Abstract
Rationale: An increased tricuspid regurgitation jet velocity (TRV > 2.5 m/s) and pulmonary hypertension defined by right heart catheterization both independently confer increased mortality in sickle cell disease (SCD).
Objectives: We explored the usefulness of peripheral blood mononuclear cell–derived gene signatures as biomarkers for an elevated TRV in SCD.
Methods: Twenty-seven patients with SCD underwent echocardiography and peripheral blood mononuclear cell isolation for expression profiling and 112 patients with SCD were genotyped for single-nucleotide polymorphisms.
Measurements and Main Results: Genome-wide gene and miRNA expression profiles were correlated against TRV, yielding 631 transcripts and 12 miRNAs. Support vector machine analysis identified a 10-gene signature including GALNT13 (encoding polypeptide N-acetylgalactosaminyltransferase 13) that discriminates patients with and without increased TRV with 100% accuracy. This finding was then validated in a cohort of patients with SCD without (n = 10) and with pulmonary hypertension (n = 10, 90% accuracy). Increased TRV-related miRNAs revealed strong in silico binding predictions of miR-301a to GALNT13 corroborated by microarray analyses demonstrating an inverse correlation between their expression. A genetic association study comparing patients with an elevated (n = 49) versus normal (n = 63) TRV revealed five significant single-nucleotide polymorphisms within GALNT13 (P < 0.005), four trans-acting (P < 2.1 × 10−7) and one cis-acting (P = 0.6 × 10−4) expression quantitative trait locus upstream of the adenosine-A2B receptor gene (ADORA2B).
Conclusions: These studies validate the clinical usefulness of genomic signatures as potential biomarkers and highlight ADORA2B and GALNT13 as potential candidate genes in SCD-associated elevated TRV.
Keywords: microarray, candidate gene approach, eQTL, pulmonary hypertension
At a Glance Commentary
Scientific Knowledge on the Subject
An elevated tricuspid regurgitation jet velocity (TRV) on transthoracic echocardiography and right heart catheterization (RHC)-defined pulmonary hypertension are both independently associated with increased mortality in sickle cell disease.
What This Study Adds to the Field
This study reports a novel gene signature for an elevated TRV, which is supported by integrated genomic and genetic approaches and validated in an independent cohort of patients with sickle cell disease and RHC-defined PH.
Cardiopulmonary complications represent the major cause of death in adult patients with sickle cell disease (SCD) with growing recognition of the role of pulmonary hypertension (PH) in poor outcomes (1). Although approximately 30% of patients with SCD have an elevated tricuspid regurgitant jet velocity (TRV) and, hence, increased right ventricular systolic pressure (RVSP) on transthoracic echocardiography (TTE), right heart catheterization (RHC)-confirmed PH is present in approximately 10% of these patients (2–4). Nonetheless, both RHC-defined PH and elevated TRV (≥ 2.5 m/s) on TTE are associated with increased mortality (1, 2). Hence, the SCD population has a predictable risk for the development of both an elevated TRV and RHC-confirmed PH and represents a group that may benefit from accurate diagnosis and treatment. Unfortunately, identification of PH in SCD frequently requires invasive RHC for confirmation, and current noninvasive tests of detecting PH lack optimal sensitivity and specificity (5).
The molecular processes influencing hemoglobin S (HbS) polymerization are well understood (6); however, the broad cardiovascular phenotypic heterogeneity of cardiovascular outcomes in SCD are less well characterized. Patients with SCD bearing the same HbS genetic mutation differ in their susceptibility to PH development (7), implicating modifier genes, single nucleotide polymorphisms (SNPs), and miRNAs that may contribute to this diversity. Prior studies have used gene expression profiles from peripheral blood mononuclear cells (PBMC) to better understand and characterize patients with SCD compared with healthy control subjects or patients with SCD without hydroxyurea treatment (8). Additionally, genome-wide association studies (GWAS) have reported significant associations of genetic markers with hemoglobin F level, overall severity in SCD, and other variables (7, 9). We hypothesized that genetic variability and PBMC-based genomic expression patterns in SCD may reveal unique expression profiles that not only provide insight into complex SCD cardiovascular complications but also could serve as important biomarkers to identify SCD populations with or at risk for PH. We report a novel gene signature that predicts an elevated TRV in SCD validated by integration of genome-wide miRNA expression and genetic data. This signature was then validated against an independent SCD cohort with both an elevated TRV and RHC-confirmed PH. These studies support the specificity of this genomic biomarker for elevated TRV in SCD and highlight the potential clinical usefulness of these translational and integrative approaches.
Methods
Subjects
Subjects from the University of Chicago (UC), the National Institutes of Health (NIH), and Howard University were recruited from adult SCD outpatient programs. Details of trial registration and TTE measurements are provided in the online supplement. Subjects provided written consent to participate in this study with the approval by the respective institutional human subjects review boards. The “discovery cohort” included 27 clinically stable African American subjects with homozygous SCD (HbSS demonstrated by high-performance liquid chromatographic separation or gel electrophoresis) recruited at the UC who all underwent prospective TTE as part of a research protocol regardless of clinical symptoms. The “validation cohort” consisted of 20 African American subjects with stable SCD (all HbSS) participating in the NIH SCD PH screening study (4) who all underwent prospective TTE, with a subset of patients with an elevated TRV (≥ 2.5 m/s with a clinical suspicion of PH [i.e., 6-min walk distance < 500 m, unexplained dyspnea or desaturation, or both]) who underwent a screening protocol for PH with RHC. For the current study, 20 patients were selected from this screening cohort, 10 of whom had a TRV less than 2.5 m/s and no clinical suspicion for PH and the remaining 10 or whom had an elevated TRV greater than or equal to 2.5 m/s and RHC-proven pulmonary arterial hypertension (n = 5, defined as a resting mean pulmonary artery pressure [mPAP] ≥ 25 mm Hg with a wedge pressure of ≤ 15 mm Hg) or pulmonary venous hypertension (n = 5, defined as PH with wedge pressure > 15 mm Hg). The former subgroup of patients with a normal TRV did not undergo RHC and hence represents a population with a low likelihood of having PH. A third subgroup (n = 112) of African American subjects with steady-state SCD (all Hb SS) from both the UC (including 27 subjects from the discovery cohort plus 23 newly recruited subjects) and the Howard University cohort (n = 62) all underwent prospective TTE and were used to perform a candidate gene SNP analysis. Subjects from all centers were excluded if they were clinically unstable, defined by having vasoocclusive crisis, acute chest syndrome, or unscheduled blood transfusions within 3 weeks of the study.
Microarray Preparation and Analysis
Both the discovery and validation cohorts underwent microarray gene and mature miRNA (isolated from PBMCs as described in the online supplement and previously [10]) expression analysis submitted to Gene Expression Omnibus (accession #: GSE38528). Platforms, labeling, hybridization, quality control, and data normalization are presented in the online supplement. In the discovery cohort, Spearman rank correlation test was first used to detect the relationship between gene (or miRNA) expression level and TRV (correlation coefficient recorded as ρT) and RVSP (correlation coefficient recorded as ρR) severity. Genes with ρR2 greater than 0.15 and ρT2 greater than 0.15 were considered as potentially differentially expressed. Differential expression of miRNAs used a threshold of ρR2 greater than 0.15 or ρT2 greater than 0.15.
Identification of a Gene Signature for Increased TRV
To identify a gene signature useful in the diagnosis of an elevated TRV, a machine learning algorithm based on support vector machine analysis (SVM) (11) was applied to further filter the differentially regulated gene subset from the discovery cohort yielding the top discriminating genes. The details of this analysis are discussed in the online supplement. The gene signature generated from the discovery cohort was then cross-examined against the validation cohort using an identical SVM-based approach to determine the predictive capacity for RHC-defined PH.
Genotypic Data and Association Analysis
We genotyped DNA samples from 112 subjects (49 with TRV ≥ 2.5 m/s, 63 with TRV < 2.5 m/s). Further details regarding genotype calls, analysis, and mapping expression quantitative trait locus mapping (eQTL) are provided in the online supplement. Briefly, TRV was analyzed as a categorical variable (elevated TRV vs. normal TRV) using classical association test (χ2 test) adjusting for sex. Given the strong evidence of association of the signature genes with an elevated TRV from expression profiling, we evaluated the genetic contribution of the signature genes and chose a relatively lenient cutoff of P < 0.005 (corresponding to a false discovery rate less than 50% after the conservative Bonferroni correction for individual genes) to identify SNPs significantly associated with the TRV phenotype. In addition, we mapped eQTLs for the signature genes to evaluate the genetic contribution to their regulation.
Reverse Transcriptase Quantitative Polymerase Chain Reaction
Selection of three differentially expressed transcripts and three miRNAs for confirmation and validation by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) are further discussed in the online supplement.
Results
Study Populations
Table 1 displays the characteristics of the discovery cohort. There were no significant differences in the majority of variables between the patients with an elevated and normal TRV. Significantly greater number of women exhibited increased TRV, as previously observed (12). Table 2 displays the distinguishing features of patients with RHC-defined PH versus those patients with low suspicion for PH in the validation cohort. Patients with PH in the validation cohort were significantly older and were associated with increased systemic blood pressures, decreased leukocytosis, increased presence of renal dysfunction, increased alkaline phosphatase levels, increased ferritin levels, and increased hemoglobin F and S levels when compared with the patients with low suspicion for PH. Many of these findings have been previously reported (12, 13) in patients with PH in SCD and are believed to represent markers for increased SCD severity and poor outcomes.
TABLE 1.
CHARACTERISTICS OF PATIENTS IN THE DISCOVERY COHORT
| Discovery Cohort (n = 27) |
|||
| Elevated TRV (n = 18) | Normal TRV (n = 9) | P Value | |
| Age, yr | 34.7 ± 8.1 | 29.5 ± 6.2 | 0.10 |
| Female/male | 13/5 | 3/6 | 0.05 |
| Genotype, Hgb SS | 18 | 9 | NA |
| BMI, kg/m2 | 21.9 ± 2 | 22.6 ± 4 | 0.64 |
| SBP, mm Hg | 119 ± 19 | 115 ± 14 | 0.60 |
| DBP, mm Hg | 69 ± 13 | 59 ± 13 | 0.06 |
| Hydroxyurea therapy, % | 88.9 | 88.9 | 1.00 |
| History of ACS, % | 100 | 77.8 | 0.10 |
| >20 Blood transfusions, % | 50 | 7 | 0.23 |
| 10–20 Blood transfusions | 4 | 1 | 0.63 |
| <10 Blood transfusions | 5 | 1 | 0.13 |
| WBC, 10−3/mm3 | 10.0 ± 3.1 | 11.1 ± 5.1 | 0.49 |
| Hemoglobin, g/dl | 8.1 ± 1.1 | 7.4 ± 1.1 | 0.17 |
| Platelets, 10−3/mm3 | 383 ± 94 | 346 ± 113 | 0.39 |
| BUN, mg/dl | 11 ± 7.4 | 10.4 ± 7.6 | 0.86 |
| Creatinine, mg/dl | 0.85 ± 0.6 | 0.73 ± 0.3 | 0.59 |
| Total bilirubin, mg/dl | 2.3 ± 1.5 | 3.5 ± 2.5 | 0.15 |
| Alkaline phosphatase, U/L | 104 ± 49 | 134 ± 89 | 0.28 |
| ALT, U/L | 27.5 ± 17 | 24.3 ± 14 | 0.63 |
| AST, U/L | 42.1 ± 18 | 38.7 ± 15 | 0.65 |
| Ferritin, mg/L | 2,204 ± 2,910 | 1,664 ± 1,546 | 0.61 |
| Hemoglobin A, % | 26 ± 26 | 11 ± 14 | 0.15 |
| Hemoglobin S, % | 63 ± 23 | 77 ± 12 | 0.12 |
| Hemoglobin F, % | 7 ± 5 | 8 ± 6 | 0.64 |
| Average TRV, m/s | 2.7 ± 0.3 | 2.1 ± 0.2 | <0.01 |
Definition of abbreviations: ACS = acute chest syndrome; ALT = alanine aminotransferase; AST = aspartate aminotransferase; BMI = body mass index; BUN = blood urea nitrogen; DBP = diastolic blood pressure; Hgb SS = hemoglobin SS; SBP = systolic blood pressure; TRV = tricuspid regurgitation jet velocity; WBC = white blood cell count.
TABLE 2.
CHARACTERISTICS OF PATIENTS IN THE VALIDATION COHORT
| Validation Cohort (n = 20) |
Discovery vs. Validation | |||
| PH (n = 10) | Normal TRV (n = 10) | P Value | P Value* | |
| Age, yr | 49.3 ± 7.4 | 30.2 ± 8.0 | <0.01 | <0.01 |
| Female/male | 4/6 | 7/3 | 0.43 | 0.14 |
| Genotype, Hgb SS | 10 | 10 | NA | NA |
| BMI, kg/m2 | 27.2 ± 13 | 20.5 ± 3 | 0.08 | 0.12 |
| SBP, mm Hg | 132 ± 19 | 114 ± 12 | 0.01 | 0.09 |
| DBP, mm Hg | 77 ± 14 | 65 ± 11 | 0.03 | 0.16 |
| Hydroxyurea therapy, % | 5 | 8 | 0.69 | 0.04 |
| History of ACS, % | 8 | 8 | 0.13 | 0.06 |
| >20 Blood transfusions, % | 1 | 2 | 0.58 | 0.02 |
| 10-20 Blood transfusions, % | 5 | 3 | 0.21 | 0.25 |
| <10 Blood transfusions, % | 3 | 1 | 0.22 | 1.00 |
| WBC, 10−3/mm3 | 6.2 ± 2.4 | 8.3 ± 2.5 | 0.03 | 0.01 |
| Hemoglobin, g/dl | 8.4 ± 1.2 | 9.1 ± 1.6 | 0.2 | 0.41 |
| Platelets, 10−3/mm3 | 315 ± 134 | 326 ± 104 | 0.79 | 0.11 |
| BUN, mg/dl | 29.9 ± 22 | 6.5 ± 1.5 | <0.01 | <0.01 |
| Creatinine, mg/dl | 2.5 ± 2.7 | 0.6 ± 0.1 | <0.01 | 0.01 |
| Total bilirubin, mg/dl | 1.5 ± 1.4 | 2.3 ± 1.1 | 0.10 | 0.14 |
| Alkaline phosphatase, U/L | 151 ± 98 | 97 ± 26 | 0.05 | 0.14 |
| ALT, U/L | 37 ± 20 | 34 ± 13 | 0.59 | 0.052 |
| AST, U/L | 48 ± 41 | 41 ± 14 | 0.54 | 0.58 |
| Ferritin, mg/L | 1,959 ± 1,518 | 718 ± 597 | 0.03 | 0.84 |
| Hemoglobin A, % | 37 ± 38 | 20 ± 25 | 0.15 | 0.34 |
| Hemoglobin S, % | 43 ± 29 | 66 ± 22 | 0.02 | 0.04 |
| Hemoglobin F, % | 19 ± 11 | 10 ± 7 | 0.02 | <0.01 |
| Average TRV, m/s | 2.9 ± 0.5 | 2.3 ± 0.1 | <0.01 | 0.25 |
Definition of abbreviations: ACS = acute chest syndrome; ALT = alanine aminotransferase; AST = aspartate aminotransferase; BMI = body mass index; BUN = blood urea nitrogen; DBP = diastolic blood pressure; Hgb SS = hemoglobin SS; PH = pulmonary hypertension; SBP = systolic blood pressure; TRV = tricuspid regurgitation jet velocity; WBC = white blood cell count.
Comparison between patients with an elevated TRV in the discovery cohort against patients with right heart catheterization–defined PH in the validation cohort.
When comparing patients with an elevated TRV in the discovery cohort against patients with RHC-defined PH in the validation cohort, patients with PH were significantly older, with fewer lifetime blood transfusions, reduced white blood cell (WBC) count, increased renal dysfunction, and increased hemoglobin F levels (Table 2). These data further provide evidence for increased overall severity of SCD in the PH subset of patients in the validation cohort compared with patients with an elevated TRV in the discovery cohort and, hence, two distinct cohorts. A comparison of echocardiographic parameters did not show significant differences between the cohorts (see Table E1 in the online supplement).
Of the 10 patients with RHC-defined PH, 5 revealed hemodynamic profiles consistent with PAH (mPAP was > 25 mm Hg and wedge pressure was < 15 mm Hg) and 5 showed evidence of pulmonary venous hypertension (PVH) (Table E2). The frequency of these subgroups is consistent with what has been previously published in the hemodynamic profiles of patients with SCD with PH (2, 14). A comparison of hemodynamic values between patients with PAH and PVH did not reveal significant differences in right atrial pressures, pulmonary artery pressure, or cardiac output/cardiac index.
Identification of a Molecular Signature Associated with an Elevated TRV
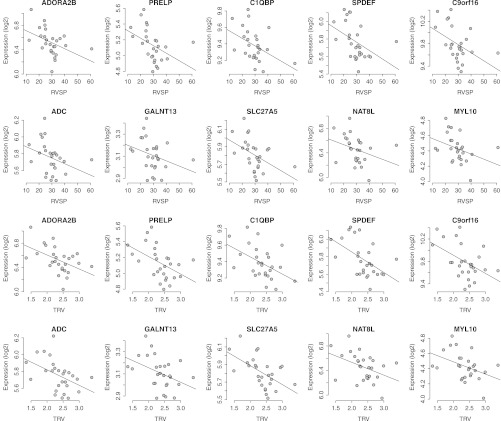
Although there was strong evidence of clustering within the elevated versus normal TRV comparison in the discovery cohort, unsupervised analysis of the microarray data was unable to distinguish elevated TRV in SCD with adequate sensitivity or specificity. We therefore performed a supervised analysis (Spearman correlation test) of the microarray data from the discovery cohort identifying the top 631 differentially expressed transcripts correlating with increasing TRV and RVSP as estimated on TTE. Applying SVM analysis to this subset of differentially regulated genes within the discovery cohort, a signature containing 10 genes distinguished subjects with an elevated TRV from those with normal TRV with 100% accuracy (Figure E1). The correlation coefficient values for each of these signature genes against RVSP or TRV are provided in Table E3. Spearman correlation confirmed down-regulation of all signature genes with increasing TRV or RVSP (Figure 1).
Figure 1.
Spearman correlation plot for genes within the signature for an elevated tricuspid regurgitation jet velocity (TRV). The correlation graphs depict the relationship between the expression values of the 10 signature genes plotted against either estimated right ventricular systolic pressure (RVSP) or TRV. These plots confirm the down-regulation of all 10 genes with increasing TRV or RVSP severity.
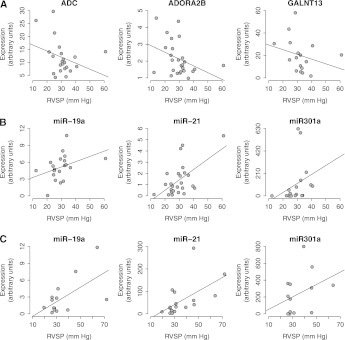
Figure 2A demonstrates the RT-qPCR results for three selected signature genes for an elevated TRV from the discovery cohort. Expression values of each gene showed reduced expression with increasing RVSP consistent with the microarray data (RT-qPCR, square of correlation coefficient ρR for ADORA2B, −0.44; GALNT13, −0.50; ADC, −0.36).
Figure 2.
Validation by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) of selected genes and miRNAs. Three signature genes (ADORA2B, C1QBP, ADC) and three miRNAs were selected for further validation by RT-qPCR. (A) Expression values of all three genes correlated with the changes in right ventricular systolic pressure (RVSP) and closely mirror the observed correlation by the microarray data. (B, C) Expression values of three miRNAs from the discovery cohort and the validation cohort, respectively, which correlated with the changes in RVSP and closely mirror the observed correlation by the microarray data.
Validation of the Molecular Signature Associated with an Elevated TRV in SCD in Patients with Pulmonary Hypertension
The gene signature for an elevated TRV generated from the discovery cohort was then assessed in the validation cohort of patients with RHC-defined PH (n = 10) and with low suspicion for PH (n = 10) using SVM analysis. The signature demonstrated a sensitivity of 90% and a specificity of 90% in this cohort, with an overall accuracy of 90% to discriminate PH in SCD. The positive and negative predictive values for the signature were also 90%.
Functional Analysis of Differentially Regulated Genes and Comparison to Published Pulmonary Hypertension Literature
Biological pathway analysis of the top 631 TRV-correlated genes from the discovery cohort reveal Wnt signaling, axon guidance, calcium signaling, neuroactive ligand–receptor interaction, vascular smooth muscle contraction, cancer, NOD-like receptor signaling, and primary bile acid biosynthesis pathways as the most significantly represented in patients with elevated TRV (Figure 3). The majority of these pathways have been linked to PH, further strengthening the association of the microarray data to PH in SCD (15, 16). Ingenuity analysis for the select elevated TRV signature genes, ADORA2B (adenosine A2B receptor) and GALNT13 (UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 or GalNAc-T13), provide further insights into potential gene–gene and pathway interactions (Figure E2). Integrating the 631-gene set for an elevated TRV, ADORA2B-related networks involved carbohydrate metabolism and cell signaling, whereas GALNT13-related networks were linked to cancer, cellular proliferation, and genetic disorders.
Figure 3.
Pathway analysis of genes associated with an elevated tricuspid regurgitation jet velocity (TRV). Pathway analysis of the 631 elevated TRV-associated genes reveals Wnt signaling, axon guidance, calcium signaling, neuroactive ligand–receptor interaction, vascular smooth muscle contraction, and cancer pathways as the most significantly represented in patients with SCD.
The top 631 correlated genes with an elevated TRV were next blasted against previously published PAH-specific microarray data (17) and against SCD-specific microarray data (distinguishing cases from African American control subjects [8]). For the latter comparison, a two-gene intersection was identified (vaccinia related kinase 2, proteasome subunit, α type 3). Although there are no prior published studies that have explored microarray expression profiles of PH or elevated TRV in SCD, comparison of the 631 dysregulated genes for an elevated TRV to published dysregulated genes in patients with PAH revealed greater than 50 common genes (17). Such a large overlap of genes provides further evidence of the genes that have surfaced from the current study for an elevated TRV and their potential for predicting PH in SCD (Table E4).
A Pubmatrix evaluation of identified signature genes against known PH search terms revealed established (ADORA2B, ADC) and potentially novel candidate genes (GALNT13, C1QBP) involved in PH pathophysiology (Table 3). For example, ADORA2B exhibited 27 citations and ADC exhibited 58 citations when cross-referenced to PH, whereas GALNT13 and C1QBP failed to have a single PH citation. The presence of signature genes for an elevated TRV that are also cited in PH literature also strengthens signature specificity for PH in SCD, whereas the presence of novel genes provides avenues for further exploration of PH development in SCD.
TABLE 3.
PUBMATRIX EVALUATION OF ELEVATED TRICUSPID REGURGITATION JET VELOCITY–DRIVEN GENES IN SICKLE CELL DISEASE ACROSS PULMONARY HYPERTENSION–RELATED SEARCH TERMS
| Gene Names | Gene Symbol | PH | Vascular Remodeling | Inflammation | Cancer | Angiogenesis | Hypoxia | NO | IR |
| Myosin light chain 10, regulatory | MYL10 | 1 | 2 | 3 | 14 | 2 | 5 | 6 | 0 |
| Adenosine A2b receptor | ADORA2B | 27 | 4 | 88 | 51 | 17 | 50 | 31 | 25 |
| Complement component 1, q subcomponent binding protein | C1QBP | 0 | 1 | 35 | 33 | 0 | 0 | 1 | 2 |
| Arginine decarboxylase | ADC | 58 | 0 | 369 | 2,030 | 16 | 231 | 58 | 86 |
| Proline/arginine-rich end leucine-rich repeat protein | PRELP | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 |
| SAM pointed domain containing ets transcription factor | SPDEF | 0 | 0 | 3 | 39 | 2 | 0 | 0 | 0 |
| Polypeptide N-acetylgalactosaminyltransferase 13 | GALNT13 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 0 |
| Solute carrier family 27, member 5 | SLC27A5 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| N-acetyltransferase 8-like | NAT8L | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chrom 9 open reading frame 16 | C9orf16 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
Definition of abbreviations: IR = ischemia reperfusion; NO = nitric oxide; PH = pulmonary hypertension.
Survey of Genetic Variants in the Molecular Signature Genes
We next queried the genetic variation of the signature genes associated with an elevated TRV phenotype by leveraging a dataset from a cohort of 112 patients with SCD. Table E5 displays the clinical characteristics of these patients. The samples were confirmed for homogeneity by performing principal component analysis using a panel of ancestry informative markers (18). We evaluated associations between 412 informative SNPs within these signature genes and the TRV phenotype. In total, five SNPs within GALNT13 and one SNP within PRELP were found to be associated with the TRV phenotype (P < 0.005, Table 4). This robust overrepresentation of GALNT13 SNPs further supports the notion of genetic contribution to the potential role of GALNT13 in determining an elevated TRV phenotype in SCD. In addition, the allele frequencies of these variants in patients with SCD without the TRV phenotype appear to be comparable to the frequencies in a panel of African American healthy individuals (ASW, African ancestry from Southwest United States) (dbSNP v131).
TABLE 4.
TOP SINGLE-NUCLEOTIDE POLYMORPHISMS WITHIN SIGNATURE GENES ASSOCIATED WITH ELEVATED TRICUSPID REGURGITATION JET VELOCITY
| SNP | Signature Gene | Ref Allele | P Value | P Value* | OR | Freq SCD High TRV Cases | Freq SCD Control | Freq ASW | Function |
| rs799813 | GALNT13 | C | 0.001 | 0.002 | 0.28 | 0.094 | 0.27 | 0.25 | Intronic |
| rs2794452 | PRELP | C | 0.001 | 0.013 | 0.42 | 0.34 | 0.55 | 0.58 | Upstream |
| rs10497120 | GALNT13 | C | 0.001 | 0.001 | 2.76 | 0.33 | 0.15 | 0.13 | Upstream |
| rs13407922 | GALNT13 | A | 0.002 | 0.003 | 3.08 | 0.24 | 0.095 | 0.14 | Intronic |
| rs16833378 | GALNT13 | G | 0.003 | 0.003 | 0.26 | 0.061 | 0.20 | 0.11 | Upstream |
| rs9808145 | GALNT13 | A | 0.004 | 0.006 | 2.97 | 0.22 | 0.089 | 0.13 | Intronic |
Definition of abbreviations: ASW = African ancestry from Southwest United States; Freq = frequency; OR = odds ratio; Ref = reference; SCD = sickle cell disease.
Adjusted for cohort.
To further assess the effects of genetic variation in SCD on gene expression (i.e., eQTL in PBMCs), we integrated data from 24 patients from the discovery cohort with available gene expression profiling and genotypic data to derive eQTLs associated with the signature genes (false discovery rate < 5% with Bonferroni correction). Notably, three trans-acting eQTLs were identified for SAM domain–containing prostate-derived Ets factor (SPDEF), and each was in proximity to calcium-sensing receptor and cystatin A genes (rs10934581, rs7633800, rs7633667, rs2268443, P = 2.1 × 10−7). One trans-acting eQTL was found to be associated with the expression of chromosome 9 open reading frame 16 (c9orf16, rs3864785, P = 4.8 × 10−8). In addition, one cis-acting eQTL residing upstream of the ADORA2B gene (an established PH and SCD candidate gene) (rs7208480, P = 0.6 × 10−4) was identified.
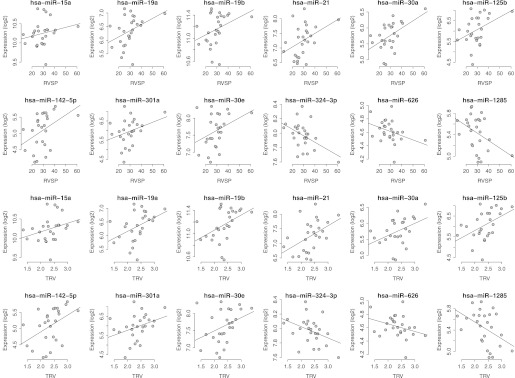
miRNA Array Analysis and Functional Profiling of Top Elevated TRV-associated miRNAs
Given the established role of miRNAs in the regulation of gene expression, we also interrogated the potential usefulness of miRNA expression profiling as a clinical tool to predict an elevated TRV in SCD. Although evidence of clustering exists within the elevated TRV comparison set in the discovery cohort, unsupervised analysis of the miRNA microarray data was unable to distinguish elevated TRV in SCD with adequate sensitivity or specificity (Figure E3). We therefore performed a supervised analysis (Spearman correlation test) of the microarray data identifying 12 unique miRNAs with the strongest correlation against TRV and RVSP severity (Figure 4). The correlation coefficient values for each of these top 12 miRNAs are listed in Table E6. Figure 2B demonstrates the RT-qPCR results for three selected signature miRNAs from the discovery cohort. Expression values of all three miRNAs were consistent with the microarray data (from RT-qPCR, correlation coefficient ρR for miR-301a, 0.64; miR-19a, 0.54; and miR-21, 0.43). To further validate these miRNA expression data, RT-qPCR of these three miRNAs was performed in the validation cohort. Figure 2C demonstrates consistent expression the three miRNAs (miR-301a, 0.35; mir-19a, 0.39; and miR-21, 0.66) in the validation cohort compared with the discovery cohort, further establishing the association of these miRNAs to both an elevated TRV and PH in SCD.
Figure 4.
Spearman correlation graphs for top miRNAs associated with an elevated tricuspid regurgitation jet velocity (TRV). The correlation graphs depict the relationship between the expression values of the top 12 miRNAs plotted against either right ventricular systolic pressure (RVSP) (top rows) or TRV (bottom rows).
miRNA-mRNA In Silico and Integrative Analysis
We next searched for a list of predicted mRNA targets for the top 12 elevated TRV-related miRNAs using data provided by www.microrna.org, with filtering based on a stringent mirSVR score threshold to select the top binding miRNA-mRNA pairs. Given the large size of predicted mRNA target lists (hundreds to thousands per miRNA), we intersected these predicted transcripts against the microarray-observed 631 transcripts best correlated with an elevated TRV revealing a total of 89 miRNA-mRNA pairs. We then further filtered these 89 pairs based on an inverse expression profile pattern as would be predicted by a conventional miRNA to mRNA relationship, yielding a total of 29 unique miRNA-mRNA pairs (positively and negatively significantly correlated from the microarray dataset) (Table 5). From these 29 filtered pairs, GALNT13, an elevated TRV signature gene, again emerged as a leading potential candidate gene demonstrating an association with miR-301a, as predicted by in silico analyses and as observed in our microarray data with significant inverse correlation with expression (Table 5).
TABLE 5.
INTEGRATED ANALYSIS OF MIRNA AND GENE EXPRESSION PROFILING
| miRNA | mirSVR Score | Gene Symbol | Gene Name | Correlation Gene vs miRNA | P Value |
| miR-21 | −0.75 | CFTR | Cystic fibrosis transmembrane conductance regulator | −0.36 | 0.026 |
| miR-21 | −0.55 | KIAA1804 | Mixed lineage kinase 4 | −0.37 | 0.019 |
| miR-21 | −0.95 | PRR18 | Proline rich 18 | −0.46 | 0.004 |
| miR-30a | −0.62 | PARG1 | Rho GTPase activating protein 29 | −0.42 | 0.007 |
| miR-301a | −0.68 | GALNT13 | Polypeptide N-acetylgalactosaminyltransferase 13 | −0.36 | 0.024 |
| miR-142-5p | −1.08 | GRM5 | Glutamate receptor, metabotropic 5 | −0.37 | 0.022 |
| miR-142-5p | −0.86 | SHC4 | SHC (Src homology 2 domain) family, member 4 | −0.42 | 0.007 |
| miR-142-5p | −0.79 | CNTN5 | Contactin 5 | −0.32 | 0.050 |
| miR-142-5p | −0.88 | ADSSL1 | Adenylosuccinate synthase like 1 | −0.43 | 0.007 |
| miR-142-5p | −1.29 | LMX1A | LIM homeobox transcription factor 1, alpha | −0.43 | 0.007 |
| miR-142-5p | −1.11 | EPDR1 | Ependymin related protein 1 (zebrafish) | −0.41 | 0.009 |
| miR-142-5p | −0.58 | TCF7L1 | Transcription factor 7-like 1 (T-cell specific) | −0.40 | 0.012 |
| miR-324-3p | −0.99 | EI24 | Etoposide induced 2.4 mRNA | −0.38 | 0.017 |
| miR-626 | −0.93 | RBM23 | RNA binding motif protein 23 | 0.33 | 0.042 |
| miR-1285 | −0.56 | PRDM10 | PR domain containing 10 | −0.36 | 0.026 |
| miR-1285 | −0.77 | ARF6 | ADP-ribosylation factor 6 | −0.39 | 0.015 |
| miR-1285 | −1.05 | GPATCH8 | G patch domain containing 8 | −0.32 | 0.047 |
| miR-1285 | −0.78 | PRMT3 | Protein arginine methyltransferase 3 | −0.35 | 0.029 |
| miR-1285 | −0.92 | PATL1 | Protein associated with topoisomerase II - 1 | −0.32 | 0.046 |
| miR-1285 | −0.76 | TFCP2 | Transcription factor CP2 | −0.37 | 0.022 |
| miR-1285 | −0.59 | PTPN2 | Protein tyrosine phosphatase, non-receptor type 2 | −0.37 | 0.021 |
| miR-1285 | −0.98 | KDM2B | lysine (K)-specific demethylase 2B | −0.37 | 0.022 |
| miR-1285 | −0.91 | TBK1 | TANK-binding kinase 1 | −0.34 | 0.032 |
| miR-1285 | −1.29 | VPS35 | Vacuolar protein sorting 35 homolog (Saccharomyces cerevisiae) | −0.37 | 0.020 |
| miR-1285 | −1.20 | SC5DL | Sterol-C5-desaturase | −0.45 | 0.004 |
| miR-1285 | −1.10 | CSTF3 | Cleavage stimulation factor, 3′ pre-RNA, subunit 3 | −0.42 | 0.007 |
| miR-1285 | −1.20 | CP110 | CP110 protein | −0.36 | 0.023 |
| miR-1285 | −0.77 | PIBF1 | Progesterone immunomodulatory binding factor 1 | −0.38 | 0.018 |
| miR-1285 | −0.56 | UHRF2 | Ubiquitin-like with PHD and ring finger domains 2 | −0.34 | 0.033 |
Definition of abbreviations: SCD = sickle cell disease; TRV = tricuspid regurgitation jet velocity.
The table reflects the top miRNA–gene pairs expressed in patients with an elevated TRV in SCD based on their individual significant correlations with TRV from expression profiling, strong in silico binding prediction scores, and demonstration of a positive or negative correlation to each other based on their observed expression from the microarray data.
Discussion
An elevated estimated pulmonary artery systolic pressure is independently associated with increased mortality in patients with SCD (1, 19), yet reliable biomarkers and novel targets are not currently available. We now report the first PBMC-derived novel gene signature for an elevated TRV in SCD derived from a discovery cohort with validation in an independent replicate cohort of patients with SCD with confirmed PH by RHC. The current study also represents the first integration of genome-wide mRNA and miRNA expression profiling and genetic data in SCD. These robust bioinformatic analyses further validate the genomic signature for both an elevated TRV and PH and identified candidate genes potentially associated with these SCD phenotypes.
Based on the evaluation of an elevated TRV phenotype as a continuous variable, the derived gene signature supports the notion of a discrete echo-defined elevated TRV phenotype in SCD and suggests potential clinical usefulness as a biomarker for this phenotype. The validation cohort was characterized by additional parameters of increased SCD severity in comparison to the discovery cohort (including renal dysfunction, relatively low WBC count, and older age), thereby further empowering the predictive capacity of the signature for an elevated TRV to broadly apply to varied SCD populations in contrast to a unique population in a single center. In addition, the RHC-defined PH phenotype in the validation cohort strengthens the potential predictability of the molecular signature for PH as well as an elevated TRV. This signature was found to be composed of both novel and established PH candidate genes as evidenced by literature mining and the PubMed search tool. For example, ADORA2B, a candidate gene known to modify the severity of SCD (20, 21), was recently identified via metabolomic profiling of a transgenic SCD mouse model with excessive adenosine signaling through the ADORA2B leading to sickling and hemolysis (22), processes known to be independently correlated with the presence of PH in SCD (13, 23). Furthermore, ADC, another signature gene, which decarboxylates l-arginine to agmatine, is an established PAH candidate gene (24) and is potentially involved in SCD-related PH (25). In fact, circulating modified forms of l-arginine, obligate substrates of the nitric oxide synthases (NOS) that disrupt NO production, are elevated with increasing severity of PH in SCD (25). Given the decreasing expression of ADC in PBMCs with increasing TRV we observed, the role of circulating PBMCs in the induction of the NOS pathway observed in patients with SCD and PH must be considered. Therefore, within the 10-gene signature, the presence of established candidate genes from both PH and SCD-related literature independently strengthen the validity of this molecular biomarker for an elevated TRV and possibly PH.
The genomic signature in PBMCs for an elevated TRV represents expression changes that may not directly reflect the dynamics of the pulmonary vascular microenvironment in PH or in patients with an elevated TRV. However, the role of these circulating cells in biomarker discovery and their potential contribution to the pathobiology of SCD and PAH have both been well established (8, 10, 26, 27). Circulating blood cells may carry disease-specific information due to genetics or due to alterations in their local environment. The gene expression of PBMCs may also represent the specificity of SCD and PH because of inflammatory and immune mechanisms that likely play important roles in their development. A relatively low WBC count as evidenced in the validation PH cohort in Table 2 may represent a cause or effect of the severity of SCD disease when associated with PH, which can result in generating a distinct population of PBMCs with a unique expression profile distinguishing those with and without PH. Although this WBC count difference is also observed between those with an elevated TRV in the discovery cohort and those with RHC-defined PH in the validation cohort, the signature remains accurate in predicting both phenotypes. This latter observation argues against the role of WBC counts in defining PBMC expression profiling and warrants further studies to determine if any mechanistic association exists.
Despite potential limitations, our studies highlight the potential for individual signature genes to participate in the pathophysiology of an elevated TRV and PH in SCD. The signature gene, GALNT13, was identified as a target for the elevated TRV-associated miRNA (miR-301a) with significant correlation between miRNA-GALNT13 expression. Furthermore, multiple GALNT13-associated SNPs were linked to an elevated TRV phenotype. GALNT13 encodes a glycosyltransferase enzyme responsible for the synthesis of O-glycan (28, 29). Given that aberrant glycosylation patterns are a hallmark of the tumor phenotype influencing proliferation, invasion, angiogenesis, and metastasis (30–33), processes known to be involved in pulmonary vascular remodeling, GALNT13 may be speculated as a novel candidate in SCD with an elevated TRV and PH.
The finding of dysregulated miRNA expression in patients with SCD and an elevated TRV suggests an elevated TRV may result in altered miRNA expression profiles. Among the 12 most dysregulated miRNAs in the discovery cohort, miR-21, miR-19a, miR-15a, and miR-125b have previously been found to be dysregulated in different cellular processes that contribute to pulmonary vascular remodeling, including apoptosis, cell proliferation, and angiogenesis (34–38). A recent report also described increased vascular cell proliferation in hereditary PAH as a consequence of decreased levels of miR-21 (39), consistent with the trends of miR-21 expression in increasing TRV observed in this study. Notably, miR-15a and miR-19a are associated with increases in hemoglobin F levels, regulation of hematopoiesis, and vascular flow processes directly involved with SCD pathophysiology (37, 40). Data from this study combined with this background highlight the potential roles for miR-15a, miR19a, and miR-21 in SCD-related PH and further validate the miRNA signature for an elevated TRV in SCD.
Pathway analyses of the differentially regulated genes for an elevated TRV comparison set revealed involvement of established PH processes, including Wnt signaling, calcium signaling, vascular smooth muscle contraction, cancer pathways, and primary bile acid biosynthesis pathways (41–44), although these have yet to be explored in PH associated with SCD. Axon guidance and NOD-like receptor signaling appear to represent novel pathways involved in the elevated TRV phenotype. Pubmatrix evaluation of the candidate genes in the molecular signature further underscored the novelty of many involved genes (MYL10 and C1QBP in PH) while validating several genes recognized to be associated with PH (ADORA2B, ADC). Beyond pathway analyses, this study is the first to integrate available published microarray analyses of human SCD and PAH studies. The presence of several overlapping genes between the current study in an elevated TRV setting and previous PH studies has further strengthened the specificity of our differentially regulated genes.
We used a candidate gene approach to SNP discovery in our signature genes. Although we studied a small cohort in this analysis, genetic variants have been found to be associated with SCD-related elevated TRV in this study, suggesting that there could be genetic contribution to this trait and further strengthening the case for association between the candidate genes that surfaced from expression profiling of circulating cells to the elevated TRV phenotype. However, the downstream mechanisms of action have not been fully elucidated. The genotyping platform captures the general genetic variation, and most of these SNPs were located either intronic or upstream from the gene. Therefore, the associated SNPs could be linked to certain untyped causal SNPs, and further fine-mapping and functional validation are warranted to elucidate their roles. Additionally, knowledge of intermediate steps can also provide perspective on disease pathogenesis, while also providing further evidence for the specificity of the molecular signature. To begin to address these questions, the effects of genetic variation on PBMC gene expression eQTL in patients with an elevated TRV were assessed. An example is the finding of the cis-acting eQTL and its association with the expression of ADORA2B, a previously established SCD candidate gene that was also a signature gene. This cis-acting eQTL may represent a novel potential intermediate gene that connects the genetic variant with the development of an elevated TRV in SCD.
Limitations to this study include the small sample size of our cohorts, which emphasizes the requirement of a larger sample cohort for translation of the molecular signature into clinical practice and raises the possibility of overfitting the gene signature data for an elevated TRV. We attempted to address this concern by validating the gene signature in a separate and distinctive SCD-PH cohort. Despite differences in characteristics observed across the discovery and validation cohorts, representing phenotypic and geographic differences as well as limited sample size, the signature remained powerfully predictive, with 90% accuracy. This validation cohort also exhibited an overrepresentation of RHC-defined PH in SCD compared with the established prevalence of PH in SCD and furthermore did not achieve an adequate sample size to interpret predictions of the signature for PAH and PVH. Therefore, given these limitations, the value of this signature for predicting PH in SCD also needs to be carefully assessed and further investigated to confirm these preliminary findings. Finally, it is unknown whether the identified gene expression differences generated by comparing patients with and without an elevated TRV are a result of an elevated TRV or if these genes are related to the pathogenesis of the underlying disease, especially given their derivation from a circulating pool of cells. Nonetheless, we can speculate a potential mechanistic role for these circulating cells given the documented roles and pathways of some of the signature genes in SCD and in PH. Ultimately, the usefulness of the signature as an independent molecular biomarker or in conjunction with TRV as a noninvasive imaging biomarker remains and demands its further clinical application in a prospective fashion.
Supplementary Material
Acknowledgments
The authors thank Zarema Arbieva, Ph.D. and the Research Resources Center at the University of Illinois at Chicago (UIC) for assistance in the microarray data, and Brandon Mapes, B.S., also at the UIC, for his general assistance. They also thank Ms. Laurel Mendelsohn and Ms. Kimberly Woodhouse at the National Heart, Lung, and Blood Institute Genomics Core Facility, National Institutes of Health, for help with sample collection and processing.
Footnotes
Funded by National Institutes of Health grants NHLBI F32HL090359 (A.A.D.), K23HL098454 (R.F.M.), 1-R0 HL079912-02 (V.R.G.), and the National Institutes of Health National Center for Research Resources/National Center for Advancing Translational Sciences grant UL1RR029879 (A.A.D.).
Author Contributions: A.A.D. designed research, performed research, contributed vital analytical tools, collected data, analyzed and interpreted data, performed statistical analysis, and wrote the manuscript. T.Z. contributed vital analytical tools, analyzed and interpreted data, performed statistical analysis, and wrote the manuscript. H.A. contributed vital analytical tools, analyzed and interpreted data. W.Z. analyzed, interpreted data, performed statistical analysis, and wrote the manuscript. W.M. analyzed and interpreted data, performed statistical analysis. S.T. designed research, performed research, collected data. M.S.W. performed research, contributed vital new reagents. N.R. collected data, analyzed and interpreted data. G.J.K. collected data, analyzed and interpreted data. M.H.P.-L. collected data. T.T. performed research, collected data. K.T. collected data. N.A. performed research, collected data. Y.H. analyzed and interpreted data. A.R.P. designed research, collected data. J.X.-J.Y. analyzed and interpreted data and wrote the manuscript. V.R.G. collected data, analyzed and interpreted data, performed statistical analysis, and wrote the manuscript. R.M.L. designed research, performed research, contributed vital analytical tools, collected data, analyzed and interpreted data. J.G.N.G. designed research, performed research, contributed vital analytical tools, collected data, analyzed and interpreted data, performed statistical analysis, and wrote the manuscript. R.F.M. designed and performed research, contributed vital analytical tools, collected, analyzed, and interpreted data, and wrote the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201201-0057OC on June 7, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Gladwin MT, Sachdev V, Jison ML, Shizukuda Y, Plehn JF, Minter K, Brown B, Coles WA, Nichols JS, Ernst I, et al. Pulmonary hypertension as a risk factor for death in patients with sickle cell disease. N Engl J Med 2004;350:886–895 [DOI] [PubMed] [Google Scholar]
- 2.Parent F, Bachir D, Inamo J, Lionnet F, Driss F, Loko G, Habibi A, Bennani S, Savale L, Adnot S, et al. A hemodynamic study of pulmonary hypertension in sickle cell disease. N Engl J Med 2011;365:44–53 [DOI] [PubMed] [Google Scholar]
- 3.Fonseca GH, Souza R, Salemi VM, Jardim CV, Gualandro SF. Pulmonary hypertension diagnosed by right heart catheterisation in sickle cell disease. Eur Respir J 2012;39:112–118 [DOI] [PubMed] [Google Scholar]
- 4.Mehari A, Gladwin MT, Tian X, Machado RF, Kato GJ. Mortality in adults with sickle cell disease and pulmonary hypertension. JAMA 2012;307:1254–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rich JD, Shah SJ, Swamy R, Kamp A, Rich S. The inaccuracy of Doppler echocardiographic estimates of pulmonary artery pressures in patients with pulmonary hypertension: implications for clinical practice. Chest 2011;139:988–993 [DOI] [PubMed] [Google Scholar]
- 6.Ferrone FA, Rotter MA. Crowding and the polymerization of sickle hemoglobin. J Mol Recognit 2004;17:497–504 [DOI] [PubMed] [Google Scholar]
- 7.Fertrin KY, Costa FF. Genomic polymorphisms in sickle cell disease: implications for clinical diversity and treatment. Expert Rev Hematol. 2010;3:443–458 [DOI] [PubMed] [Google Scholar]
- 8.Jison ML, Munson PJ, Barb JJ, Suffredini AF, Talwar S, Logun C, Raghavachari N, Beigel JH, Shelhamer JH, Danner RL, et al. Blood mononuclear cell gene expression profiles characterize the oxidant, hemolytic, and inflammatory stress of sickle cell disease. Blood 2004;104:270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashley-Koch AE, Elliott L, Kail ME, De Castro LM, Jonassaint J, Jackson TL, Price J, Ataga KI, Levesque MC, Weinberg JB, et al. Identification of genetic polymorphisms associated with risk for pulmonary hypertension in sickle cell disease. Blood 2008;111:5721–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoryev DN, Mathai SC, Fisher MR, Girgis RE, Zaiman AL, Housten-Harris T, Cheadle C, Gao L, Hummers LK, Champion HC, et al. Identification of candidate genes in scleroderma-related pulmonary arterial hypertension. Transl Res 2008;151:197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vapnik V. Statistical Learning Theory. New York: John Wiley & Sons; 1998 [Google Scholar]
- 12.De Castro LM, Jonassaint JC, Graham FL, Ashley-Koch A, Telen MJ. Pulmonary hypertension associated with sickle cell disease: clinical and laboratory endpoints and disease outcomes. Am J Hematol 2008;83:19–25 [DOI] [PubMed] [Google Scholar]
- 13.Machado RF, Gladwin MT. Pulmonary hypertension in hemolytic disorders: pulmonary vascular disease: the global perspective. Chest 2010;137:30S–38S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anthi A, Machado RF, Jison ML, Taveira-Dasilva AM, Rubin LJ, Hunter L, Hunter CJ, Coles W, Nichols J, Avila NA, et al. Hemodynamic and functional assessment of patients with sickle cell disease and pulmonary hypertension. Am J Respir Crit Care Med 2007;175:1272–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machado RD, Eickelberg O, Elliott CG, Geraci MW, Hanaoka M, Loyd JE, Newman JH, Phillips JA, III, Soubrier F, Trembath RC, et al. Genetics and genomics of pulmonary arterial hypertension. J Am Coll Cardiol 2009;54(1, Suppl)S32–S42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuder RM, Abman SH, Braun T, Capron F, Stevens T, Thistlethwaite PA, Haworth SG. Development and pathology of pulmonary hypertension. J Am Coll Cardiol 2009;54(1, Suppl)S3–S9 [DOI] [PubMed] [Google Scholar]
- 17.Rajkumar R, Konishi K, Richards TJ, Ishizawar DC, Wiechert AC, Kaminski N, Ahmad F. Genomewide RNA expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. Am J Physiol Heart Circ Physiol 2010;298:H1235–H1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng CY, Reich D, Haiman CA, Tandon A, Patterson N, Elizabeth S, Akylbekova EL, Brancati FL, Coresh J, Boerwinkle E, et al. African ancestry and its correlation to type 2 diabetes in african americans: a genetic admixture analysis in three U.S. population cohorts. PLoS ONE 2012;7:e32840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machado RF. Sickle cell anemia-associated pulmonary arterial hypertension. J Bras Pneumol 2007;33:583–591 [DOI] [PubMed] [Google Scholar]
- 20.Haynes J, Jr, Obiako B, Thompson WJ, Downey J. Adenosine-induced vasodilation: receptor characterization in pulmonary circulation. Am J Physiol 1995;268:H1862–H1868 [DOI] [PubMed] [Google Scholar]
- 21.Pearl RG. Adenosine produces pulmonary vasodilation in the perfused rabbit lung via an adenosine A2 receptor. Anesth Analg 1994;79:46–51 [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Dai Y, Wen J, Zhang W, Grenz A, Sun H, Tao L, Lu G, Alexander DC, Milburn MV, et al. Detrimental effects of adenosine signaling in sickle cell disease. Nat Med 2011;17:79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aliyu ZY, Gordeuk V, Sachdev V, Babadoko A, Mamman AI, Akpanpe P, Attah E, Suleiman Y, Aliyu N, Yusuf J, et al. Prevalence and risk factors for pulmonary artery systolic hypertension among sickle cell disease patients in Nigeria. Am J Hematol 2008;83:485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reis DJ, Li G, Regunathan S. Endogenous ligands of imidazoline receptors: classic and immunoreactive clonidine-displacing substance and agmatine. Ann N Y Acad Sci 1995;763:295–313 [DOI] [PubMed] [Google Scholar]
- 25.Kato GJ, Wang Z, Machado RF, Blackwelder WC, Taylor JG, 6th, Hazen SL. Endogenous nitric oxide synthase inhibitors in sickle cell disease: abnormal levels and correlations with pulmonary hypertension, desaturation, haemolysis, organ dysfunction and death. Br J Haematol 2009;145:506–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asosingh K, Aldred MA, Vasanji A, Drazba J, Sharp J, Farver C, Comhair SA, Xu W, Licina L, Huang L, et al. Circulating angiogenic precursors in idiopathic pulmonary arterial hypertension. Am J Pathol 2008;172:615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frid MG, Brunetti JA, Burke DL, Carpenter TC, Davie NJ, Reeves JT, Roedersheimer MT, van Rooijen N, Stenmark KR. Hypoxia-induced pulmonary vascular remodeling requires recruitment of circulating mesenchymal precursors of a monocyte/macrophage lineage. Am J Pathol 2006;168:659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berois N, Blanc E, Ripoche H, Mergui X, Trajtenberg F, Cantais S, Barrois M, Dessen P, Kagedal B, Benard J, et al. ppGalNAc-T13: a new molecular marker of bone marrow involvement in neuroblastoma. Clin Chem 2006;52:1701–1712 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Iwasaki H, Wang H, Kudo T, Kalka TB, Hennet T, Kubota T, Cheng L, Inaba N, Gotoh M, et al. Cloning and characterization of a new human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, designated pp-GalNAc-T13, that is specifically expressed in neurons and synthesizes GalNAc alpha-serine/threonine antigen. J Biol Chem 2003;278:573–584 [DOI] [PubMed] [Google Scholar]
- 30.Cheng L, Wang X, Tan M, Zhou X, Wang Q. Expression of polypeptide GalNAc-transferases in hematopoietic stem/progenitor cells. Cell Biol Int 2004;28:635–640 [DOI] [PubMed] [Google Scholar]
- 31.Gill DJ, Clausen H, Bard F. Location, location, location: new insights into O-GalNAc protein glycosylation. Trends Cell Biol 2011;21:149–158 [DOI] [PubMed] [Google Scholar]
- 32.Ishikawa M, Kitayama J, Kohno K, Nagawa H. The expression pattern of UDP-N-acetyl-alpha-D-galactosamine-polypeptide N-acetyl-galactosaminyl transferase-3 in squamous cell carcinoma of the esophagus. Pathobiology 2005;72:139–145 [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Tachibana K, Zhang Y, Iwasaki H, Kameyama A, Cheng L, Guo J, Hiruma T, Togayachi A, Kudo T, et al. Cloning and characterization of a novel UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase, pp-GalNAc-T14. Biochem Biophys Res Commun 2003;300:738–744 [DOI] [PubMed] [Google Scholar]
- 34.Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, Urbich C, Hofmann WK, Zeiher AM, Dimmeler S. Members of the microRNA-17–92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 2010;115:4944–4950 [DOI] [PubMed] [Google Scholar]
- 35.Fabbri M, Bottoni A, Shimizu M, Spizzo R, Nicoloso MS, Rossi S, Barbarotto E, Cimmino A, Adair B, Wojcik SE, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA 2011;305:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrisey EE. The magic and mystery of miR-21. J Clin Invest 2010;120:3817–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin X, Wang X, Wang Y, Tang Z, Cui Q, Xi J, Li YS, Chien S, Wang N. MicroRNA-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells. Proc Natl Acad Sci USA 2010;107:3240–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surdziel E, Cabanski M, Dallmann I, Lyszkiewicz M, Krueger A, Ganser A, Scherr M, Eder M. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood 2011;117:4338–4348 [DOI] [PubMed] [Google Scholar]
- 39.Drake KM, Zygmunt D, Mavrakis L, Harbor P, Wang L, Comhair SA, Erzurum SC, Aldred MA. Altered microRNA processing in heritable pulmonary arterial hypertension: an important role for Smad-8. Am J Respir Crit Care Med 2011;184:1400–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sankaran VG, Menne TF, Scepanovic D, Vergilio JA, Ji P, Kim J, Thiru P, Orkin SH, Lander ES, Lodish HF. MicroRNA-15a and -16–1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc Natl Acad Sci USA 2011;108:1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kochar R, Fallon MB. Pulmonary diseases and the liver. Clin Liver Dis 2011;15:21–37 [DOI] [PubMed] [Google Scholar]
- 42.Kochar R, Nevah Rubin MI, Fallon MB. Pulmonary complications of cirrhosis. Curr Gastroenterol Rep 2011;13:34–39 [DOI] [PubMed] [Google Scholar]
- 43.Laumanns IP, Fink L, Wilhelm J, Wolff JC, Mitnacht-Kraus R, Graef-Hoechst S, Stein MM, Bohle RM, Klepetko W, Hoda MA, et al. The noncanonical WNT pathway is operative in idiopathic pulmonary arterial hypertension. Am J Respir Cell Mol Biol 2009;40:683–691 [DOI] [PubMed] [Google Scholar]
- 44.Moreno-Vinasco L, Gomberg-Maitland M, Maitland ML, Desai AA, Singleton PA, Sammani S, Sam L, Liu Y, Husain AN, Lang RM, et al. Genomic assessment of a multikinase inhibitor, sorafenib, in a rodent model of pulmonary hypertension. Physiol Genomics 2008;33:278–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.