Abstract
Dimeric intercellular adhesion molecule-1 (ICAM-1) binds more efficiently to lymphocyte function-associated antigen-1 (LFA-1) than monomeric ICAM-1. However, it is unknown whether dimerization enhances binding simply by providing two ligand-binding sites and thereby increasing avidity, or whether it serves to generate a single “fully competent” LFA-1-binding surface. Domain 1 of ICAM-1 contains both the binding site for LFA-1, centered on residue E34, and a homodimerization interface. Whether the LFA-1-binding site extends across the homodimerization interface has not been tested. To address this question, we constructed four different heterodimeric soluble forms of ICAM-1 joined at the C terminus via an α-helical coiled coil (ACID-BASE). These heterodimeric ICAM-1 constructs include, (i) E34/E34 (two intact LFA-1-binding sites), (ii) E34/K34 (one disrupted LFA-1-binding site), (iii) E34/ΔD1–2 (one deleted LFA-1-binding site), and (iv) K34/K34 (two disrupted LFA-1-binding sites). Cells bearing activated LFA-1 bound similarly to surfaces coated with either E34/K34 or E34/ΔD1–2 and with an ≈2-fold reduction in efficiency compared with E34/E34, suggesting that D1 dimerization, which is precluded in E34/ΔD1-D2, is not necessary for optimal LFA-1 binding. Furthermore, BIAcore (BIAcore, Piscataway, NJ) affinity measurements revealed that soluble open LFA-1 I domain bound to immobilized soluble ICAM-1, E34/E34, E34/K34, and E34/ΔD1-D2 with nearly identical affinities. These studies demonstrate that a single ICAM-1 monomer, not dimeric ICAM-1, represents the complete, “fully competent” LFA-1-binding surface.
Intercellular adhesion molecule-1 (ICAM-1) is a cell-surface glycoprotein with five extracellular Ig-like domains (domains 1–5, D1–5), a hydrophobic transmembrane domain, and a short cytoplasmic domain. ICAM-1 is an inducible ligand for at least two members of the β2 family of leukocyte integrins, lymphocyte function-associated antigen-1 (LFA-1) (αLβ2) and Mac-1 (αMβ2) (1–3), and is important for granulocyte extravasation (4–8), lymphocyte-mediated cytotoxicity (9, 10), and the development of specific immunologic responses involving cell–cell interactions (6, 7, 11, 12). Antibodies to ICAM-1 inhibit leukocyte adhesion to endothelial cells, granulocyte migration through endothelium, mitogen- and Ag-induced lymphocyte proliferation, and mixed lymphocyte reactions (4, 5, 13, 14). Furthermore, crosslinking of ICAM-1 activates signaling pathways in monocytes and endothelial cells (15, 16). Given the potential clinical importance of ICAM-1/LFA-1-mediated adhesion (17), it is important to understand this receptor–ligand interaction at a fundamental level.
Integrins are large heterodimeric membrane glycoproteins composed of combinations of various α and β subunits, the N-terminal regions of which possess ligand-binding sites. The N terminus of all integrin α subunits is composed of seven 60-aa repeats predicted to fold into a β-propeller structure (18). In some integrins, including the four β2 integrins, a structurally characterized inserted (I) domain (19) of ≈200 residues exists between repeats 2 and 3, positioned on top of the β-propeller (18). In LFA-1 (αLβ2), the I domain has been shown to directly mediate conformation- and cation-dependent ICAM-1-binding through its metal ion-dependent adhesion site (20–23).
ICAM-1 has been shown to exist as a dimer and larger multimers on the cell surface, and dimerization appears to enhance binding to LFA-1 (24, 25). Indeed, LFA-1-expressing lymphoblasts bind to cells expressing wild-type ICAM-1, which is largely dimeric, more efficiently than cells expressing an equal amount of glycosylphosphatidylinositol-linked ICAM-1, which is largely monomeric (24). Recombinant soluble ICAM-1 (sICAM-1), lacking the transmembrane and cytoplasmic domains, exists as a monomer in solution (26, 27). Engineered dimerization of sICAM-1 leads to significantly enhanced binding to LFA-1 compared with monomeric sICAM-1 (24, 28, 29).
The overall topology of the ICAM-1 dimer on the cell surface has not yet been fully defined. However, data suggest that D5 and/or the transmembrane domain are important for dimerization (24, 25). Additionally, an x-ray crystal structure of ICAM-1 domains 1–2 revealed a hydrophobic dimerization interface in domain 1 on the β-sheet containing β-strands B, E, and D, suggesting this domain may also mediate dimerization (30). Indeed, we have recently provided experimental evidence confirming that such a domain 1 dimerization interface exists in solution (C.-D.J., C.V.C., S. D. Redick, and M.S., unpublished results). Significantly, the ligand-binding surface for LFA-1 in the dimeric ICAM-1 crystal structure was found on the face of domain 1 opposite the dimerization interface (30). The ligand-binding site is centered on Glu-34 (E34), which is the single most important residue for ligand binding and is hypothesized to ligate the Mg2+ in the metal ion-dependent adhesion site of the I domain. E34 is located near the middle of the domain in β-strand C, on the edge of the β-sandwich. In the domain 1 dimer, the two E34 residues point away from one another and are separated by ≈42 Å (30). With this geometry, simultaneous binding of two LFA-1 molecules seemed plausible. However, it would also be possible for a single molecule of LFA-1 to bind across the dimer interface and contact residues in both monomers. For example, E34 in one monomer is only 20 Å away from residue L43 in the other monomer.
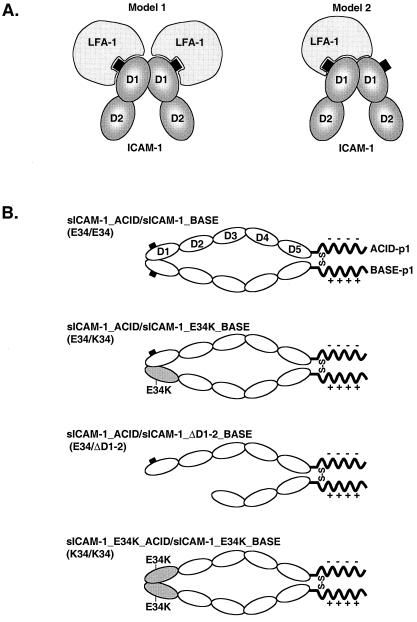
Given the proximity of the dimerization interface and the LFA-1-binding interface in domain 1 of ICAM-1, we propose two basic models for the greater LFA-1 binding observed to dimeric ICAM-1 (Fig. 1A). In model 1, all of the ICAM-1 contacts of LFA-1 occur within a single monomer, centered around E34. In this model, the increased binding of the dimer is derived from increased avidity, which by definition includes binding of two LFA-1 molecules to a single ICAM-1 dimer. In model 2, LFA-1 binds to the ICAM-1 dimer through both contacts centered around E34 of one monomer and contacts across the dimer interface to the second monomer (Fig. 1A, model 2). In this model, the dimer represents a complete or “fully competent” LFA-1-binding surface, whereas the monomer contains only a partial LFA-1-binding surface.
Figure 1.
Binding models and ICAM-1 constructs. (A) Models of LFA-1 binding to ICAM-1 dimers. In model 1, all contacts with LFA-1 occur within domain 1 of a single monomer, centered around E34 (black square). In model 2, contacts occur around E34 in domain 1 of one monomer and extend across the domain interface to include contacts with domain 1 of the second monomer. Formally, this model does not preclude binding of two LFA-1 molecules to an ICAM-1 dimer. (B) Schematic representation of ICAM-1 ACID/BASE heterodimers. Diagram shows intact or mutated ectodomains (D1-D5; ovals) of ICAM-1 fused at the C terminus to either ACID-p1 (−) or BASE-p1 (+) peptides. The leucines in the “d” position in the first heptad repeat of ACID-p1 and BASE-p1 peptides were mutated to cysteine to introduce an intersubunit disulfide bond (S-S) on dimerization.
To differentiate between these mechanisms, we have created sICAM-1 constructs C-terminally fused to ACID and BASE α-helical coiled-coil peptides to drive the formation of specific ICAM-1 ACID–BASE coiled-coil heterodimers (Fig. 1B). We have designed ICAM-1 heterodimers in which one of the LFA-1-binding sites was disrupted by mutation of E34 or completely deleted. Functional tests on these heterodimers clearly demonstrate that a monomer of ICAM-1 bears a complete set of LFA-1-binding determinants.
Methods
Cells and Antibodies.
JY (Epstein–Barr virus-transfected B-cell), SKW3 (T cell lymphoma), 293T, and Chinese hamster ovary (CHO)-K1 cell lines were maintained as previously described (31–33). CHO-K1 cells were maintained in Ham's F12K medium supplemented with 2 mM l-glutamine, 10% FBS, and 50 μg/m penicillin/streptomycin. Peripheral blood T cells were prepared as described (34). ICAM-1 mAbs R6.5 (35), CA-7 (27), CL203 (36), and CBRIC1/11 (37), and the LFA-1 blocking mAb TS1/22 (38) have been described. Within ICAM-1, mAb R6.5 maps to domain 2 (26), mAb CBRIC1/11 maps to domain 3 (37), mAb CL203 maps to domain 4 (26), and mAb CA-7 maps to domain 5 (27). mAb 2H11 was a generous gift from Ellis L. Reinherz (Dana–Faber Institute, Boston) (39).
cDNA Constructions.
The human wild-type ICAM-1 cDNA (40) was subcloned into the HindIII and NotI restriction sites of pAprM8 vector to generate ICAM-1/pAprM8. A DNA construct encoding IgSF domains 1–5 of ICAM-1 fused to the ACID-p1 peptide (sICAM-1_ACID) was prepared by using a three-round PCR protocol. In the first PCR reaction, by using ICAM-1/pAprM8 as a template, an ≈260-bp fragment was generated that spanned from an internal ICAM-1 BglII site through codons for the last amino acids of the ICAM-1 ectodomain (SPRYE) fused with the first amino acids (AQCEKELQALEKENAQLE) of the ACID-p1 sequence. Also, by using the ICAM-1/pAprM8 as a template, an ≈500-bp fragment was generated, beginning with a short sequence encoding the last C-terminal amino acids (KENAQLEWELQALEKELAQ) of the ACID-p1 sequence followed by a stop codon, ≈470 bp of nontranslated sequence, and a NotI site. In the final PCR reaction, the 260- and ≈500-bp products were used together as an overlapping template to generate an ≈760-bp product. After digestion with BglII and NotI, this product was used to replace the respective wild-type sequence in ICAM-1/pAprM8, generating sICAM-1_ACID/pAprM8. By using a similar cloning strategy, a DNA construct encoding the ICAM-1 ectodomain fused to the α-helical BASE-p1 peptide (sICAM-1_BASE) was prepared. The final amino acid sequences of the ACID-p1 and BASE-p1 peptides in sICAM-1_ACID and sICAM-1_BASE, respectively, were as follows: ACID-p1:
AQCEKELQALEKENAQLEWELQALEKELAQ; BASE-p1:
AQCKKKLQALKKKNAQLKWKLQALKKKLAQ (41).
By using sICAM-1_ACID/pAprM8 and sICAM-1_BASE/pAprM8 as templates, site-directed mutagenesis was performed to mutate E34 to lysine(K) resulting in sICAM-1_E34K_ACID/pAprM8 and sICAM-1_E34K_BASE/pAprM8. For the plasmid encoding only domains 3–5 of ICAM-1 fused to the BASE-p1 peptide (sICAM-1_ΔD1–2_BASE/pAprM8), domains 1 and 2 of ICAM-1 (184 residues) were deleted by using a long (45-bp) mutant oligonucleotide such that codons for the end of the signal sequence and F185 would be joined. Wild-type and mutant sICAM-1_BASE chimeras were further subcloned into the BamHI and NotI sites of pEF1/V5_puro vector, whereas wild-type and mutant sICAM-1_ACID constructs were subcloned into the SpeI and NotI sites of pEF1/V5_neo vector (Invitrogen).
cDNA Transfections.
Transient transfection of 293T cells was performed as described (33). CHO-K1 cells that stably express sICAM-1_ACID/BASE heterodimers were generated by fugene 6 (Boehringer Mannheim) transfection of sICAM-1_ACID and BASE constructs followed by selection with, and maintenance in, 3 μg/ml of puromycin and 1 μg/ml neomycin beginning at 48 h.
Radiolabeling and Immunoprecipitation.
Metabolic labeling and immunoprecipitation have been described (42). Briefly 5 × 106 293T cells in 4 ml of labeling medium (cysteine/methionine-free RPMI medium containing 15% dialyzed FBS) were labeled with 0.5 mCi of [35S]cysteine-methionine (ICN) overnight at 37°C. Labeled cell culture supernantants (500 μl) were incubated with R6.5-, 2H11-, or CA-7-Sepharose beads (50 μl of a 1:1 slurry coupled at 3 mg/ml) for 3 h at 4°C. The immunoprecipitates were subjected to SDS/10% PAGE and fluorography.
Protein Purification.
ICAM-1 heterodimers were purified at 4°C. Culture supernatants containing sICAM-1_ACID and BASE proteins were passed through a 2H11 mAb affinity column (20 ml coupled at 2 mg/ml), followed by extensive washing with 10 mM Tris·HCl, pH 8.0/0.15 M NaCl. Bound proteins were eluted with 50 mM triethylamine, pH 11.5/0.15 M NaCl and fractions were collected in test tubes containing 1/10 volume 1 M Tris·HCl, pH 6.5. Pooled fractions were then subjected to Superdex 200 size exclusion chromatography in PBS (0.15 M NaCl/2.7 mM KCl/1.47 mM KH2PO4/4.86 mM Na2HPO4, pH 7.4) to remove aggregated materials. Monomeric sICAM-1 lacking the transmembrane and cytoplasmic domains was purchased from Boehringer Mannheim. Fc-ICAM-1, Fc-ICAM-2, and Fc-ICAM-3 chimeric proteins were purchased from R & D Systems.
Cell-Binding Assay.
2H11 mAb (50 μl of 20 μg/ml in PBS) was adsorbed to each well of flat-bottom 96-well polystyrene plates (Flow Laboratories) by incubation overnight at 4°C. Nonspecific binding sites were blocked with 1% heat-treated BSA for 1 h at 37°C. Purified ICAM-1 heterodimers (50 μl of a range of concentrations from 1.25 to 320 nM) were then incubated for 3 h at 37°C to allow binding to immobilized 2H11 mAb. The ICAM-1 density was determined by saturation binding with [125I]-CBRIC1/11 mAb, as described (31). BCECF-AM-labeled cells (43) were resuspended (1 × 106cells/ml) in L15 medium supplemented with 5% FBS (L15/FBS). Fifty microliters of cell suspension was added to ICAM-1-coated wells with an equal volume of L15/FBS containing phorbol 12-myristate 13-acetate (PMA) (100 ng/ml). In some experiments, L15/FBS and PMA were substituted with Hepes-buffered saline (20 mM Hepes, pH 7.5/140 mM NaCl) supplemented with 2 mg/ml of glucose and indicated cations or cation chelators. The 96-well plates were centrifuged at 200 × g for 2 min at 4°C and incubated at 37°C for 30 min. Unbound cells were then removed by using a Microplate Autowasher (Bio-Tek Instruments, Winooski, VT). The fluorescence signal of bound cells (after washing) was expressed as a percentage of the fluorescence of total input cells (before washing) as quantitated on a fluorescent concentration analyzer (IDEXX, Westbrook, ME). The washing procedure was programmed (43) such that binding of mock-transfected cells or binding in the presence of ICAM-1-blocking mAb was below 5% of total input.
Antibody-Binding Assay.
mAbs R6.5, CA-7, or 2H11 (50 μl of 10 μg/ml PBS) were adsorbed to each well of flat-bottom 96-well polystyrene plates (Flow Laboratories) by incubation overnight at 4°C. Nonspecific binding sites were blocked with 1% heat-treated BSA for 1 h at 37°C. sICAM-1 or ICAM-1 heterodimers (E34/E34 and E34/ΔD1-D2) (500 ng/ml in PBS) were then added to the wells and incubated for 30 min at 37°C followed by washing 3 × with PBS. Binding of ICAM-1 was detected by incubation with biotin-conjugated CBRIC1/11 mAb followed by washing with PBS and addition of streptavidin-conjugated horseradish peroxidase and 2,2′-azinobis[3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt as substrate.
Binding of Open LFA-1 I Domain to Monomeric and Heterodimeric ICAM-1 by Surface Plasmon Resonance.
The binding of a designed mutant (K287C/K294C) of the LFA-1 I domain, which is locked in the high-affinity open conformation (open LFA-1 I domain) to ICAM-1 proteins, was monitored with a BIAcore 1000 instrument (BIAcore, Piscataway, NJ), as previously described (23). Briefly, sICAM-1, heterodimeric ICAM-1 or BSA (control) were covalently immobilized onto sensor chips (ligands), and open LFA-1 I domain (analyte) was flowed over the sensor chips. kon and koff values were obtained by curve fitting of the association and dissociation phases of sensorgrams, respectively, with a 1:1 binding model by using BIAevaluation software (BIAcore). KD was then calculated from kon and koff (KD = koff/kon).
Results
Expression of ICAM-1 ACID/BASE Heterodimers.
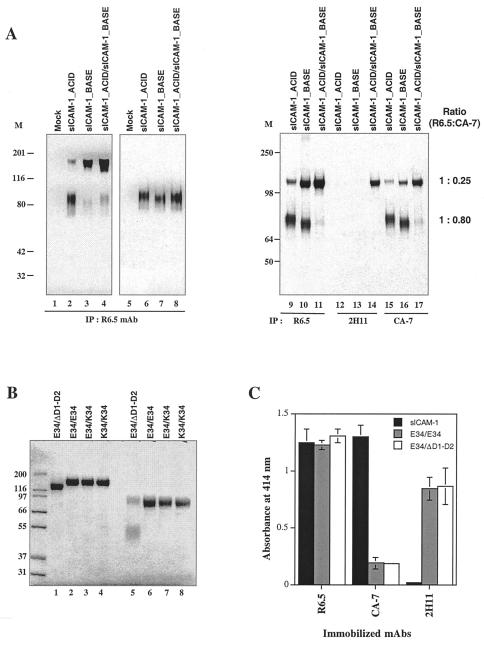
To produce soluble ICAM-1 heterodimers, the transmembrane and cytoplasmic domains of ICAM-1 were replaced with peptides termed “ACID” and “BASE” that form α-helical coiled coils, with a strong preference for ACID-BASE heterodimers (39, 41) (Fig. 1B). Cysteines were introduced in the “d” position of the first heptad repeat to covalently link the coiled coils (44). Proteins from metabolically labeled 293T cells expressing sICAM-1_ACID, sICAM-1_BASE, or sICAM-1_ACID and sICAM-1_BASE together were immunoprecipitated with the ICAM-1-specific mAb R6.5 and subjected to SDS/PAGE and fluorography (Fig. 2A). Disulfide-linked dimers were formed, as shown by SDS/PAGE under nonreducing conditions (Fig. 2A, lanes 2–4 compared with 6–8). Cells doubly transfected with sICAM-1_ACID and sICAM-1_BASE formed dimers, as expected (Fig. 2A, lanes 4 and 11). Dimers were also formed by cells transfected with sICAM-1_ACID alone and sICAM-1_BASE alone (Fig. 2A, lanes 2, 3, 9, and 10); however, the ratio of dimers to monomers was markedly higher for cells coexpressing sICAM-1_ACID and sICAM_BASE (Fig. 2A, lanes 4 and 11). The mAb 2H11 specifically recognizes the heterodimeric, coiled-coil state of the ACID-p1 and BASE-p1 peptides (39). Immunoprecipitation with 2H11 mAb demonstrated that ACID/BASE α-helical coiled-coil heterodimers were, indeed, formed by coexpression of the sICAM-1_ACID and sICAM-1_BASE chimeras (Fig. 2A, lane 14). Furthermore, mAb 2H11 did not immunoprecipitate material from cells expressing sICAM-1_ACID or sICAM-1_BASE alone (Fig. 2A, lanes 12 and 13), demonstrating that ACID/BASE heterodimers could be isolated from ACID and BASE homodimers.
Figure 2.
Expression and characterization of ICAM-1_ACID and BASE chimeras. (A) Secreted material from metabolically labeled 293T cells transfected with the indicated constructs was immunoprecipitated with the indicated mAbs. Samples were subjected to SDS/10% PAGE in the absence (lanes 1–4 and 9–17) and presence (lanes 5–8) of 10 mM DTT. 125I-labeled ICAM-1 bands were quantified by using a Storm 660 PhosphorImager (Molecular Dynamics). (B) Material from CHO-K1 cells expressing E34/E34, E34/K34, E34/ΔD1-D2, or K34/K34 ICAM-1 heterodimers was purified with a 2H11 mAb column, subjected to SDS/10% PAGE in the absence (lanes 1–4) and presence (lanes 5–8) of 10 mM DTT, and stained with Coomassie blue. (C) Purified ICAM-1 preparations (500 ng/ml) were tested for binding to immobilized R6.5, CA7, or 2H11 mAb. Bound ICAM-1 was detected with biotin-conjugated-CBRIC1/11 mAb followed by streptavidin-conjugated horseradish peroxidase and ELISA.
Stable transfectants of CHO-K1 cells were established that expressed sICAM-1_ACID/sICAM-1_BASE (E34/E34), sICAM-1_ACID/sICAM-1_E34K_BASE (E34/K34), sICAM-1_ ACID/sICAM-1_ΔD1–2_BASE (E34/ΔD1–2), and sICAM-1_E34K_ACID/sICAM-1_E34K_BASE (K34/K34) (Fig. 1B). Heterodimers were specifically isolated by immunoaffinity chromatography with the 2H11 mAb specific for ACID/BASE α-helical coiled-coil heterodimers. E34/E34, E34/K34 and K34/K34 all migrated as dimers of ≈160 kDa on nonreducing SDS/PAGE (Fig. 2B, lanes 2–4) and as monomers of 80 kDa on reducing SDS/PAGE (Fig. 2B, lanes 6–8). E34/ΔD1–2 ran as a single band of 135 kDa under nonreducing SDS/PAGE (Fig. 2B, lane 1), and as two bands of 80 and 55 kDa on reduction (Fig. 2B, lane 5). As expected after isolation with 2H11 mAb affinity chromatography, homodimer bands of 160 and 110 kDa were absent from the E34/ΔD1–2 preparation (Fig. 2B, lane 1).
The mAb CA-7 has previously been shown to recognize D5 of ICAM-1 (27) in monomeric ICAM-1, while recognizing native cell surface-expressed dimeric ICAM-1 relatively poorly (24). Thus, to confirm appropriate formation of ICAM-1 heterodimers, we assessed binding of mAbs CA-7, R6.5, and 2H11 to immobilized ICAM-1. R6.5 to domain 2 of ICAM-1 recognized sICAM-1, E34/E34 and E34/ΔD1–2 proteins similarly, and 2H11 bound E34/E34 and E34/ΔD1–2 similarly, but failed to bind sICAM-1 (Fig. 2C). By contrast, CA-7 bound monomeric sICAM-1 efficiently but bound heterodimeric ICAM-1 (E34/E34 and E34/ΔD1–2) relatively weakly (Fig. 2C). Thus, the soluble ICAM-1 heterodimers resembled cell surface ICAM-1 in masking of the CA-7 epitope. We further compared immunoprecipitation of monomeric and dimeric forms of ICAM-1 by CA-7 mAb (Fig. 2A, lanes 15–17). Compared with R6.5 mAb (Fig. 2A, lanes 9–11), CA-7 mAb precipitated similar amounts of monomer (80% as much as R6.5 mAb), but markedly lesser amounts of dimer (25% as much as R6.5 mAb). Therefore, the differential CA-7 binding to dimers likely reflects proper dimer formation, as opposed to masking of the CA-7 epitope by ACID and BASE peptides.
Binding of LFA-1-Bearing Cells to Immobilized ICAM-1 Heterodimers.
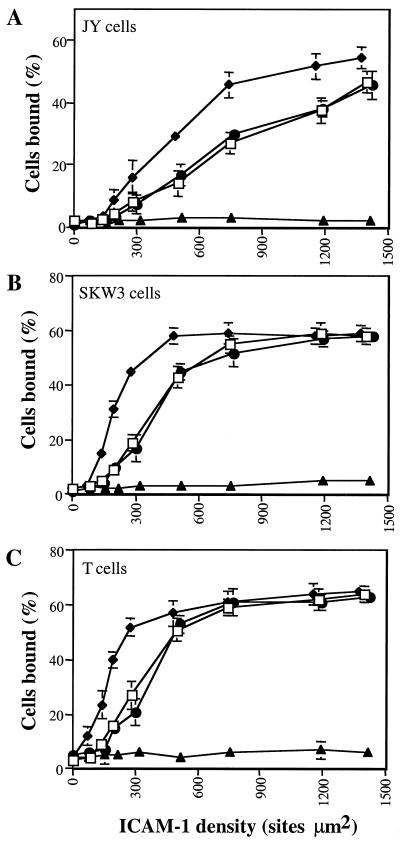
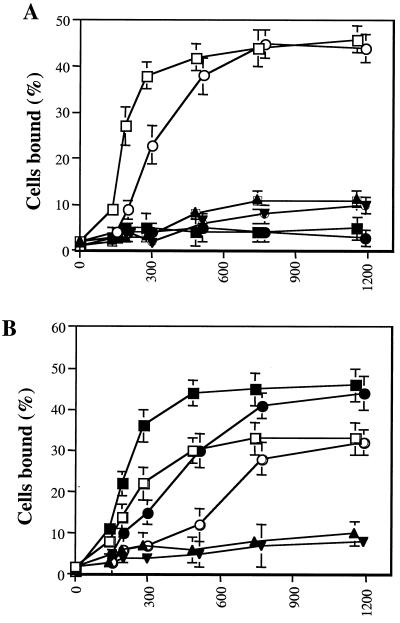
To assess function, ICAM-1 heterodimers were immobilized on 2H11-coated 96-well plates and assessed for their ability to support PMA-stimulated adhesion of JY B lymphoblastoid cells, SKW3 T lymphoma cells, and resting peripheral blood T cells. A range of heterodimer concentrations was used, and the density of captured ICAM-1 was determined by saturation binding with 125I-CBRIC1/11 mAb. Binding of all three cell types depended on LFA-1 and ICAM-1, as shown by ≥97% inhibition with mAbs TS1/22 and RR1/1, respectively. The E34/E34, E34/K34, and E34/ΔD1–2 dimers all supported binding that depended on their density on the substrate (Fig. 3). By contrast, the K34/K34 dimer was completely inactive for all three cell types. The ability of the E34/K34 and E34/ΔD1-D2 dimers to support LFA-1-dependent adhesion was reduced compared with the E34/E34 dimer (Fig. 3). Maximal binding was similar; however, a 2-fold higher density of dimer was required for half-maximal binding to E34/K34 and E34/ΔD1-D2 compared with E34/E34. Similar results were obtained when SKW3 cell binding was stimulated by either Mn2+ (Fig. 4A) or Mg2+/EGTA (Fig. 4B) rather than PMA, or when binding was in the presence of Mg2+ (Fig. 4B). In all cases, a 2-fold higher density of E34/K34 than E34/E34 was required to support the same amount of adhesion. PMA and Mn2+ or Mg2+/EGTA have been reported to stimulate LFA-1-dependent binding to ICAM-1 by avidity and affinity modulation, respectively (45). Because binding to E34/K34 and E34/ΔD1-D2 is equivalent, and because dimerization of domain 1 is precluded in E34/ΔD1-D2, these data demonstrate that an individual monomer of ICAM-1 possesses all of the necessary LFA-1-binding determinants.
Figure 3.
PMA-stimulated binding of LFA-1-bearing cells to ICAM-1 heterodimers. ICAM-1 heterodimers (□, E34/ΔD1-D2; ⧫, E34/E34; ●, E34/K34; ▴, K34/K34) were bound at different concentrations to 2H11 mAb absorbed to plastic wells, as described in Materials and Methods. Binding of JY, SKW3, and resting T cells was performed in L15/FBS at 37°C for 30 min in the presence of 100 ng/ml of PMA. Binding of LFA-1-bearing cells was inhibited almost completely by LFA-1- or ICAM-1-blocking mAbs TS1/22 or RR1/1, respectively (≥97% inhibition). Results are expressed as mean ± SD of triplicate samples and are representative of three independent experiments.
Figure 4.
Metal ion-stimulated binding of LFA-1-bearing cells to ICAM-1 heterodimers. ICAM-1 heterodimers (E34/E34 and E34/K34) were bound at different concentrations to 2H11 mAb absorbed to plastic wells, as described in Materials and Methods. Binding of SKW3 cells was performed in Hepes-buffered saline supplemented with glucose (2 mg/ml) at 37°C for 30 min in the presence of various cations or cation chelators. (A) Binding of SKW3 cells was performed in the presence of either 2 mM Mn2+ (□, E34/E34; ○, E34/K34), 1 mM Ca2+ (■, E34/E34; ●, E34/K34), or no divalent cations (▴, E34/E34; ▾, E34/K34). (B) Binding was performed in the presence of 2 mM Mg2+ (□, E34/E34; ○, E34/K34), 2 mM Mg2+ + 1 mM EGTA (●, E34/E34; ●, E34/K34), or 2 mM Mg2+ + 10 mM EDTA (▴, E34/E34; ▾, E34/K34). Results are expressed as mean ± SD of triplicate samples and are representative of two independent experiments.
Interaction of ICAM-1 Heterodimers with the Open LFA-1 I Domain in BIAcore.
To more directly characterize the ligand-binding properties of the ICAM-1 heterodimers, we performed BIAcore studies in which ICAM-1 proteins were immobilized on the surface of a BIAcore sensor chip. Binding of a purified soluble I domain mutant that is locked in the open conformation by introduction of two cysteines that form a conformation-selective disulfide bond (22, 23) was measured. In initial experiments, specificity was demonstrated in that the open LFA-1 I domain exhibited cation-dependent binding to chips with immobilized sICAM-1 but not to chips with immobilized BSA (data not shown). Kinetics were measured for binding of the open LFA-1 I domain to monomeric sICAM-1 and heterodimeric E34/E34, E34/K34 and E34/ΔD1-D2 ICAM-1 (Table 1). Whereas K34/K34 exhibited very weak binding affinity (KD ≈ 0.5 mM), sICAM-1, E34/E34, E34/K34, and E34/ΔD1-D2 all bound the open LFA-1 I domain with high affinity and strikingly similar kon, koff and KD values (Table 1). The KD values ranged from 167 to 181 nM. These data are consistent with the cell-binding experiments shown in Figs. 3 and 4, and demonstrate that LFA-1 binds to individual ICAM-1 monomers independently of dimerization state.
Table 1.
The kinetics and affinity of the open LFA-1 I domain for monomeric and dimeric ICAM-1
| Immobilized ligand | Analyte | kon (M−1s−1 × 10−4) | koff (s−1 × 103) | KD, nM |
|---|---|---|---|---|
| E34/E34 | open-I | 13.5 ± 0.5 | 23.1 ± 1.4 | 171 ± 4.4 |
| E34/K34 | open-I | 14.4 ± 0.6 | 25.9 ± 1.3 | 181 ± 3.7 |
| E34/ΔD1-D2 | open-I | 14.9 ± 1.4 | 24.6 ± 1.0 | 167 ± 14.4 |
| K34/K34 | open-I | 0.106 ± 0.04 | 503 ± 143 | 514,000 ± 49,100 |
Purified sICAM-1 or sICAM-1 heterodimers were immobilized on a BIAcore sensor chip. The purified soluble locked open LFA-1 I domain was injected and passed over the ICAM-1 surface at a constant flow rate of 10–60 μl/min in TBS containing 1 mM MgCl2. Curve fitting of the association and dissociation phases with BIAevaluation 3.1 software was used to calculate kon, koff, and KD values. All values are expressed as mean ± SEM for three separate experiments.
Discussion
Many previous studies have shown that dimeric ICAM-1 is more effective than momeric ICAM-1 as a ligand for LFA-1 (24, 25, 28, 29). The LFA-1-binding site is located entirely within domain 1 of ICAM-1 (26, 46). Given the finding that ICAM-1 dimerizes through an interface in domain 1 (ref. 30; C.-D.J., C.V.C., S. D. Redick, and M.S., unpublished results), we hypothesized two possible mechanisms for the enhanced LFA-1-binding activity of dimeric ICAM-1 (Fig. 1A). Fundamentally, these models differ in the question of whether LFA-1 recognizes an individual monomer as a complete binding surface or if it requires additional binding determinants, supplied by the second monomer of a dimer, for optimal binding. By creating ICAM-1 heterodimers of defined composition, where one of the two LFA-1-binding sites was either disrupted or completely deleted, we have established that all of the LFA-1-binding determinants exist within domain 1 of a single ICAM-1 monomer. In other words, the binding site does not extend across a dimer interface.
To assay adhesion through LFA-1, equal amounts of ICAM-1 heterodimers were immobilized and binding of LFA-1-bearing cells was assessed. The heterodimers were presented in a uniform manner on the substrate by capture with a mAb to the ACID/BASE α-helical coiled coil. Because the heterodimers were not free in solution but immobilized on a surface, monovalency or bivalency of the dimers per se was irrelevant. Rather the avidity of the LFA-1-bearing cells for the ICAM-1 bound surface, a function of the total number of intact, “fully competent” binding sites present in the cell/substrate contact region, was important. We found that the E34/E34 dimer exhibited ≈2-fold greater cell-binding efficiency than the E34/K34 dimer, an equal amount of which contains only half the number of active LFA-1-binding sites. Significantly, binding of cells to E34/ΔD1–2, in which dimerization of domain 1 is precluded, was not further reduced compared with E34/K34, demonstrating that LFA-1 binding is not enhanced by or dependent on an ability to form a domain 1 dimerization interface. Furthermore, these results demonstrate that dimerization of ICAM-1 does not enhance adhesiveness, because the dimers that contained only one active binding site were as effective as dimers with two active binding sites when the total number of active binding sites was the same. Support for these conclusions was provided by direct measurement of affinity for open LFA-1 I domains. The affinity and binding kinetics of the open LFA-1 I domain for immobilized monomeric sICAM-1, and dimeric E34/E34, E34/K34 and E34/ΔD1–2 were nearly identical (Table 1). This could occur only if all of the LFA-1-binding determinants exist within a single ICAM-1 monomer.
These data suggest that the previously reported higher effectiveness of the soluble ICAM-1 dimer is likely because of its bivalency. It is noteworthy that most previous studies of dimeric ICAM-1 function have been performed with soluble ICAM-1 binding to an LFA-1-bearing surface (24, 25, 28, 29). In solution, the bivalent nature of an ICAM-1 dimer will cause a significant suppression of koff yielding a greater overall affinity. Indeed, we have previously demonstrated this for ICAM-1 dimer binding to rhinovirus (47) and have confirmed it for ICAM-1 dimer binding to the immobilized open LFA-1 I domain (data not shown).
Although this finding is clearly relevant to the design of anti-integrin-based therapies, it raises the question what, then, is the physiological role of ICAM-1 dimerization on the cell surface? In fact, functional advantages of dimerization have been observed with cell-surface ICAM-1 (≈2-fold increased adhesion to dimer compared with monomer), although more modest than those seen for soluble ICAM-1 dimers (10- to 100-fold increased binding of dimer compared with monomer) (24, 25, 28, 29). However, on the cell surface, native dimeric ICAM-1 was compared with monomeric ICAM-1 with an artificial glycosylphosphatidyl inositol membrane anchor. Binding of the CA-7 mAb was greatly enhanced to the glycosylphosphatidyl inositol anchored ICAM-1, suggesting that a putative dimerization interface in domain 5 was unmasked. The ICAM-1 dimers studied here, including those containing only one active LFA-1-binding site per dimer, demonstrated a masking of the CA-7 epitope similar to that seen for cell surface dimers. The epitope was not masked in monomers fused to the α-helical peptides, suggesting that dimerization at domain 5, and not fusion to the α-helical peptide, resulted in masking. Therefore, the orientational influence of dimerization in the C-terminal portion of ICAM-1 is retained in the molecules containing one and two binding sites for LFA-1 in this study, but not in the study of Miller et al. (24). In the present study, we have ruled out an effect of dimerization on the nature of the LFA-1-binding site in domain 1 of ICAM-1. Moreover, we have demonstrated that when present on a surface and at the same density of total active binding sites, dimers that contain a single LFA-1-binding site are as effective as dimers that contain two binding sites. Thus, one likely functional role for ICAM-1 dimerization on the cell surface as demonstrated by Miller et al. (24) is to properly orient ICAM-1 and present it for binding to LFA-1. Putative dimerization sites at the C terminus and in domain 5 appear sufficient for this orienting function, because we observed no difference in efficacy when the domain 1 dimerization site was eliminated in the E34/ΔD1–2 heterodimer. Another possible role for dimerization is in signaling through ICAM-1 (15, 16).
Overall, the observations presented here demonstrate that each individual ICAM-1 monomer is fully competent to bind LFA-1 and that ICAM-1 dimerization, although functionally important for orientation on the cell surface, is not required to form a complete LFA-1-binding site.
Acknowledgments
We thank Michael Dustin and Robert Rothlein for reviewing the manuscript. This work was supported by National Institutes of Health Grant CA31798.
Abbreviations
- ICAM-1
intercellular adhesion molecule-1
- sICAM
soluble ICAM
- LFA-1
lymphocyte function-associated antigen-1
References
- 1.Springer T A. Nature (London) 1990;346:425–433. doi: 10.1038/346425a0. [DOI] [PubMed] [Google Scholar]
- 2.Springer T A. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 3.Dustin M L, Springer T A. In: Guidebook to the Extracellular Matrix and Adhesion Proteins. Kreis T, Vale R, editors. New York: Sambrook and Tooze; 1999. [Google Scholar]
- 4.Smith C W, Rothlein R, Hughes B J, Mariscalco M M, Schmalstieg F C, Anderson D C. J Clin Invest. 1988;82:1746–1756. doi: 10.1172/JCI113788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith C W, Marlin S D, Rothlein R, Toman C, Anderson D C. J Clin Invest. 1989;83:2008–2017. doi: 10.1172/JCI114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu H, Gonzalo J A, St. Pierre Y, Williams I R, Kupper T S, Cotran R S, Springer T A, Gutierrez-Ramos J-C. J Exp Med. 1994;180:95–109. doi: 10.1084/jem.180.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sligh J E, Jr, Ballantyne C M, Rich S, Hawkins H K, Smith C W, Bradley A, Beaudet A L. Proc Natl Acad Sci USA. 1993;90:8529–8533. doi: 10.1073/pnas.90.18.8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond M S, Staunton D E, Marlin S D, Springer T A. Cell. 1991;65:961–971. doi: 10.1016/0092-8674(91)90548-d. [DOI] [PubMed] [Google Scholar]
- 9.Makgoba M W, Sanders M E, Ginther Luce G E, Gugel E A, Dustin M L, Springer T A, Shaw S. Eur J Immunol. 1988;18:637–640. doi: 10.1002/eji.1830180423. [DOI] [PubMed] [Google Scholar]
- 10.Mentzer S J, Rothlein R, Springer T A, Faller D V. J Cell Physiol. 1988;137:173–178. doi: 10.1002/jcp.1041370121. [DOI] [PubMed] [Google Scholar]
- 11.Dougherty G J, Murdoch S, Hogg N. Eur J Immunol. 1988;18:35–39. doi: 10.1002/eji.1830180107. [DOI] [PubMed] [Google Scholar]
- 12.Altmann D M, Hogg N, Trowsdale J, Wilkinson D. Nature (London) 1989;338:512–514. doi: 10.1038/338512a0. [DOI] [PubMed] [Google Scholar]
- 13.Dustin M L, Springer T A. J Cell Biol. 1988;107:321–331. doi: 10.1083/jcb.107.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd A W, Wawryk S O, Burns G F, Fecondo J V. Proc Natl Acad Sci USA. 1988;85:3095–3099. doi: 10.1073/pnas.85.9.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothlein R, Kishimoto T K, Mainolfi E. J Immunol. 1994;152:2488–2495. [PubMed] [Google Scholar]
- 16.Etienne-Manneville S, Manneville J B, Adamson P, Wilbourn B, Greenwood J, Couraud P O. J Immunol. 2000;165:3375–3383. doi: 10.4049/jimmunol.165.6.3375. [DOI] [PubMed] [Google Scholar]
- 17.Rothlein R, Jaeger J R. Ciba Found Symp. 1995;189:200–211. doi: 10.1002/9780470514719.ch14. [DOI] [PubMed] [Google Scholar]
- 18.Springer T A. Proc Natl Acad Sci USA. 1997;94:65–72. doi: 10.1073/pnas.94.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu A, Leahy D J. Proc Natl Acad Sci USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang C, Springer T A. J Biol Chem. 1995;270:19008–19016. doi: 10.1074/jbc.270.32.19008. [DOI] [PubMed] [Google Scholar]
- 21.Edwards C P, Fisher K L, Presta L G, Bodary S C. J Biol Chem. 1998;273:28937–28944. doi: 10.1074/jbc.273.44.28937. [DOI] [PubMed] [Google Scholar]
- 22.Lu C, Shimaoka M, Ferzly M, Oxvig C, Takagi J, Springer T A. Proc Natl Acad Sci USA. 2001;98:2387–2392. doi: 10.1073/pnas.041606398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimaoka M, Lu C, Palframan R T, von Andrian U H, McCormack A, Takagi J, Springer T A. Proc Natl Acad Sci USA. 2001;98:6009–6014. doi: 10.1073/pnas.101130498. . (First Published May 15, 2001; 10.1073/pnas.101130498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J, Knorr R, Ferrone M, Houdei R, Carron C P, Dustin M L. J Exp Med. 1995;182:1231–1241. doi: 10.1084/jem.182.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reilly P L, Woska J R, Jr, Jeanfavre D D, McNally E, Rothlein R, Bormann B-J. J Immunol. 1995;155:529–532. [PubMed] [Google Scholar]
- 26.Staunton D E, Dustin M L, Erickson H P, Springer T A. Cell. 1990;61:243–254. doi: 10.1016/0092-8674(90)90805-o. [DOI] [PubMed] [Google Scholar]
- 27.Rothlein R, Mainolfi E A, Czajkowski M, Marlin S D. J Immunol. 1991;147:3788–3793. [PubMed] [Google Scholar]
- 28.Woska J R, Jr, Morelock M M, Jeanfavre D D, Bormann B J. J Immunol. 1996;156:4680–4685. [PubMed] [Google Scholar]
- 29.Labadia M E, Jeanfavre D D, Caviness G O, Morelock M M. J Immunol. 1998;161:836–842. [PubMed] [Google Scholar]
- 30.Casasnovas J M, Stehle T, Liu J-h, Wang J-h, Springer T A. Proc Natl Acad Sci USA. 1998;95:4134–4139. doi: 10.1073/pnas.95.8.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dustin M L, Springer T A. Nature (London) 1989;341:619–624. doi: 10.1038/341619a0. [DOI] [PubMed] [Google Scholar]
- 32.de Fougerolles A R, Stacker S A, Schwarting R, Springer T A. J Exp Med. 1991;174:253–267. doi: 10.1084/jem.174.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oxvig C, Lu C, Springer T A. Proc Natl Acad Sci USA. 1999;96:2215–2220. doi: 10.1073/pnas.96.5.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puri K D, Finger E B, Springer T A. J Immunol. 1997;158:405–413. [PubMed] [Google Scholar]
- 35.Rothlein R, Czajkowski M, O'Neil M M, Marlin S D, Mainolfi E, Merluzzi V J. J Immunol. 1988;141:1665–1669. [PubMed] [Google Scholar]
- 36.Maio M, Tessitori G, Pinto A, Temponi M, Colombatti A, Ferrone S. J Immunol. 1989;143:181–188. [PubMed] [Google Scholar]
- 37.Parkos C A, Colgan S P, Diamond M S, Nusrat A, Liang T W, Springer T A, Madara J L. Mol Med. 1996;2:489–505. [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez-Madrid F, Krensky A M, Ware C F, Robbins E, Strominger J L, Burakoff S J, Springer T A. Proc Natl Acad Sci USA. 1982;79:7489–7493. doi: 10.1073/pnas.79.23.7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang H-C, Bao Z-z, Yao Y, Tse A G D, Goyarts E C, Madsen M, Kawasaki E, Brauer P P, Sacchettini J C, Nathenson S G, Reinherz E L. Proc Natl Acad Sci USA. 1994;91:11408–11412. doi: 10.1073/pnas.91.24.11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staunton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer T A. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 41.O'Shea E K, Lumb K J, Kim P S. Curr Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 42.Casasnovas J M, Springer T A. J Virol. 1994;68:5882–5889. doi: 10.1128/jvi.68.9.5882-5889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu C, Springer T A. J Immunol. 1997;159:268–278. [PubMed] [Google Scholar]
- 44.Wagschal K, Tripet B, Hodges R S. J Mol Biol. 1999;285:785–803. doi: 10.1006/jmbi.1998.2284. [DOI] [PubMed] [Google Scholar]
- 45.Stewart M, Hogg N. J Cell Biochem. 1996;61:554–561. doi: 10.1002/(sici)1097-4644(19960616)61:4<554::aid-jcb8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 46.Fisher K L, Lu J, Riddle L, Kim K J, Presta L G, Bodary S C. Mol Biol Cell. 1997;8:501–515. doi: 10.1091/mbc.8.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casasnovas J M, Springer T A. J Biol Chem. 1995;270:13216–13224. doi: 10.1074/jbc.270.22.13216. [DOI] [PubMed] [Google Scholar]