytotoxicity. To ensure that cells with similar expression levels are assayed, cells showing moderate levels of expression were systematically chosen using identical 488 nm laser settings.
The association and dissociation kinetics of YFP-tagged Alc1 at sites of laser micro-irradiation were monitored on the same microscope by measuring YFP fluorescence over time in the damaged region using the 488-nm argon laser. To correct for overall bleaching of the signal due to repetitive imaging, fluorescence intensities were normalized against intensities measured in an undamaged nucleus in the same field after background substraction.
Variations in fluorescence intensity (I) were plotted as a function of time (t) using Microsoft Excel software. Plotted data are averaged values of a minimum of 10 cells from at least two independent experiments. To compare between different experimental conditions, data were normalized against the fluorescence intensity at the time of micro-irradiation (10).
For mathematical modeling of protein mobility, the association kinetics was determined from (It-10)/(Imax-10) plotted as a function of time, and the dissociation kinetics from (It-Imin)/(Imax-Imin) plotted as a function of time, where Imax and Imin are the maximum and mimimum values measured, respectively. Curve fitting and determination of the time constant (τ) for each kinetics was carried out with Prism4 sofware, based on a first order exponential model using the following equations: f(t)=1-exp(-t/τ) and f(t)=exp(-t/τ) for association and dissociation kinetics, respectively.
Sensitivity to DNA damage monitored by MTS
shControl and shALC1 cells were seeded into 96-well plates (3000 cells/well), treated with indicated concentrations of DNA damaging agents and grown for 3 days. CellTiter 96 AQueous One Solution reagent (Promega) was then added and incubated with cells for approximately 2 h. Proliferation was determined by measuring OD at 490 nm.
Quantitative H2AX activation
Flp-In FLAG, ALC, C1 and KR cell lines were untreated or treated with 0.5 mg/ml phleomycin for 1 h. Cells were then trypsinised, washed in PBS and processed by using H2A.X Phosphorylation Assay Kit (Flow Cytometry) (Millipore) according to specifications. Briefly, cells were fixed for 20 min, washed in PBS, permeabilized and incubated with FITC-conjugated γH2AX antibody for 20 min. The samples are then scanned in a flow cytometer to quantitate the number of cells staining positive for phosphorylated histone H2AX.
Comet assay
Flp-In FLAG and ALC cells were trypsinised and treated with indicated concentrations of phleomycin. CometAssay Single Cell Gel Electrophoresis kit (R&D systems) was used to prepare samples as specified by the manufacturer. Cells (approximately 1 × 105/ml) were combined with molten LMAgarose (at 37 °C) at a ratio of 1:10 (v/v). 75 μl of the mixture was immediately pipetted onto a CometSlide. The slides were left at 4 °C for 10 min and then immersed in lysis solution for 30 min. They were subsequently incubated in alkaline solution for 20 min and subjected to electrophoresis at 300 mA for 20 min. Following electrophoresis, slides were washed in 70% ethanol and left to dry overnight. Samples were stained by SYBR Green I and scored by image analysis software (Comet IV, Perceptive Instruments).
Supplementary Material
Acknowledgements
Research in the SJB, SPJ, SCW, IA and MS labs is supported by Cancer Research UK. The lab of TO-H is funded by a Wellcome Trust Senior Fellowship 064414. SPJ is also funded by BBSRC, MRC, The Wellcome Trust and the European Union (EU Projects GENICA and DNA Repair). DA is supported by EMBO. SEP is supported by Human Frontier Science Program Organization. SCW and IA are also funded by the Louis-Jeantet Foundation. The authors declare no conflict of interest.
References
- 1.Li B, Carey M, Workman JL. Cell. 2007 Feb 23;128:707. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Osley MA, Tsukuda T, Nickoloff JA. Mutat Res. 2007 May 1;618:65. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Downs JA, Nussenzweig MC, Nussenzweig A. Nature. 2007 Jun 21;447:951. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 4.Malanga M, Althaus FR. Biochem Cell Biol. 2005 Jun;83:354. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 5.Hassa PO, Haenni SS, Elser M, Hottiger MO. Microbiol Mol Biol Rev. 2006 Sep;70:789. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindahl T, Satoh MS, Dianov G. Philos Trans R Soc Lond B Biol Sci. 1995 Jan 30;347:57. doi: 10.1098/rstb.1995.0009. [DOI] [PubMed] [Google Scholar]
- 7.Satoh MS, Lindahl T. Nature. 1992 Mar 26;356:356. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 8.de Murcia G, et al. J Biol Chem. 1986 May 25;261:7011. [PubMed] [Google Scholar]
- 9.Leduc Y, de Murcia G, Lamarre D, Poirier GG. Biochim Biophys Acta. 1986 Mar 14;885:248. doi: 10.1016/0167-4889(86)90239-9. [DOI] [PubMed] [Google Scholar]
- 10.Frechette A, et al. Can J Biochem Cell Biol. 1985 Jul;63:764. doi: 10.1139/o85-096. [DOI] [PubMed] [Google Scholar]
- 11.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Proc Natl Acad Sci U S A. 1982 Jun;79:3423. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Realini CA, Althaus FR. J Biol Chem. 1992 Sep 15;267:18858. [PubMed] [Google Scholar]
- 13.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Biochem J. 1999 Sep 1;342(Pt 2):249. [PMC free article] [PubMed] [Google Scholar]
- 14.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. Nucleic Acids Res. 2003 Oct 1;31:5526. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Nucleic Acids Res. 2006;34:2887. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karras GI, et al. Embo J. 2005 Jun 1;24:1911. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Nat Struct Mol Biol. 2005 Jul;12:624. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 18.Ahel I, et al. Nature. 2008 Jan 3;451:81. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira H, Flaus A, Owen-Hughes T. J Mol Biol. 2007 Nov 30;374:563. doi: 10.1016/j.jmb.2007.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamiche A, Kang JG, Dennis C, Xiao H, Wu C. Proc Natl Acad Sci U S A. 2001 Dec 4;98:14316. doi: 10.1073/pnas.251421398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gagne JP, et al. Nucleic Acids Res. 2008 Dec;36:6959. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma NF, et al. Hepatology. 2008 Feb;47:503. doi: 10.1002/hep.22072. [DOI] [PubMed] [Google Scholar]
- 23.Kuo MT. Cancer Res. 1981 Jun;41:2439. [PubMed] [Google Scholar]
- 24.Kouzarides T. Cell. 2007 Feb 23;128:693. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Jenuwein T, Allis CD. Science. 2001 Aug 10;293:1074. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 26.Eberharter A, Becker PB. J Cell Sci. 2004 Aug 1;117:3707. doi: 10.1242/jcs.01175. [DOI] [PubMed] [Google Scholar]
- 27.Gangaraju VK, Bartholomew B. Mutat Res. 2007 May 1;618:3. doi: 10.1016/j.mrfmmm.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohrmann L, Verrijzer CP. Biochim Biophys Acta. 2005 Jan 11;1681:59. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Horn PJ, Peterson CL. Front Biosci. 2001 Aug 1;6:D1019. doi: 10.2741/horn. [DOI] [PubMed] [Google Scholar]
- 30.Marfella CG, Imbalzano AN. Mutat Res. 2007 May 1;618:30. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marmorstein R, Berger SL. Gene. 2001 Jul 11;272:1. doi: 10.1016/s0378-1119(01)00519-4. [DOI] [PubMed] [Google Scholar]
- 32.Pal S, Sif S. J Cell Physiol. 2007 Nov;213:306. doi: 10.1002/jcp.21180. [DOI] [PubMed] [Google Scholar]
- 33.Eberharter A, Ferreira R, Becker P. Biol Chem. 2005 Aug;386:745. doi: 10.1515/BC.2005.087. [DOI] [PubMed] [Google Scholar]
- 34.Sala A, et al. PLoS Biol. 2008 Oct 14;6:e252. doi: 10.1371/journal.pbio.0060252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tjeertes JV, Miller KM, Jackson SP. EMBO J. 2009 Apr 30; doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.