Abstract
A recently developed anti-KIR2DL5 (CD158f) antibody has demonstrated KIR2DL5 expression on the surface of NK and T lymphocytes, making it the last functional KIR identified in the human genome. KIR2DL5 belongs to an ancestral lineage of KIR with Ig-like domains of the D0-D2 type, of which KIR2DL4, an HLA-G receptor, is the only other human member. Despite KIR2DL4 and KIR2DL5 being encoded by genes with similar domain usage, several KIR2DL5 functions resemble more closely those of KIR recognizing classical HLA class I molecules – surface-expressed KIR2DL5 inhibits NK cells through the SHP-2 phosphatase and displays a clonal distribution on NK and T lymphocytes. No activating homolog of KIR2DL5 has been described in any species. The genetics of KIR2DL5 is complicated by duplication of its gene in an ancestor of modern humans living ∼1.7 million years ago. Both KIR2DL5 paralogs have undergone allelic diversification; the centromeric gene is most often represented by alleles whose expression is silenced epigenetically through DNA methylation, thus providing a natural system to investigate the regulation of KIR transcription. The role of KIR2DL5 in immunity is not completely understood, in spite of different attempts to define its ligand. Here we revisit the most relevant characteristics of KIR2DL5, an NK-cell receptor possessing a unique combination of genetic, structural, and functional features.
Keywords: gene polymorphism, human NK cells, inhibitory receptors, KIR, NK cell receptors, transcription
Introduction
KIR2DL5 (CD158f) is the most recently described human KIR expressed on NK and T lymphocytes (Estefanía et al., 2007), for which no ligands have yet been identified. It belongs to an ancestral lineage of KIR with Ig-like domains of the D0-D2 type, whose only other member is KIR2DL4, an HLA-G receptor (Rajagopalan, 2010; Rajagopalan and Long, 2012). Although the KIR2DL5 and KIR2DL4 genes encode proteins with a similar domain organization, distinct structural features make several KIR2DL5 functions resemble more closely those of KIR recognizing classical HLA class I molecules (Table 1).
Table 1.
Structural, genetic, and functional features of KIR2DL5 in comparison with other human KIR.
| KIR2DL5 | KIR2DL4 | KIR2DL1 | KIR3DL1 | |
|---|---|---|---|---|
| Ig-like domains | D0-D2 | D0-D2 | D1-D2 | D0-D1-D2 |
| No. of exons encoding Ig-like domains | 2 | 2 | 2+ pseudoexon | 3 |
| Charged residue in transmembrane | No | Yes | No | No |
| Tyrosine-based signaling motifs | 1 ITIM, 1 ITSM | 1 ITIM | 2 ITIM | 2 ITIM |
| Signaling molecules | SHP-2 > 1 | FcεRγ, DNA-PKcs* | SHP-1 > 2 | SHP-1 > 2 |
| Function | Inhibition | IFNγ secretion, inhibition? | Inhibition | Inhibition |
| Transcription in NK cells | Clonal | Ubiquitous | Clonal | Clonal |
| Ligand | Unknown | HLA-G | HLA-C | HLA-A/B |
| Copy number variation | ++ | ± | + | ±** |
| Conservation in primates | ++ | +++ | − | ± |
The 9.3-kbp KIR2DL5 gene was identified in 2000 by amplification of genomic DNA with oligonucleotide primers recognizing conserved KIR regions (Vilches et al., 2000b) and analysis of the first sequenced KIR haplotype (Wilson et al., 2000). Exon-walking and RACE strategies isolated the complete KIR2DL5 coding region, an open reading frame of 1128 bp encompassing eight exons organized similarly to those of KIR2DL4 – they both lack the fourth exon coding for the D1 Ig-like domain in all other KIR, and encode cytoplasmic tails 20–39 amino acids longer than other human inhibitory KIR (Vilches and Parham, 2002).
This structure is conserved in KIR2DL5 orthologs identified in common and pigmy chimpanzees, gorillas, and orangutans (Khakoo et al., 2000; Rajalingam et al., 2004; Guethlein et al., 2012). Genomic and complementary DNA clones isolated from other Old World primates resemble human KIR2DL5 in part of its sequence or in the domain organization, but true functional orthologs appear to be restricted to hominoids (Hershberger et al., 2001, 2005; Sambrook et al., 2005; Bimber et al., 2008; Abi-Rached et al., 2010; Palacios et al., 2011). No activating homolog of KIR2DL5 has been described in any species, human KIR2DS5 being homologous to HLA-C-specific KIR.
Genetic Organization: Two KIR2DL5 Genes Subjected to Extensive Copy Number Variation
KIR2DL5 is highly polymorphic, like other KIR, and it epitomizes the copy number variation that is a hallmark of the KIR complex. Non-mendelian inheritance and different relative locations of the two most common variants seen in Caucasoids demonstrated that KIR2DL5 alleles belong to two series encoded by different loci (Vilches et al., 2000a; Gómez-Lozano et al., 2002). These loci, designated officially KIR2DL5A and KIR2DL5B, are now often referred to with the suffixes T and C, for their location in the telomeric and the centromeric intervals of the KIR complex, respectively (Marsh et al., 2003; Pyo et al., 2010; Parham et al., 2012).
Linkage to KIR2DS3S5 in KIR-B haplotypes
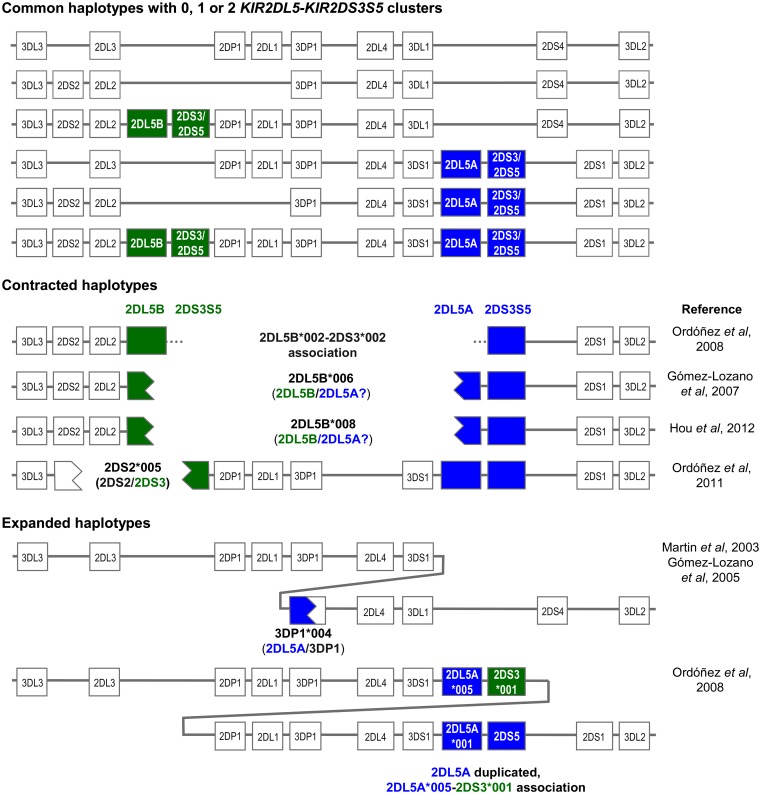
Both the centromeric and the telomeric KIR2DL5 loci are followed by the paralogs of a duplicated KIR2DS3S5 gene, each of which encodes different alleles of the activating KIR 2DS3 and 2DS5, now considered allotypes of each other (Ordóñez et al., 2008; Hou et al., 2010; Pyo et al., 2010). Thus, the centromeric and the telomeric parts of many KIR-B haplotypes are marked by different KIR2DL5-KIR2DS3S5 clusters (Figure 1). The common centromeric sequence KIR2DL5B*002 is associated with KIR2DS3*001, whereas other KIR2DL5B alleles (see below) tend to associate in Black populations with several KIR2DS5 alleles (Hou et al., 2010). On the telomeric side, the predominant KIR2DL5A alleles, *001, and *005, are linked with KIR2DS5*002 and KIR2DS3*002, respectively. At its 5’ end, KIR2DL5B is normally flanked by KIR2DL2, whereas KIR2DL5A is preceded by KIR3DS1 (Vilches et al., 2000a; Pyo et al., 2010).
Figure 1.
Copy number variation and allelic polymorphism of KIR2DL5-KIR2DS3S5 clusters contribute substantial diversity to KIR haplotypes.
Duplication of KIR2DL5 is specific to humans
The KIR2DL5 duplication has not been seen in other primates, and is possibly specific to humans. Pyo et al. (2010) estimated that an ancestral KIR2DL5-KIR2DS3S5 group duplicated ca. 1.7 million years ago, and proposed several models for subsequent diversification through point mutation and recombination. The duplication, seen in all races, is now fixed in our species. However, not every human carries two (or one) KIR2DL5-KIR2DS3S5 clusters, because each is subjected to presence/absence variation, with all A haplotypes and one centromeric B haplotype lacking these genes (Figure 1).
Expanded and contracted KIR haplotypes generated by recombination in the KIR2DL5-KIR2DS3S5 cluster
On the other hand, presence of two highly homologous sequence segments in two different parts of the KIR complex has facilitated subsequent asymmetric (i.e., non-allelic) homologous recombination resulting in contracted and expanded haplotypes (one of them with a third KIR2DL5 locus), often carrying fusion genes or alleles, as represented in Figure 1 (Gómez-Lozano et al., 2003, 2005, 2007; Martin et al., 2003; Ordóñez et al., 2008, 2011; Hou et al., 2012). In contracted haplotypes lacking the central framework KIR genes, assignment of KIR2DL5 and KIR2DS3S5 to the centromeric or the telomeric sides is somewhat arbitrary.
KIR2DL5 Allelic Polymorphism
The KIR2DL5 coding region
KIR2DL5 is represented in the Immuno Polymorphism Database (v2.4.0) by 15 KIR2DL5A and 25 KIR2DL5B alleles (Robinson et al., 2010). Nineteen polymorphic sites have been found within the 1125-bp coding region, of which 11 are non-synonymous. Twelve nucleotide substitutions occurring in exons 3 and 5 create seven amino acid replacements in the extracellular Ig-like domains (Table 2), which may reflect balancing selection having favored polymorphisms that could modulate avidity or specificity in the interaction of KIR2DL5 with unknown ligands. Du et al. (2008) pointed out, however, that many polymorphisms fall out of predicted ligand-interacting loops of the Ig-like domains. Of note, a single polymorphism in exon 1 distinguishes all KIR2DL5A from all KIR2DL5B alleles, whilst many substitutions are shared by alleles of both loci (Table 2). An extensive exchange of genetic material between the centromeric and the telomeric KIR2DL5 loci has taken place during human evolution, as eloquently illustrated by two allele pairs (one from each locus) and a four-allele group (two from each locus) encoding identical mature polypeptides and differing only in their signal peptides. Among 65 additional polymorphisms occurring in KIR2DL5 introns (not shown), none alters its splicing sites.
Table 2.
Comparison of the deduced primary structures and surface expression profiles of KIR2DL5 alleles.
| 2DL5 allele | Leader peptide exons 1–2 |
D0 domain exon 3 |
D2 domain exon 5 |
Stem exon 7 |
Cyt. exon 9 |
Surface expression detectable with UP-R1* | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −16 Ile | −1 Thr | 16 Ala | 78 His | 95 Val | 114 Arg | 152 Asn | 167 Gly | 174 Gly | 215 Arg | 284 Val | ||
| A*001 | – | – | – | – | – | – | – | – | – | – | – | Yes |
| B*008 | Val | – | – | – | – | – | – | – | – | – | – | na |
| A*012 | – | Pro | – | – | – | – | – | – | – | – | – | (Yes) |
| B*006 | Val | Pro | – | – | – | – | – | – | – | – | – | |
| A*015 | – | – | – | – | – | Cys | – | – | – | – | – | ? |
| B*013 | Val | – | – | – | – | Cys | – | – | – | – | – | na |
| A*005 | – | – | – | – | – | – | Asp | – | Ser | – | – | No |
| B*002 | Val | – | – | – | – | – | Asp | – | Ser | – | – | na |
| A*014 | – | – | – | – | – | – | – | Arg | – | – | – | ? |
| B*004 | Val | – | Thr | – | – | – | – | – | – | – | – | na |
| B*007 | Val | – | – | – | Met | – | – | – | – | – | – | na |
| B*003 | Val | Pro | – | – | Met | – | – | – | – | – | – | ? |
| B*011 | Val | Pro | – | – | – | – | – | – | – | – | Ile | na |
| B*009 | Val | – | – | – | – | – | Asp | – | Ser | Leu | – | na |
| B*010 | Val | Pro | – | – | – | – | Asp | – | Ser | – | – | na |
| B*016 | Val | – | – | Tyr | – | – | Asp | – | Ser | – | – | na |
*Yes: surface expression demonstrated by flow cytometry with UP-R1; No: lack of staining with UP-R1 demonstrated in KIR2DL5A*005 cells; (Yes): surface expression and staining with UP-R1 is predictable, according to the allele primary structure and promoter type, but has not been assessed; ?: surface expression and staining with UP-R1 can not be predicted in a transcribed allele, due to polymorphisms in the ectodomain; na: not applicable due to demonstrated or likely lack of transcription; (Yes)/na: different variants of allele KIR2DL5B*006 have functional or non-functional promoters (Table 3). KIR2DL5A alleles are shown in  , and KIR2DL5B ones, in
, and KIR2DL5B ones, in  . Allele order has been arranged to highlight patterns of homology between alleles.
. Allele order has been arranged to highlight patterns of homology between alleles.
Polymorphism in the KIR2DL5 proximal promoter region
The regulatory regions upstream of the KIR2DL5 genes are even more polymorphic – the three first known KIR2DL5 alleles are distinguished by 20–32 nucleotide substitutions in the 1.2-Kbp region immediately 5′ of their start codon (1.6–2.5% variation). A neighbor-joining phylogenetic tree based on the nucleotide sequences of this region sorts KIR2DL5 alleles into three well-differentiated lineages. One of them includes all and only KIR2DL5A alleles; a second lineage comprises multiple KIR2DL5B alleles, of which 2DL5B*0020101 is the prototype; and the third cluster is formed by KIR2DL5B*003 and *00602 (Du et al., 2008). We will refer herein to these clusters as promoters of types I, II, and III. The origin of this divergence, of profound functional importance (alleles controlled by type II promoter are not transcribed), has not been explained.
Distribution of KIR2DL5 alleles
KIR2DL5 is present in all human populations at frequencies ranging from 26 to 86%, but the distributions of the two paralogs and their allotypes are uneven. Whereas KIR2DL5A and KIR2DL5B predominate in Mongoloid and Black populations, respectively, they have similar frequencies in Caucasoids. Alleles KIR2DL5A*001, B*002, and A*005 are widely distributed, accompanied by B*006 in Blacks, who retain the highest KIR2DL5 diversity, and constitute the only human group in which KIR2DL5 alleles controlled by the third type of promoter are not rare (Vilches et al., 2000a; Gómez-Lozano et al., 2007; Du et al., 2008; Middleton et al., 2008; Mulrooney et al., 2008; Hou et al., 2010; González-Galarza et al., 2011 and our own unpublished results).
KIR2DL5 and disease
Data on possible implication of KIR2DL5 copy number variation and polymorphism in susceptibility to disease are scarce. Complex polymorphism and strong linkage disequilibrium with neighboring KIR genes complicates evaluating the individual role of KIR2DL5 as a risk or a protective factor. Search of the PubMed database with the term “KIR2DL5” in June 2012 retrieved 16 citations describing significant deviations of the gene frequency in different diseases and clinical situations (not shown). Among them, only an association between ankylosing spondylitis and presence of KIR2DL5 in the genome of Asian patients has been replicated (Díaz-Peña et al., 2008; Jiao et al., 2008, 2010).
KIR2DL5 genotyping
KIR2DL5 polymorphism has been explored using PCR with sequence-specific primers (SSP) or oligonucleotide-probe hybridization (2008), methods that reliably identify common alleles (Gómez-Lozano and Vilches, 2002; Gómez-Lozano et al., 2007; González et al., 2008). Sequence-based typing (SBT) and mass spectrometry methods that enable studying the entire KIR2DL5 sequence have led to identification of multiple new alleles (Houtchens et al., 2007; Du et al., 2008; Mulrooney et al., 2008; Hou et al., 2010). However, existence of two KIR2DL5 loci poses extra difficulties to genotyping: firstly, because a person having the two loci on both chromosomes may have up to four different KIR2DL5 sequences; secondly, because the alleles of each locus share many single-nucleotide polymorphisms (SNPs). Knowing the phase of KIR2DL5 SNPs is essential for locus/allele assignment, but this is hindered by the hundreds or thousands of base-pairs separating many individual polymorphism (e.g., the only locus-specific SNP in exon 1 is ca. 3 Kbp apart from those in exon 5). The published methods can make tentative assignments of reasonable reliability on samples derived from populations in which the KIR2DL5 allele distribution has been previously investigated in depth, but none of them can assign unambiguously all possible KIR2DL5 genotypes. Separation of KIR2DL5 alleles by locus-specific long-range PCR, followed by probe hybridization or enzymatic sequencing, and long reads of individual DNA molecules by second-generation sequencing are promising strategies for accurate KIR2DL5 genotyping, which remains currently a challenge.
KIR2DL5 Expression
Gene transcription
The fact that highly similar KIR2DL5 coding sequences are controlled by three structurally divergent forms of promoter has profound functional consequences, constituting a valuable natural experiment that provides major insight into the complex regulation of KIR transcription. Of the KIR2DL5 alleles whose transcription has been investigated, those controlled by type I or type III promoters feature variegated patterns of expression; whilst mRNA of alleles controlled by type II promoters is undetectable (Vilches et al., 2000a; Gómez-Lozano et al., 2007). No single exception to this rule has ever been described; furthermore, the KIR3DP1 pseudogene, also controlled by a type II promoter, is transcriptionally silent too, with a key exception: as the empirical rule predicted, KIR3DP1*004, which gained a type I promoter through recombination with KIR2DL5A, is an expressed allele (Vilches et al., 2000b; Gómez-Lozano et al., 2005).
Consistent with the epigenetic regulation of KIR genes (Santourlidis et al., 2002; Chan et al., 2003), lack of transcription of the silent KIR2DL5B*002 allele correlates with a hypermethylated status of CpG islands in its promoter. Furthermore, pharmacological DNA demethylation of cultured NK cells suffices for restoring KIR2DL5B*002 transcription, demonstrating that only an epigenetic mechanism prevents its expression (Gómez-Lozano et al., 2007). In agreement with this are studies of transiently transfected promoters controlling a reporter gene, an in vitro situation in which epigenetic regulation is not relevant. In this setting, the promoters of naturally silent KIR2DL5 alleles tend to show similar or higher activities than functional KIR alleles (Gómez-Lozano et al., 2007; Mulrooney et al., 2008).
Among the sequence patterns that distinguish the three types of KIR2DL5 promoter, only two linked SNPs at nucleotides 97 and 84 upstream of the start codon correlate completely with the expression pattern: GA is seen in transcribed alleles, and AG in silent ones (Table 3). Nucleotide −97G lies within a TGTGGT motif that provides a core binding site for the RUNX family of transcription factors (Vilches et al., 2000a). RUNX3 is recruited from nuclear extracts of NK cells by probes derived from KIR2DL5 alleles having an intact motif, but not by those carrying the −97G > A mutation (Gómez-Lozano et al., 2007). In support of an essential role for RUNX in KIR expression is conservation of its binding motif in all human KIR with clonal transcription (Trompeter et al., 2005; van Bergen et al., 2005; Presnell et al., 2006); and demonstration that two redundant RUNX binding sites, highly conserved in primates, are possibly essential for expression of KIR2DL4, a gene that is transcribed ubiquitously in NK cells (Presnell et al., 2012).
Table 3.
Comparison of the proximal promoter sequences and transcription profiles of KIR2DL5 alleles.
| KIR2DL5 allele | Promoter region |
Promoter type | Transcription | |||||
|---|---|---|---|---|---|---|---|---|
| −104 G | −97 G | −84 A | −27 C | −23 C | −10 C | |||
| A*0010101–00105 | – | – | – | – | – | – | I | + |
| A*0050101/03–04 | – | – | – | – | – | – | I | + |
| A*01201/02 | – | – | – | – | – | – | I | (+) |
| A*0050102 | A | – | – | – | – | – | I | (+) |
| B*0020101–03/05/07, *0070101 | A | A | G | – | T | – | II | − |
| B*004, *0080101/00802, *009, *01301/02 | A | A | G | – | T | – | II | − |
| B*00601/03 | A | A | G | – | – | – | II | − |
| B*0020104, *00202, *0080102, *010, *011 | A | A | G | – | – | – | II | (−) |
| B*003 | A | – | – | T | – | T | III | + |
| B*00602 | A | – | – | T | – | T | III | (+) |
Transcription of the alleles shown in bold face has been assessed experimentally; in parentheses are shown predictions for groups of alleles of which no member has been studied by RT-PCR. The promoter regions of KIR2DL5 alleles A*014, A*015, B*0020106, B*0070102, B*01303, and B*016 have not been sequenced.
KIR transcription is controlled not only by a proximal promoter, but also by the complex interaction of additional regulatory elements (Cichocki et al., 2011). In brief, a distal, non-tissue-specific promoter element located ∼1.1 Kbp upstream of the KIR start codon has been suggested to induce histone modifications that facilitate subsequent function of the proximal promoter. The latter is actually bidirectional – reverse transcripts derived from it have been proposed to repress KIR expression and favor epigenetic silencing, whilst predominance of forward transcription would result in KIR expression. Finally, an additional reverse promoter element in intron 2 appears to function in early NK-cell progenitors. It has been suggested that the RUNX role might be to down-regulate the antisense promoter activity during NK-cell ontogeny, thus favoring a local open chromatin conformation at the bound KIR gene, and its subsequent expression in the mature cell (Davies et al., 2007; Cichocki et al., 2011). Consistent with this hypothesis is that the only reverse KIR transcripts detected in CD56bright NK cells (possible precursors of KIR+CD56dim cells) are those derived from genes with promoters lacking the RUNX binding site – KIR2DL5B*002 and KIR3DP1 (Davies et al., 2007).
Other locus- and allele-specific polymorphisms of these regulatory elements influence KIR transcription and may also help us understand mechanisms controlling KIR2DL5 expression. For instance, a Ying Yang-1 (YY1) binding site conserved in many proximal KIR promoters is mutated in KIR 2DL1, 2DS1/S3/S5, and all KIR2DL5 alleles, which correlates with enhanced reverse transcription (Davies et al., 2007; Li et al., 2008). This phenomenon may induce low forward activity of the KIR2DL5 promoter, which might be associated with the receptor being generally expressed at low levels on the surface of only small proportions of NK cells (Estefanía et al., 2007). Likewise, disruption of the Sp1 site in the promoter of the expressed allele KIR2DL5B*003 (−27C > T, Table 3) decreases its forward activity in vitro and has been proposed to reduce its expression on NK cells (Li et al., 2008), which needs experimental confirmation.
Of possible interest, none of the transcriptionally silent KIR2DL5B alleles bear structural abnormalities in their reading frames (in contrast with other human KIR, no null KIR2DL5 alleles have yet been identified; Vilches et al., 2000a; Robinson et al., 2010). The fact that KIR2DL5B generally retains an intact structure could mean that inactivation of its expression is evolutionarily recent (the mutated RUNX site is not seen in other hominoids, personal communication of Libby Guethlein, Stanford University); or that the gene still serves an unknown function.
Cell surface expression
Generation of a specific monoclonal antibody (clone UP-R1) enabled us to characterize KIR2DL5 surface expression (Estefanía et al., 2007). Like most other KIR (and contrasting with the non-clonal expression of KIR2DL4), KIR2DL5 features a variegated pattern on the surface of CD56dim NK cells and on T lymphocytes from peripheral blood, in agreement with the clonal distribution seen by reverse transcription (RT) PCR in mRNAs isolated from NK- and T-cell clones (Vilches et al., 2000b; Estefanía et al., 2007). The proportion of NK cells expressing KIR2DL5 tends to be lower than 10% in most healthy individuals. That proportion is even lower in T lymphocytes, of the CD8 subset in their vast majority. The receptor density on the surface, as assessed by the median fluorescence intensity (MFI) value in flow cytometry with mAb UP-R1, is also lower than for several other KIR in resting lymphocytes, but it increases, to a lesser extent, upon expansion in presence of IL-2 and lymphoblastoid cell lines (our own observation). These features might owe to weak promoters controlling the transcription of functional KIR2DL5 alleles.
Interestingly, higher numbers of KIR2DL5+ cells (20% of total NK lymphocytes) have been reported in a TAP-deficient woman. Furthermore, the phenotype of this patient also differed from that of most TAP-deficient individuals in her resting NK cells retaining cytotoxic capacity against allogeneic targets without pre-activation (Zimmer et al., 2009). The exact mechanisms determining this behavior remain to be ascertained.
Analysis of bulk and cloned NK cells by flow cytometry and RT-PCR reveals no coordinated expression of KIR2DL5 with other KIR, but rather an apparently random distribution (Vilches et al., 2000b; Estefanía et al., 2007). Importantly, a minority of NK cells expresses KIR2DL5 but neither other inhibitory KIR, nor the inhibitory lectin-like receptor NKG2A. Existence of this subpopulation is consistent with a capacity of KIR2DL5 to license NK cells, but this has not been demonstrated functionally. Also lacking are studies on possible patterns of co-expression of KIR2DL5 and LILRB1, the third lineage of inhibitory MHC class I receptors expressed by human NK cells.
Allelic polymorphism is essential for understanding the different patterns of KIR2DL5 expression (Table 2). Transcriptionally silent alleles are, obviously, undetectable on the cell surface by definition. Furthermore, only allele KIR2DL5A*001 has been formally demonstrated to be expressed on the cell surface. In contrast, NK cells transcribing allele KIR2DL5A*005 are not stained by mAb UP-R1 in flow cytometry (Gómez-Lozano et al., 2007). Whether this is due to a lack of surface expression, to the UP-R1 epitope being altered in KIR2DL5A*005 by its D2-domain polymorphisms, or to a combination of both factors, has not yet been elucidated. Surface expression and recognition by UP-R1 of other transcribed KIR2DL5 alleles has, to the best of our knowledge, never been evaluated. Amongst other transcriptionally active KIR2DL5 alleles, A*012 and B*00602 code for mature polypeptides identical to A*001, therefore they are predictably surface-expressed and detected by UP-R1; whereas expression and UP-R1 recognition of KIR2DL5B*003, A*014, and A*15 (each bearing one amino acid replacement in the Ig-like domains in comparison with A*001) needs to be tested empirically (Table 2).
KIR2DL5 Function
KIR2DL5 inhibits NK cells
KIR2DL5 is predominantly expressed on the cell surface as a glycosylated monomer of ∼60 kDa (Estefanía et al., 2007). Its cytoplasmic tail contains one canonical (VxYxxL) immunoreceptor tyrosine-based inhibitory motif (ITIM) separated by 24 amino acid residues from an atypical ITIM sequence (TxYxxL) similar to the immunoreceptor tyrosine-based switch motifs (ITSM) seen in 2B4, SLAM, and other receptors (Vilches et al., 2000b; Yusa et al., 2004). The latter motif, not seen in other human KIR, does not confer upon KIR2DL5 the capacity to recruit and signal through the SLAM-associated protein (SAP), but it is conserved in KIR2DL5 orthologs of other hominoids (Rajalingam et al., 2004; Yusa et al., 2004).
Since the KIR2DL5 ligand is unknown, its actual inhibitory character in physiological conditions has not been explored. Cross-linkage of naturally expressed KIR2DL5 inhibits NK-cell cytotoxicity against mAb-coated P815 target cells to an extent comparable to that seen with the “classical” KIR 3DL1 (Estefanía et al., 2007). This result is in agreement with that obtained previously using NK92 cells transduced with a chimera containing a KIR3DL1 ectodomain fused to the KIR2DL5 cytoplasmic tail; such chimera, however, displayed a lower capacity to inhibit NK92-target conjugation than full-length KIR3DL1 (Yusa et al., 2004). Based on the results obtained with a mutated KIR3DL1/2DL5 chimera, Yusa et al. (2004) proposed that the canonical KIR2DL5 ITIM, but not its ITSM-like motif, is essential for its inhibitory capacity in transduced NK92 cells.
Experiments performed independently on transduced NK92 cells and on NK cells expressing endogenous KIR2DL5 demonstrated that the phosphorylated receptor recruits the Src homology region 2-containing protein tyrosine phosphatase 2 (SHP-2) preferentially over SHP-1 in comparison with other KIR (Yusa et al., 2004; Estefanía et al., 2007). Furthermore, the inhibitory effect of the KIR2DL5 tail in transduced NK92 cells was prevented by a dominant-negative (DN) SHP-2, but only to a lesser extent by DN SHP-1 (Yusa et al., 2004). The importance of KIR2DL5 using predominantly a SHP-2-dependent pathway for its function has not been explored in depth.
KIR2DL5, an orphan receptor
Demonstration that KIR2DL5 is a surface-expressed glycoprotein capable of inhibiting cytotoxic lymphocytes suggested that this molecule participated in NK-cell mediated defense according to the missing-self model. Such a possibility was reinforced by identification of NK cells which express KIR2DL5 and lack all other detectable inhibitory KIR and NKG2A, and it implies existence of a cellular ligand, possibly expressed in physiological conditions. Enhanced KIR2DL5 expression and retention of NK-cell cytotoxicity in a TAP-deficient patient (Zimmer et al., 2009) suggest that she possibly expressed a ligand capable of licensing KIR2DL5+ NK cells.
As a first approach to investigate expression of a KIR2DL5 ligand, we made a fusion protein containing the KIR2DL5 ectodomains and the Fc of human IgG1. The fusion protein, along with positive and negative controls (KIR2DL1-, KIR2DL2-, and non-fused Fc constructs kindly donated by Dr. Eric Long), was produced in human embryonic kidney (HEK)-293T cells, and used in indirect flow cytometry experiments on multiple cell lines grown in vitro. In these experiments, we observed a dull staining of, essentially, every human cell line of hematopoietic origin. Such staining seemed independent of the cells HLA allotypes; furthermore, it was apparently not affected by lack of surface HLA expression in mutant cell lines (results not shown). However, the variably low signal-to-noise ratios with which the positive controls often stained cells expressing their known ligands, and the variable behavior of different batches of fusion proteins of known specificity indicated that the method did not attain sufficient sensitivity and consistency in our hands to allow screening for an unknown ligand in a series of heterogeneous cell types.
As an alternative approach of possibly higher sensitivity, we tried to apply the MHC-tetramer technology to build KIR2DL5 forms of higher avidity. The first codons of a KIR2DL5 cDNA were adapted by site-directed mutagenesis to the codon usage bias of Escherichia coli (Nakamura et al., 2000), for higher protein yield; and the construct encoding the Ig-like and stem regions of KIR2DL5 was subcloned into the pGMT7 plasmid (a kind gift of Dr. Veronique Braud), which provided an in-frame recognition sequence for the BirA biotinylase at the carboxy-terminal end of the construct. Upon IPTG induction, the recombinant KIR2DL5 protein was efficaciously produced in strain BL21(DE3)pLysS. After purification from inclusion bodies, the KIR2DL5 ectodomain was solubilized in concentrated urea, refolded in an arginine/gluthation buffer, and biotinylated with BirA. However, the labeled KIR2DL5 protein could not be quantitatively recovered after molecular exclusion chromatography, apparently due to aggregation, even in presence of mild detergents like Chaps and Octyl-b-d-glycopyranoside.
The ability to identify KIR2DL5 with a novel specific monoclonal antibody opened new possibilities for studying the outcome of the interaction of KIR2DL5-positive NK lymphocytes with potential target cells. For instance, we attempted to study differential degranulation (assessed by CD107a expression) of KIR2DL5-positive and -negative NK cells against different target cells. However, several hindrances made this approach unpractical, including: low levels of degranulation induced in freshly isolated NK cells by many targets, which made it difficult to evaluate any further reduction attributable to inhibition through KIR2DL5; the low proportions of NK cells expressing KIR2DL5 in most donors, which do not readily increase during in vitro NK-cell expansion in response to lymphoblastoid cell lines. These studies indicated that use of cells homogeneously expressing KIR2DL5, and of a positive readout (rather than inhibition of another signal) are more promising approaches for screening the interaction of KIR2DL5 with potential ligand molecules. Knowing such interactions is essential for understanding the role of KIR2DL5 in immunity, and its importance for human health.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are indebted to Drs. Peter Parham, Frances Brodsky, Alessandro Moretta, and Campbell Witt for helpful discussions on strategies for investigating KIR2DL5 ligands. We thank Ernesto Estefanía and David Ordóñez for previous contributions to those investigations. Research on KIR2DL5 was supported by successive grants from the Spanish Instituto de Salud Carlos III and Plan Nacional de I + D, including SAF2010-22153-C03.
References
- Abi-Rached L., Kuhl H., Roos C., ten Hallers B., Zhu B., Carbone L., de Jong P. J., Mootnick A. R., Knaust F., Reinhardt R., Parham P., Walter L. (2010). A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons. J. Immunol. 184, 1379–1391 10.4049/jimmunol.0903016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimber B. N., Moreland A. J., Wiseman R. W., Hughes A. L., O’Connor D. H. (2008). Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J. Immunol. 181, 6301–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H. W., Kurago Z. B., Stewart C. A., Wilson M. J., Martin M. P., Mace B. E., Carrington M., Trowsdale J., Lutz C. T. (2003). DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J. Exp. Med. 197, 245–255 10.1084/jem.20021127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki F., Miller J. S., Anderson S. K. (2011). Killer immunoglobulin-like receptor transcriptional regulation: a fascinating dance of multiple promoters. J. Innate Immun. 3, 242–248 10.1159/000323929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G. E., Locke S. M., Wright P. W., Li H., Hanson R. J., Miller J. S., Anderson S. K. (2007). Identification of bidirectional promoters in the human KIR genes. Genes Immun. 22, 22. [DOI] [PubMed] [Google Scholar]
- Díaz-Peña R., Blanco-Gelaz M. A., Suarez-Alvarez B., Martinez-Borra J., Lopez-Vazquez A., Alonso-Arias R., Bruges-Armas J., Vidal-Castineira J. R., Lopez-Larrea C. (2008). Activating KIR genes are associated with ankylosing spondylitis in Asian populations. Hum. Immunol. 69, 437–442 10.1016/j.humimm.2008.04.012 [DOI] [PubMed] [Google Scholar]
- Du Z., Sharma S. K., Spellman S., Reed E. F., Rajalingam R. (2008). KIR2DL5 alleles mark certain combination of activating KIR genes. Genes Immun. 9, 470–480 10.1038/gene.2008.39 [DOI] [PubMed] [Google Scholar]
- Estefanía E., Flores R., Gómez-Lozano N., Aguilar H., López-Botet M., Vilches C. (2007). Human KIR2DL5 is an inhibitory receptor expressed on the surface of NK and T lymphocyte subsets. J. Immunol. 178, 4402–4410 [DOI] [PubMed] [Google Scholar]
- Gómez-Lozano N., de Pablo R., Puente S., Vilches C. (2003). Recognition of HLA-G by the NK cell receptor KIR2DL4 is not essential for human reproduction. Eur. J. Immunol. 33, 639–644 10.1002/eji.200323741 [DOI] [PubMed] [Google Scholar]
- Gómez-Lozano N., Estefanía E., Williams F., Halfpenny I., Middleton D., Solís R., Vilches C. (2005). The silent KIR3DP1 gene (CD158c) is transcribed and might encode a secreted receptor in a minority of humans, in whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 genes are duplicated. Eur. J. Immunol. 35, 16–24 10.1002/eji.200425493 [DOI] [PubMed] [Google Scholar]
- Gómez-Lozano N., Gardiner C. M., Parham P., Vilches C. (2002). Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics 54, 314–319 10.1007/s00251-002-0476-2 [DOI] [PubMed] [Google Scholar]
- Gómez-Lozano N., Trompeter H. I., de Pablo R., Estefanía E., Uhrberg M., Vilches C. (2007). Epigenetic silencing of potentially functional KIR2DL5 alleles: implications for the acquisition of KIR repertoires by NK cells. Eur. J. Immunol. 37, 1954–1965 10.1002/eji.200737277 [DOI] [PubMed] [Google Scholar]
- Gómez-Lozano N., Vilches C. (2002). Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: an update. Tissue Antigens 59, 184–193 10.1034/j.1399-0039.2002.590302.x [DOI] [PubMed] [Google Scholar]
- González A., Meenagh A., Sleator C., Middleton D. (2008). Investigation of killer cell immunoglobulin-like receptor (KIR) gene diversity: KIR2DL2, KIR2DL5 and KIR2DS5. Tissue Antigens 72, 11–20 10.1111/j.1399-0039.2008.01050.x [DOI] [PubMed] [Google Scholar]
- González-Galarza F. F., Christmas S., Middleton D., Jones A. R. (2011). Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 39, D913–D919 10.1093/nar/gkq1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guethlein L. A., Abi-Rached L., Norman P. J., Babrzadeh F., de Groot N. G., Bontrop R. E., Parham P. (2012). Orangutan KIR haplotype variation (abstract). Tissue Antigens 79, 420 [Google Scholar]
- Hershberger K. L., Kurian J., Korber B. T., Letvin N. L. (2005). Killer cell immunoglobulin-like receptors (KIR) of the African-origin sabaeus monkey: evidence for recombination events in the evolution of KIR. Eur. J. Immunol. 35, 922–935 10.1002/eji.200425408 [DOI] [PubMed] [Google Scholar]
- Hershberger K. L., Shyam R., Miura A., Letvin N. L. (2001). Diversity of the killer cell Ig-like receptors of rhesus monkeys. J. Immunol. 166, 4380–4390 [DOI] [PubMed] [Google Scholar]
- Hou L., Chen M., Jiang B., Wu D., Ng J., Hurley C. K. (2010). Thirty allele-level haplotypes centered around KIR2DL5 define the diversity in an African American population. Immunogenetics 62, 491–498 10.1007/s00251-010-0458-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L., Chen M., Ng J., Hurley C. K. (2012). Conserved KIR allele-level haplotypes are altered by microvariation in individuals with European ancestry. Genes Immun. 13, 47–58 10.1038/gene.2011.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtchens K. A., Nichols R. J., Ladner M. B., Boal H. E., Sollars C., Geraghty D. E., Davis L. M., Parham P., Trachtenberg E. A. (2007). High-throughput killer cell immunoglobulin-like receptor genotyping by MALDI-TOF mass spectrometry with discovery of novel alleles. Immunogenetics 59, 525–537 10.1007/s00251-007-0222-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y. L., Ma C. Y., Wang L. C., Cui B., Zhang J., You L., Chen Z. J., Li J. F., Zhao Y. R. (2008). Polymorphisms of KIRs gene and HLA-C alleles in patients with ankylosing spondylitis: possible association with susceptibility to the disease. J. Clin. Immunol. 28, 343–349 10.1007/s10875-008-9183-6 [DOI] [PubMed] [Google Scholar]
- Jiao Y. L., Zhang B. C., You L., Li J. F., Zhang J., Ma C. Y., Cui B., Wang L. C., Chen Z. J., Zhao Y. R. (2010). Polymorphisms of KIR gene and HLA-C alleles: possible association with susceptibility to HLA-B27-positive patients with ankylosing spondylitis. J. Clin. Immunol. 30, 840–844 10.1007/s10875-010-9444-z [DOI] [PubMed] [Google Scholar]
- Khakoo S. I., Rajalingam R., Shum B. P., Weidenbach K., Flodin L., Muir D. G., Canavez F., Cooper S. L., Valiante N. M., Lanier L. L., Parham P. (2000). Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity 12, 687–698 10.1016/S1074-7613(00)80219-8 [DOI] [PubMed] [Google Scholar]
- Kikuchi-Maki A., Catina T. L., Campbell K. S. (2005). Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor gamma protein. J. Immunol. 174, 3859–3863 [DOI] [PubMed] [Google Scholar]
- Li H., Pascal V., Martin M. P., Carrington M., Anderson S. K. (2008). Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 4, e1000254. 10.1371/journal.pgen.1000254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh S. G., Parham P., Dupont B., Geraghty D. E., Trowsdale J., Middleton D., Vilches C., Carrington M., Witt C., Guethlein L. A., Shilling H., Garcia C. A., Hsu K. C., Wain H. (2003). Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Tissue Antigens 62, 79–86 10.1034/j.1399-0039.2003.00072.x [DOI] [PubMed] [Google Scholar]
- Martin M. P., Bashirova A., Traherne J., Trowsdale J., Carrington M. (2003). Cutting edge: expansion of the KIR locus by unequal crossing over. J. Immunol. 171, 2192–2195 [DOI] [PubMed] [Google Scholar]
- Middleton D., Meenagh A., Moscoso J., Arnaiz-Villena A. (2008). Killer immunoglobulin receptor gene and allele frequencies in Caucasoid, Oriental and Black populations from different continents. Tissue Antigens 71, 105–113 10.1111/j.1399-0039.2007.00973.x [DOI] [PubMed] [Google Scholar]
- Mulrooney T. J., Hou L., Steiner N. K., Chen M., Belle I., Ng J., Hurley C. K. (2008). Promoter variants of KIR2DL5 add to diversity and may impact gene expression. Immunogenetics 60, 287–294 10.1007/s00251-008-0273-7 [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Gojobori T., Ikemura T. (2000). Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28, 292. 10.1093/nar/28.1.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordóñez D., Gómez-Lozano N., Rosales L., Vilches C. (2011). Molecular characterisation of KIR2DS2*005, a fusion gene associated with a shortened KIR haplotype. Genes Immun. 12, 544–551 10.1038/gene.2011.35 [DOI] [PubMed] [Google Scholar]
- Ordóñez D., Meenagh A., Gómez-Lozano N., Castaño J., Middleton D., Vilches C. (2008). Duplication, mutation and recombination of the human orphan gene KIR2DS3 contribute to the diversity of KIR haplotypes. Genes Immun. 9, 431–437 10.1038/gene.2008.34 [DOI] [PubMed] [Google Scholar]
- Palacios C., Cuervo L. C., Cadavid L. F. (2011). Evolutionary patterns of killer cell Ig-like receptor genes in old world monkeys. Gene 474, 39–51 10.1016/j.gene.2010.12.006 [DOI] [PubMed] [Google Scholar]
- Parham P., Norman P. J., Abi-Rached L., Guethlein L. A. (2012). Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 800–811 10.1098/rstb.2011.0266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnell S. R., Zhang L., Chlebowy C. N., Al-Attar A., Lutz C. T. (2012). Differential transcription factor use by the KIR2DL4 promoter under constitutive and IL-2/15-treated conditions. J. Immunol. 188, 4394–4404 10.4049/jimmunol.1103352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnell S. R., Zhang L., Ramilo C. A., Chan H. W., Lutz C. T. (2006). Functional redundancy of transcription factor-binding sites in the killer cell Ig-like receptor (KIR) gene promoter. Int. Immunol. 18, 1221–1232 10.1093/intimm/dxl043 [DOI] [PubMed] [Google Scholar]
- Pyo C. W., Guethlein L. A., Vu Q., Wang R., Abi-Rached L., Norman P. J., Marsh S. G., Miller J. S., Parham P., Geraghty D. E. (2010). Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE 5, e15115. 10.1371/journal.pone.0015115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S. (2010). Endosomal signaling and a novel pathway defined by the natural killer receptor KIR2DL4 (CD158d). Traffic 11, 1381–1390 10.1111/j.1600-0854.2010.01112.x [DOI] [PubMed] [Google Scholar]
- Rajagopalan S., Long E. O. (2012). KIR2DL4 (CD158d): an activation receptor for HLA-G. Front. Immun. 3:258. 10.3389/fimmu.2012.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam R., Parham P., Abi-Rached L. (2004). Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J. Immunol. 172, 356–369 [DOI] [PubMed] [Google Scholar]
- Robinson J., Mistry K., McWilliam H., Lopez R., Marsh S. G. (2010). IPD – the Immuno Polymorphism Database. Nucleic Acids Res. 38, D863–D869 10.1093/nar/gkp879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. G., Bashirova A., Palmer S., Sims S., Trowsdale J., Abi-Rached L., Parham P., Carrington M., Beck S. (2005). Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 15, 25–35 10.1101/gr.2381205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santourlidis S., Trompeter H. I., Weinhold S., Eisermann B., Meyer K. L., Wernet P., Uhrberg M. (2002). Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J. Immunol. 169, 4253–4261 [DOI] [PubMed] [Google Scholar]
- Trompeter H. I., Gomez-Lozano N., Santourlidis S., Eisermann B., Wernet P., Vilches C., Uhrberg M. (2005). Three structurally and functionally divergent kinds of promoters regulate expression of clonally distributed killer cell Ig-like receptors (KIR), of KIR2DL4, and of KIR3DL3. J. Immunol. 174, 4135–4143 [DOI] [PubMed] [Google Scholar]
- van Bergen J., Stewart C. A., van den Elsen P. J., Trowsdale J. (2005). Structural and functional differences between the promoters of independently expressed killer cell Ig-like receptors. Eur. J. Immunol. 35, 2191–2199 10.1002/eji.200526201 [DOI] [PubMed] [Google Scholar]
- Vilches C., Gardiner C. M., Parham P. (2000a). Gene structure and promoter variation of expressed and non-expressed variants of the KIR2DL5 gene. J. Immunol. 165, 6416–6421 [DOI] [PubMed] [Google Scholar]
- Vilches C., Rajalingam R., Uhrberg M., Gardiner C. M., Young N. T., Parham P. (2000b). KIR2DL5, a novel killer-cell receptor with a D0-D2 configuration of Ig-like domains. J. Immunol. 164, 5797–5804 [DOI] [PubMed] [Google Scholar]
- Vilches C., Parham P. (2002). KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol. 20, 217–251 10.1146/annurev.immunol.20.092501.134942 [DOI] [PubMed] [Google Scholar]
- Wilson M. J., Torkar M., Haude A., Milne S., Jones T., Sheer D., Beck S., Trowsdale J. (2000). Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. U.S.A. 97, 4778–4783 10.1073/pnas.080278897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa S., Catina T. L., Campbell K. S. (2004). KIR2DL5 can inhibit human NK cell activation via recruitment of Src homology region 2-containing protein tyrosine phosphatase-2 (SHP-2). J. Immunol. 172, 7385–7392 [DOI] [PubMed] [Google Scholar]
- Zimmer J., Poli A., Villa-Forte A., Hentges F., Vilches C., de la Salle H. (2009). A new case of TAP deficiency with cytotoxic NK cells (abstract). Tissue Antigens 73, 386–386 [Google Scholar]