Abstract
The collection of components required to carry out the intricate processes involved in generating and maintaining a living, breathing and, sometimes, thinking organism is staggeringly complex. Where do all of the parts come from? Early estimates stated that about 100,000 genes would be required to make up a mammal; however, the actual number is less than one-quarter of that, barely four times the number of genes in budding yeast. It is now clear that the ‘missing’ information is in large part provided by alternative splicing, the process by which multiple different functional messenger RNAs, and therefore proteins, can be synthesized from a single gene.
Alternative splicing of precursor messenger RNA (pre-mRNA) was first described almost 30 years ago, when it was discovered that membrane-bound and secreted antibodies are encoded by the same gene1,2. Soon after, more examples of this phenomenon were described: for instance, the calcitonin/calcitonin-related polypeptide gene encodes two peptide hormones, which are expressed in a tissue-specific manner3. In both of these examples, the differential inclusion and exclusion of exonic sequences through alternative splicing (and alternative polyadenylation) is responsible for doubling the number of proteins encoded by the genes. Although it was at one time considered unusual, the estimated number of genes that encode more than one protein (or protein isoform) as a result of alternative splicing of a pre-mRNA has steadily risen over time. Recent studies using high-throughput sequencing indicate that 95–100% of human pre-mRNAs that contain sequence corresponding to more than one exon are processed to yield multiple mRNAs4,5. And not only do most genes encode pre-mRNAs that are alternatively spliced, but also the number of mRNA isoforms encoded by a single gene can vary from two to several thousand. One spectacular example is the Drosophila melanogaster gene Down syndrome cell adhesion molecule (Dscam), which can generate 38,016 distinct mRNA isoforms6, a number far in excess of the total number of genes (~14,500) in the organism.
If the enormous diversity of proteins that can be generated by alternative splicing is considered together with other processes such as the use of alternative transcription start sites, alternative polyadenylation and RNA editing, as well as post-translational modification (such as phosphorylation, ubiquitylation and SUMOylation; a largely unexplored area), the number of functionally distinct proteins that could be encoded by the genome is staggering. Although the relative extent to which these and other mechanisms and modifications contribute to increasing protein diversity is unclear, alternative splicing is one of the main sources of proteomic diversity in multicellular eukaryotes.
In this Review, we discuss the complexity of alternative splicing and consider the mechanisms that are known, or suspected, to be involved in regulating the choice of splice site. We then outline the important outstanding questions in the field and speculate on the evolutionary implications of expanded proteomic diversity.
Diverse manifestations of alternative splicing
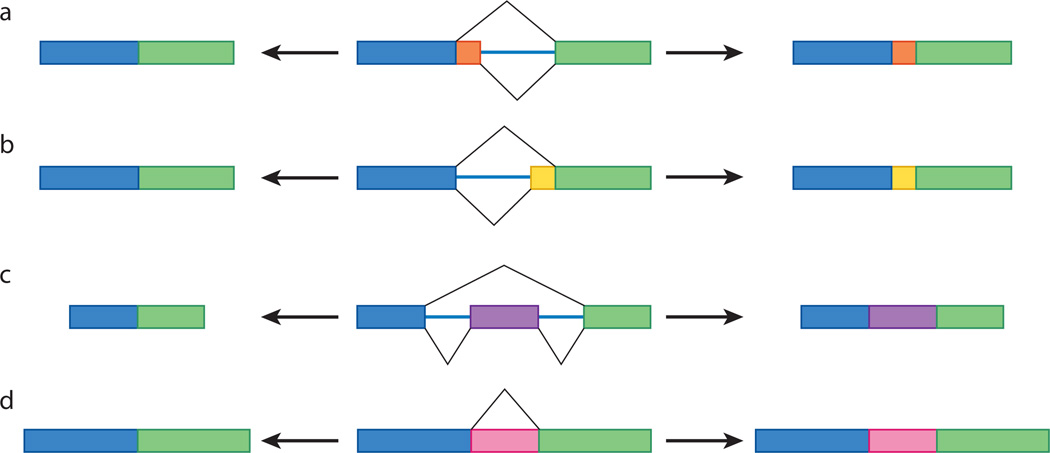
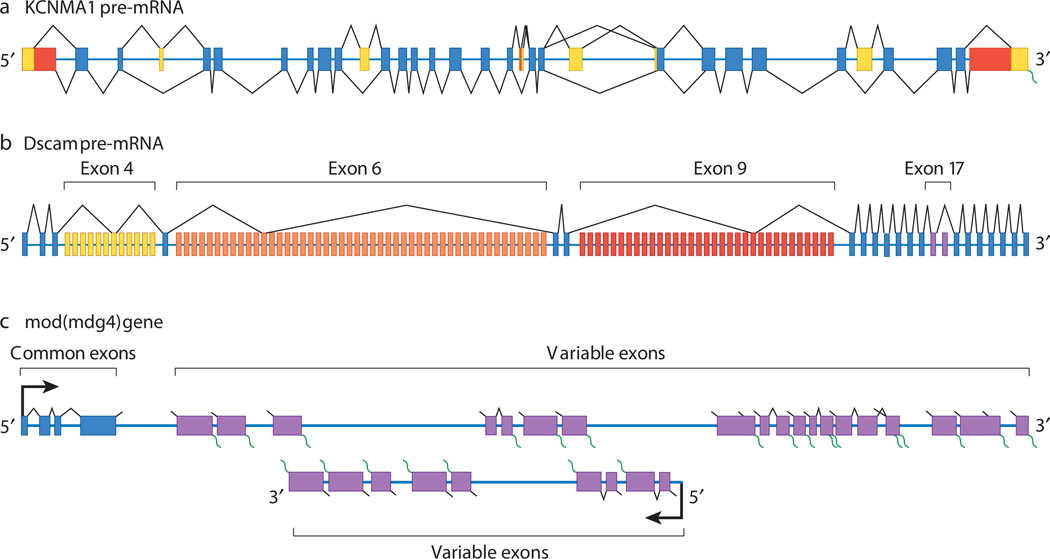
Alternative splicing involves the differential use of splice sites to create protein diversity. Nearly all instances of alternative splicing result from the use of one or more of four basic modules: alternative 5′ splice-site choice, alternative 3′ splice-site choice, cassette-exon inclusion or skipping, and intron retention (Fig. 1). Genes that encode a small number of mRNA isoforms typically contain only one or two of these modules. For instance, two mRNAs could be synthesized from a gene containing a single cassette exon: one mRNA that contains the exonic sequence, and one that lacks it. By contrast, tremendously diverse mRNA repertoires can be generated from genes constructed with many modules. For example, the human gene KCNMA1 (also known as SLO) contains numerous modules, including alternative 5′ splice sites, alternative 3′ splice sites and cassette exons, and more than 500 mRNA isoforms of KCNMA1 can be generated7,8 (Fig. 2a). Similarly, the 38,016 potential Dscam mRNA isoforms are synthesized from a gene containing 95 cassette exons, which are spliced from the pre-mRNA in a mutually exclusive manner (that is, each mRNA isoform contains sequence corresponding to only one cassette exon)6 (Fig. 2b). An even more elaborate manifestation of alternative splicing is exemplified by the D. melanogaster gene modifier of mdg4 (mod(mdg4)), which encodes 28 mRNAs. These are generated by trans-splicing of sequence corresponding to a group of common exons, present on one pre-mRNA, to sequence corresponding to one set of 28 variable exons, present on separate pre-mRNAs9,10 (Fig. 2c). It is not known whether similar trans-splicing events occur in other multicellular eukaryotes.
Figure 1. Types of alternative splicing.
There are four basic types of alternative splicing: alternative 5′ splice-site selection (a), alternative 3′ splice-site selection (b), cassette-exon inclusion or skipping (c) and intron retention (d). The rectangles in the centre represent pre-mRNAs. For each pre-mRNA, the black lines span the regions that can be spliced out, with the lines above corresponding to the mature mRNA shown on the left and the lines below to the mRNA on the right. That is, the mRNA that is synthesized when the central exon (or intron in d) is skipped is shown on the left, and the mRNA that is synthesized when this sequence is included is shown on the right. In d, the pink portion is considered an exon when included (right) and an intron when skipped (left).
Figure 2. The generation of diverse mRNA repertoires.
a, Human KCNMA1 pre-mRNA is depicted, with constitutive exons in blue and alternative exons in yellow and red. The possible splicing patterns for each exon are indicated above and below the pre-mRNA (black lines).The KCNMA1 pre-mRNA contains multiple alternative 5′ splice sites, alternative 3′ splice sites and sequence corresponding to cassette exons. Together, these allow more than 500 mRNA isoforms to be synthesized from a single pre-mRNA. b, Drosophila melanogaster Dscam pre-mRNA is depicted, with constitutive exons in blue and alternative exons in the exon 4, 6, 9 and 17 clusters shown in yellow, orange, red and purple, respectively. The splicing pattern for one mRNA isoform is shown as an example. The Dscam gene contains 95 alternative exons organized into four clusters of mutually exclusive exons (that is, only exon from each cluster is transcribed). These four clusters are at exon 4, exon 6, exon 9 and exon 17, which contain 12, 48, 33 and 2 variable exons, respectively. In combination with the 20 constitutive exons of Dscam, this structure allows 38,016 mRNAs to be synthesized from a single pre- mRNA. c, The organization of the D. melanogaster gene mod(mdg4) is depicted, with common exons in blue and variable exons in purple. The common exons and some of the variable exons are present on one strand of the DNA and are transcribed from left to right as shown, whereas another set of the variable exons is present on the opposite strand and is transcribed from right to left as illustrated. The precise number, and the precise positions of the beginnings and ends, of the common exon pre-mRNA and the variable exon pre-mRNAs is unknown. The possible splicing patterns for each exon in the corresponding pre-mRNA are indicated by black lines, with the possible positions of poly(A)+ tails indicated by green wavy lines. A total of 28 mod(mdg4) mRNAs are synthesized, and this occurs through the trans-splicing of the common exons to one set of variable exons.
Mechanisms of alternative splicing
It is unclear how much of alternative splicing is ‘constitutive’ (that is, the extent to which multiple isoforms are produced at the same ratios in all or most cell types). This is because the most extensively studied cases of alternative splicing are regulated alternative splicing events. Regulation in this context means that distinct splicing patterns are observed in different cellular environments. Such regulation can be tissue specific (for example a sequence corresponding to a particular exon is included in muscle but not brain) or dictated by developmental11 or differentiation-specific12,13 cues. In addition, some alternative splicing patterns have been shown to be modulated in response to external stimuli, such as depolarization of neurons14 or activation of signal transduction cascades15,16.
The biochemical mechanisms that control splice-site usage, and therefore alternative splicing, are complex and in large part remain poorly understood17. It is clear that there cannot be specific and distinct factors dedicated to each of the more than 100,000 alternative splicing decisions that occur in human cells; several genomes worth of regulatory proteins would be required if this were the case. Therefore, it is expected that only a small number of factors are specifically dedicated to one or a few alternative splicing events, such as the factors that dictate the sex-determination cascade in D. melanogaster11. Accordingly, it is not surprising that a small number of proteins have repeatedly been found to be responsible (at least in part) for the regulation of a large number of alternative splicing events17,18 (Table 1). Indeed, few tissue-specific RNA-binding proteins have been identified. Instead, most known regulators of alternative splicing, such as SR proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs), are ubiquitously expressed, although their relative abundances can fluctuate in different tissues. Notable exceptions are NOVA19, nPTB (also known as PTBP2)20,21, FOX1 and FOX2 (also known as A2BP1 and A2BP2)22,23, ESRP1 and ESRP2 (ref. 24), and nSR100 (also known as SRRM4)25, the genes encoding which are all expressed in a tissue-specific manner. How can this handful of splicing regulators be responsible for controlling the plethora of alternative splicing events that occur?
Table 1.
Types and examples of splicing regulatory protein
| Class | Function | Examples* |
|---|---|---|
| SR and SR-related proteins | Typically activate splicing, by recruiting components of the splicing machinery | nSR100 (SRRM4), SC35, SF2 (ASF), SRM160 (SRRM1), SRp30c, SRp38, SRp40, SRp55, SRp75, TRA2α, TRA2β |
| hnRNPs | Typically repress splicing, by a variety of poorly understood mechanisms | hnRNP A1, hnRNP A2/B1, hnRNP C, hnRNP F, hnRNP G (RBMX), hnRNP H, hnRNP L, nPTB (PTBP2), PTB (PTB1) |
| Other RNA-binding proteins | Activate or repress splicing U1 snRNP is essential for constitutive splicing but can also repress splicing |
CELF4 (BRUNOL4), CUGBP, ESRP1, ESRP2, FOX1 (A2BP1), FOX2 (A2BP2), HuD, MBNL1, NOVA1, NOVA2, PSF (SPFQ), quaking, SAM68 (KHDRBS1), SLM2 (KHDRBS3), SPF45 (RBM17), TIA1, TIAR (TIAL1), U1 snRNP |
Synonyms are listed in parentheses
The most studied splicing regulators so far are members of the SR-protein and hnRNP families. SR proteins (so named because they contain domains composed of extensive repeats of serine and arginine) are RNA-binding proteins that are generally thought to activate splicing by binding to the sequences corresponding to exons and recruiting components of the core splicing machinery (the spliceosome)26. By contrast, hnRNPs, which are also RNA-binding proteins, are generally thought to repress splicing by binding to sequences corresponding to exons or introns and interfering with the ability of the core splicing machinery to engage splice sites27; however, the mechanisms by which hnRNPs negatively regulate splicing are far from understood.
The existence of ‘opposing’ classes of splicing regulatory factors, and the abundant evidence that these factors are involved in a large number of splicing decisions, has led to the elaboration of a straightforward ‘yin–yang’ model of alternative splicing28. In this model, splice-site usage is determined by the number of positively acting sites (splicing enhancers that are bound by SR proteins) and negatively acting sites (splicing silencers bound by hnRNPs). When positive sites outnumber negative sites, splicing occurs; when negative sites predominate, splicing is inhibited. Although this model is comforting in its simplicity and has some predictive value, it vastly understates the biochemical complexity of splicing regulation29,30.
The number of mechanisms that are known to be involved in splicing regulation approximates the number of specific splicing decisions that have been analysed in any detail. These mechanisms range from straightforward ones, such as steric blocking of splice sites31 or positive recruitment of the splicing machinery32, to more complicated ones, such as formation of ‘dead-end’ complexes33–35, blocking of splice-site communication36 or facilitation of splice-site communication37. Even these mechanisms are poorly understood at a detailed biochemical level (for example, what distinguishes dead-end complexes from productive complexes remains unclear).
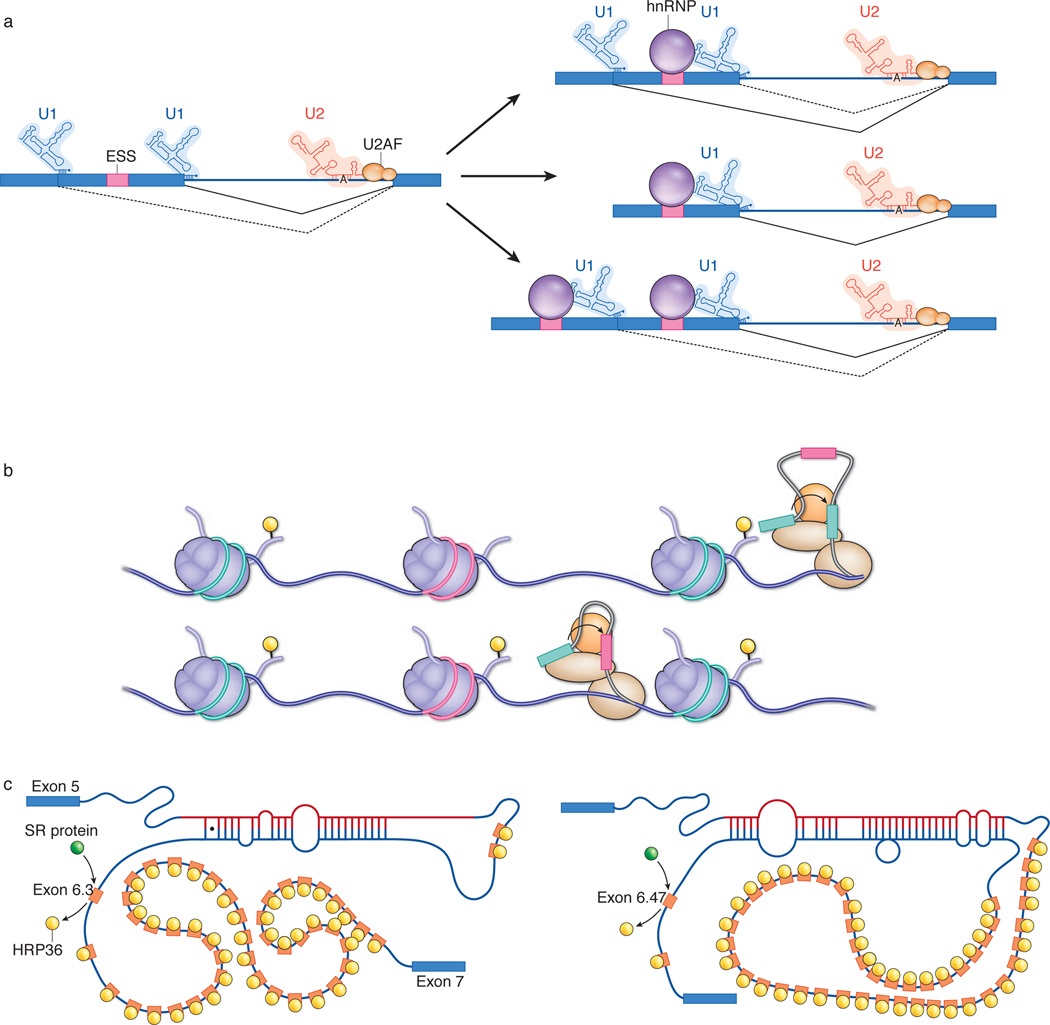
The picture becomes even cloudier when splicing (and alternative splicing) is viewed not as a static process but as a highly dynamic process encompassing a large (yet to be defined) number of kinetic steps38. It is now clear that many factors can have marked effects on splicing patterns; these include transcription rate39, core-splicing-machinery levels40,41, intron size42 and competition between splice sites43. In terms of competition, it was recently shown that the effects of some strong splicing control elements (strong splicing silencers) are apparent only when a competing splice site is present upstream of the affected site43 (Fig. 3a). These studies also showed that a ‘classical’ silencer (to which an hnRNP binds) did not function by occluding, or otherwise inactivating, the affected site but rather by impairing a non-rate-limiting step in the overall splicing reaction. Importantly, when both competing sites were ‘silenced’, the original pattern of splicing was restored43, with no reduction in the extent of splicing (Fig. 3a, bottom right). These results indicate that kinetic parameters have determining roles in splice-site choice.
Figure 3. Alternative splicing regulatory mechanisms.
a, Kinetic effect of splicing silencers on alternative splicing. Left, when two competing 5′ splice sites are present in a hypothetical pre-mRNA and the appropriate silencer is absent (that is, the hnRNP, which silences transcription by binding to the exonic splicing silencer (ESS)), the U1 small nuclear RNP (U1 snRNP) binds to both 5′ splice sites (while U2 snRNP and U2AF bind to the single 3′ splice site), and the proximal 5′ splice site is preferentially used (solid black line; dashed black line indicates splicing to the distal 5′ splice site). Top right, by comparison, when the hnRNP is present, the rate at which the proximal 5′ splice site can be used decreases, promoting splicing to the distal 5′ splice site. Centre right, in comparison with the case on the left, in the absence of a competing 5′ splice site upstream, the presence of the hnRNP does not significantly decrease the rate of splicing. Bottom right, in comparison with the case on the left, when silencer sequences are present near both 5′ splice sites, the hnRNP has no detectable impact on the ratio at which either splice site is used. b, Interplay between transcription elongation rate, chromatin structure and histone modifications, and their impact on alternative splicing. A hypothetical gene is depicted; three exons are shown packaged into nucleosomes. The constitutively transcribed exons (green) are packaged into nucleosomes that constitutively contain histone H3 that is trimethylated (yellow) at lysine residue 36 (H3K36me3). In cells that do not include the alternative exon (pink) in the mRNA, the nucleosomes packaging this exonic DNA do not contain H3K36me3 (top). We propose that when RNA polymerase (brown ovals; spliceosome, orange circle) transcribes a gene with this chromatin configuration (with the pre-mRNA shown here in grey), it traverses the alternative exon rapidly, and this exon is not tethered to the RNA polymerase and, accordingly, sequence corresponding to this exon is not included in the mature mRNA (the splicing of exonic sequences is indicated by curved arrows). By contrast, when the nucleosome that is packaging the alternative exon contains H3K36me3, this slows the progress of the RNA polymerase, allowing it to capture the exon, resulting in the inclusion of sequence corresponding to this sequence in the mature mRNA (bottom).
c, Kinetic model of mutually exclusive splicing of Dscam pre-mRNA. Mutually exclusive splicing of the sequence corresponding to the exon 6 cluster of the Dscam gene involves the formation of secondary structures in the RNA. These structures form between the docking site in the intronic sequence downstream of the sequence corresponding to exon 5 and a selector sequence located upstream of each sequence corresponding to an exon 6 variant. In addition, the hnRNP HRP36 binds to, and represses, all 48 exonic sequences in the cluster. Which exonic sequence is spliced seems to be a function of kinetic competition between the 48 potential docking-site–selector sequence structures and between SR proteins (not shown) and HRP36. On the left, the selector sequence upstream of the sequence corresponding to exon 6.3 in the pre-mRNA interacts with the docking site. HRP36 is then displaced from this exonic sequence, allowing an SR protein to bind in its place. This results in the inclusion of sequence corresponding to exon 6.3 in the mRNA. On the right, the formation of a secondary structure with a more distal sequence means that exon 6.47 is included in the mRNA instead.
When viewed from this kinetic perspective, the effects of transcription rate on alternative splicing become straightforward to rationalize. The folding of the nascent transcript, and therefore the accessibility of binding sites to the spliceosome and splicing regulators, could be determined by the rate at which the nascent transcript is exposed. In this way, ‘small’ changes could render certain splice sites at a competitive advantage or disadvantage relative to other splice sites. Recent studies have also indicated that exons in the nascent transcript become tethered to the elongating transcription complex as they emerge from RNA polymerase44. Although it seems plausible that this tethering is required for the exonic sequence to be included in the mRNA — or else become lost in the nuclear environment — the mechanisms involved in tethering exonic RNA are unknown. We propose that when RNA polymerase quickly traverses exons in the gene, the transcribed exons evade capture and are skipped; however, when they are transcribed slowly, they are efficiently tethered and the corresponding sequence is included in the mRNA (Fig. 3b).
It is now well established that chromatin structure is highly dynamic and can affect the rate of transcription, which in turn can influence the pattern of alternative splicing. Indeed, in a study in which a short interfering RNA was used to induce heterochromatin formation, this change in chromatin structure correlated with a reduced ability of the transcription elongation complex to proceed along the DNA and with corresponding changes in the patterns of alternative splicing45. Moreover, it has recently been shown that exons are specifically marked by histone H3 trimethylated at lysine residue 36 (H3K36me3) in a transcription-dependent manner. This modification of the histone was found to occur more frequently on histones that are packaged with constitutive exons (that is, those that are constitutively present in mRNA) rather than alternative exons (those that are not always present in the mRNA)46–48.
Accordingly, we speculate that a large proportion of regulated alternative splicing events might be determined not by tissue-specific or developmental-stage-specific splicing regulators but rather by tissue-specific or stage-specific differences in transcription rates, which could be dictated by local chromatin modifications (Fig. 3b). It will be interesting to explore whether transcription rates differ markedly between brain and muscle, which have particularly high levels of alternative splicing, and tissues where alternative splicing is less prevalent.
The effects on alternative splicing of depleting core spliceosome proteins40,41 also become interpretable when splice-site choice is viewed in terms of kinetic competition. If the splicing machinery is present at a saturating concentration, its recruitment would not be a rate-limiting step, so other parameters would determine splicing patterns. But, if the concentration of components of the core splicing machinery becomes limiting, it seems certain that specific splice-site pairs would be more effective at recruiting constituents of the spliceosome, resulting in altered patterns of splicing. As is the case for transcription, it will be interesting to uncover the relative concentrations of spliceosomal components in different cell types and at different stages of organism development.
A final example of the importance of kinetics in alternative splicing comes from studies of the D. melanogaster gene Dscam (Fig. 3c), which (as indicated earlier) encodes more than 38,000 mRNA isoforms. This remarkable diversity is mainly generated by mutually exclusive splicing of pre-mRNA sequence corresponding to four large, independently controlled clusters of alternative exons6,49. The mutually exclusive splicing of one of these exonic sequence clusters, that corresponding to exon 6, is mediated by competing RNA secondary structures that form between a single docking site and one of the selector sequences located upstream of each of the 48 alternative exonic sequences50,51 (Fig. 3c) in the mRNA. But it is not understood how the selector sequence is chosen to be the one that will interact with the docking site. In this case, every aspect of the process from the strength of each docking-site–selector sequence pair to the rate of transcription through the exon 6 cluster and the distance from each selector sequence to the docking site is likely to have a role. To further complicate matters, the hnRNP HRP36 (also known as HRB87F) actively represses all 48 exonic sequences (Fig. 3c), and depletion of this protein results in the inclusion of multiple exonic sequences52. Unexpectedly, however, when an SR protein was depleted together with HRP36, correct splicing was found to be restored52. These results not only show that SR proteins and hnRNPs have balancing roles but also indicate that the exonic sequences that are not selected are not ‘dormant’. The simplest explanation for these findings is that the selected exonic sequence has a competitive advantage over the non-selected exonic sequences such that its splicing to the exonic sequences that flank the cluster proceeds faster than the joining of exonic sequences within the cluster. It seems probable that many, if not all, cases of mutually exclusive splicing are determined by such kinetic parameters, although the details will undoubtedly differ.
Last, before leaving the mechanistic aspects of alternative splicing, it should be noted that we have understated the complexity of the mechanisms involved. Nearly all ‘activators’ of splicing can, in some cases, function as repressors, and nearly all ‘repressors’ have been shown to function as activators. Particularly striking examples are the U1 small nuclear RNP (U1 snRNP) and NOVA. U1 snRNP is part of the core splicing machinery and is essential for recognition of the 5′ splice site. Nevertheless, in certain contexts, U1 snRNP is a required component of potent splicing silencers35. NOVA53, a neuron-specific splicing factor, functions as a positive regulator when positioned upstream of splice sites but a negative regulator when positioned downstream of affected splice sites. The generalities of this directionality and the detailed biochemical mechanisms underlying the opposing functions of both U1 snRNP and NOVA (as well as many other splicing regulators) remain to be elucidated. But it is clear that context affects function, and this adds a layer of complexity to the already complex field of alternative and regulated splicing.
Bioinformatics and alternative splicing
One of the most useful tools for studying alternative splicing over the past decade has been bioinformatics. Computational approaches have become increasingly powerful and sophisticated with each genome that has been completely sequenced and with each advance in the technologies used for genomic analysis. The ultimate goal of using bioinformatics in the splicing field is to be able to identify which exons in a genome are alternatively spliced from pre-mRNAs, to predict accurately the spatial and temporal pattern in which the exons are regulated, and to predict which splicing factors are involved in regulating the exons. These goals have yet to be reached, but there has been considerable progress in these areas, particularly in the past five years.
One of the most powerful methods in the bioinformatics toolbox is comparative genomics, which uses the degree of similarity between two or more genomes to infer functional information about regions of interest. For instance, this approach revealed that alternative exons are typically more conserved across species than are constitutive exons54–56, presumably because of the information content needed to facilitate regulation. This observation, in turn, has been used to predict whether a particular exon is more likely to be constitutive or alternative and to identify previously unrecognized alternative exons57. Comparative genomics has also been used to characterize splicing enhancers or silencers by identifying sequences that are enriched or depleted in exons or introns58 and by pinpointing sequences within coding exons (those translated into protein) that are more highly conserved than expected given the flexibility of the genetic code59. Similarly, comparative genomics has facilitated the identification of sequences that participate in the formation of the RNA structures involved in the control of alternative splicing50,51,60.
Nevertheless, comparative genomic approaches have limitations. Although evolutionary conservation provides strong evidence for the functional significance of a particular splicing pattern, the converse is not always true. A particularly notable example is the alternative splicing cascade that controls sex determination in D. melanogaster11. Although this elegant pathway is crucial for survival of the species, it shows rapid evolutionary flux. Even among the closely related Drosophila species, there are subtle but important differences in the organization and expression of the genes that are involved in this cascade11. Furthermore, slightly more distantly related insects, such as the housefly (Musca domestica) and the Mediterranean fruitfly (Ceratitis capitata), do not splice the pre-mRNA corresponding to the first gene in the sex-determination pathway (Sex lethal) in a sex-specific manner11. Even more strikingly, almost every component of the sex-determination cascade of D. melanogaster and the honeybee, Apis mellifera, is different, although the process relies on alternative splicing in both species61,62.
These examples show the high level of evolutionary plasticity that alternative splicing provides. Because small changes (that is, point mutations) in either exons or introns can create or destroy splicing control elements63, it is easy to envisage that splicing patterns are constantly evolving: advantageous mutations would rapidly be selected for, and deleterious mutations would be selected against64. Indeed, we speculate that ‘non-conserved’ changes in splicing patterns might underlie the observed phenotypic variations between species and between individuals within species. Recent studies have provided insight into the way in which human exons have evolved65 and the extent of alternative splicing differences between humans and chimpanzees66. Additional studies along these lines are likely to improve the understanding of how alternative splicing contributes to speciation and phenotypic diversity67.
Outstanding questions
In addition to the broad questions that we have outlined, numerous issues remain unresolved. Foremost among these is whether the extent of alternative splicing can account for organismal complexity. It is noteworthy that Caenorhabditis elegans, D. melanogaster and mammals have about 20,000 (ref. 68), 14,000 (ref. 69) and 20,000 (ref. 70) genes, respectively, but mammals are clearly much more complex than nematodes or flies. In the nervous system alone, nematodes have only a few hundred neurons, whereas mammals have several billion. Indeed, at present, the absolute amount of alternative splicing that takes place in mammals4,5 seems to be much higher than in nematodes71 or flies72. These measures, however, are based on far fewer data in nematodes and flies than in humans. Thus, completion of the model organism Encyclopedia of DNA Elements (modENCODE) project73, which seeks to identify all of the sequence-based functional elements in C. elegans and D. melanogaster, should help to uncover the true extent of alternative splicing in these organisms. Nonetheless, mammalian neuronal tissue is enriched for alternatively spliced products compared with other mammalian tissues and, at least in this case, the extent of alternative splicing correlates with greater complexity.
Another crucial question is how many mRNA isoforms are functionally relevant? Teleology suggests that if an isoform exists, it is important (similarly to the way in which ‘junk’ DNA is now considered to be treasure). But this idea is hard to prove and is difficult for some to accept. Unfortunately, the function of alternative mRNA isoforms has been carefully analysed for only a small proportion of genes. Such studies have, however, provided numerous examples in which alternative splicing clearly gives rise to functionally distinct isoforms (Table 2). But just as discernible changes in phenotype are frequently not observed even when entire genes are deleted, there are also many cases in which functional distinctions between mRNA isoforms have not been observed. One example of this is the D. melanogaster gene Cadherin-N (CadN), which contains two mutually exclusive versions of exon 18 (exon 18A and exon 18B). These exons are dynamically regulated throughout development, expressed in different tissues, and highly conserved throughout evolution. However, although mutations in exon 18A disrupt development of the R7 photoreceptor, suggesting that the alternative splicing of CadN pre-mRNA is functionally relevant, this phenotype can be rescued by expressing an mRNA isoform that contains sequence corresponding to the other exon, exon 18B74. Therefore, it is formally possible that alternative splicing of CadN pre-mRNA does not generate functionally distinct isoforms but ensures that one isoform is expressed at the correct time and place. However, it is also possible that there are functional distinctions between the isoforms that cannot be detected in the assays used. Thus, the question of how many alternative splicing events are functionally relevant is destined to remain unanswered for some time.
Table 2.
Examples of functionally relevant alternative splicing events
| Gene* | Description | Reference |
|---|---|---|
| Drosophila melanogaster | ||
| Dscam | Alternative splicing has the potential to generate 19,008 extracellular domains that show isoform-specific homophilic binding, regulating neural wiring and immune-system interactions with pathogens† | 76 |
| fru | Sex-specific splicing controls courtship behaviour | 77 |
| Nurf301 (E(bx)) | Alternative splicing generates three protein isoforms, each of which assembles into a functionally distinct form of the chromatin-remodelling complex NURF | 78 |
| Caenorhabditis elegans | ||
| egl-15 | Mutually exclusive splicing generates two isoforms of a growth factor with distinct ligand specificities | 79 |
| fbl-1 | Alternative splicing generates two isoforms of a basement-membrane protein that have distinct but complementary roles in tissue assembly and organization | 80 |
| unc-60 | Alternative splicing generates two isoforms of actin-depolymerizing factor (cofilin) that have distinct actin-filament-severing activities | 81 |
| Homo sapiens | ||
| NLGN1 | Alternative splicing modulates the interactions between the neuronal synaptic cell-adhesion molecules neuroligin 1 and neurexins | 82 |
| SLACK (KCNT1) | Alternative splicing alters the amino-terminal domain of SLACK isoforms, regulating interactions with SLICK to form a heteromeric sodium-activated potassium channel | 83 |
| Mus musculus | ||
| Snap25 | Alternative splicing generates two protein isoforms that differ in their ability to stabilize synaptic vesicles in the primed state | 84 |
Synonyms are listed in parentheses.
There are 19,008 combinations of the extracellular portion (encoded by exons 4, 6 and 9) and two transmembrane domains encoded by the two variants of exon 17, making 38,016 isoforms.
Another outstanding question is whether there is a decipherable ‘splicing code’. Will a computer be able to predict reliably the splicing patterns in a cell or organism? Despite the numerous variables (known and unknown) involved in splice-site choice, rapid progress has been made in this area75. But it is not clear when or whether this Rosetta stone of splicing will emerge.
It also remains unclear to what extent elements deep within sequences corresponding to introns (and the factors that bind to them) influence alternative splicing decisions. Most investigations have focused on elements close to splice sites, but intronic regions that are distant from the splice sites themselves could almost certainly affect alternative splicing.
Much remains to be learned about the mechanisms of alternative splicing and the regulatory networks of alternative splicing. It is clear that researchers are only beginning to understand the diversity and details of the mechanisms that are used to regulate alternative splicing, as well as the factors involved. Recent technological advances, particularly in genomic analysis, suggest that the next few years are likely to be filled with many exciting and unanticipated discoveries that could rapidly reveal the mysteries of the field.
Acknowledgements
We apologize to the many authors whose publications are not cited directly because of space limitations. Research in our laboratories is supported by grants from the National Institutes of Health (T.W.N. and B.R.G.), the Raymond and Beverly Sackler Fund for the Arts and Sciences (B.R.G.) and the State of Connecticut’s Stem Cell Research Fund (B.R.G.).
Footnotes
The authors declare no competing financial interests.
References
- 1.Alt FW, et al. Synthesis of secreted and membrane-bound immunoglobulin μ heavy chains is directed by mRNAs that differ at their 3’ ends. Cell. 1980;20:293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- 2.Early P, et al. Two mRNAs can be produced from a single immunoglobulin μ gene by alternative RNA processing pathways. Cell. 1980;20:313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld MG, et al. Calcitonin mRNA polymorphism: peptide switching associated with alternative RNA splicing events. Proc. Natl Acad. Sci. USA. 1982;79:1717–1721. doi: 10.1073/pnas.79.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 5. Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. References 4 and 5 provide detailed views of the human transcriptome as determined by using deep-sequencing data. The authors conclude that the pre-mRNAs from all multi- exon genes are alternatively spliced.
- 6.Schmucker D, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 7.Navaratnam DS, Bell TJ, Tu TD, Cohen EL, Oberholtzer JC. Differential distribution of Ca2+-activated K+ channel splice variants among hair cells along the tonotopic axis of the chick cochlea. Neuron. 1997;19:1077–1085. doi: 10.1016/s0896-6273(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 8.Rosenblatt KP, Sun ZP, Heller S, Hudspeth AJ. Distribution of Ca2+-activated K+ channel isoforms along the tonotopic gradient of the chicken’s cochlea. Neuron. 1997;19:1061–1075. doi: 10.1016/s0896-6273(00)80397-9. [DOI] [PubMed] [Google Scholar]
- 9.Dorn R, Reuter G, Loewendorf A. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc. Natl Acad. Sci. USA. 2001;98:9724–9729. doi: 10.1073/pnas.151268698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Labrador M, et al. Protein encoding by both DNA strands. Nature. 2001;409:1000. doi: 10.1038/35059000. References 9 and 10 show that trans-splicing of two separate pre-mRNAs can generate a single protein-coding mRNA.
- 11.Sánchez L. Sex-determining mechanisms in insects. Int. J. Dev. Biol. 2008;52:837–856. doi: 10.1387/ijdb.072396ls. [DOI] [PubMed] [Google Scholar]
- 12.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie J, Black DL. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature. 2001;410:936–939. doi: 10.1038/35073593. [DOI] [PubMed] [Google Scholar]
- 15.Lynch KW. Regulation of alternative splicing by signal transduction pathways. Adv. Exp. Med. Biol. 2007;623:161–174. doi: 10.1007/978-0-387-77374-2_10. [DOI] [PubMed] [Google Scholar]
- 16.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nature Rev. Mol. Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 17.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nature Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 18.Gabut M, Chaudhry S, Blencowe BJ. The splicing regulatory machinery. Cell. 2008;133:192. doi: 10.1016/j.cell.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Buckanovich RJ, Posner JB, Darnell RB. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- 20.Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron- specific alternative splicing. Proc. Natl Acad. Sci. USA. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markovtsov V, et al. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin Y, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calarco JA, et al. Regulation of vertebrate nervous system-specific alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Contreras R, et al. hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 28.Fu XD. Towards a splicing code. Cell. 2004;119:736–738. doi: 10.1016/j.cell.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XH, Arias MA, Ke S, Chasin LA. Splicing of designer exons reveals unexpected complexity in pre-mRNA splicing. RNA. 2009;15:367–376. doi: 10.1261/rna.1498509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchette M, et al. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Mol. Cell. 2009;33:438–449. doi: 10.1016/j.molcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valcárcel J, Singh R, Zamore PD, Green MR. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- 32.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein–protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- 33.House AE, Lynch KW. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nature Struct. Mol. Biol. 2006;13:937–944. doi: 10.1038/nsmb1149. [DOI] [PubMed] [Google Scholar]
- 34.Giles KE, Beemon KL. Retroviral splicing suppressor sequesters a 3’ splice site in a 50S aberrant splicing complex. Mol. Cell. Biol. 2005;25:4397–4405. doi: 10.1128/MCB.25.11.4397-4405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siebel CW, Fresco LD, Rio DC. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5’ splice site control U1 snRNP binding. Genes Dev. 1992;6:1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- 36.Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nature Struct. Mol. Biol. 2008;15:183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Contreras R, et al. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith DJ, Query CC, Konarska MM. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol. Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kornblihtt AR. Coupling transcription and alternative splicing. Adv. Exp. Med. Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 40.Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl Acad. Sci. USA. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fox-Walsh KL, et al. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl Acad. Sci. USA. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Y, et al. Dynamic regulation of alternative splicing by silencers that modulate 5’ splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol. Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 45.Alló M, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nature Struct. Mol. Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 46.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon–intron architecture. Nature Struct. Mol. Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 48.Tilgner H, et al. Nucleosome positioning as a determinant of exon recognition. Nature Struct. Mol. Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 49.Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nature Genet. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- 50.Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anastassiou D, Liu H, Varadan V. Variable window binding for mutually exclusive alternative splicing. Genome Biol. 2006;7:R2. doi: 10.1186/gb-2006-7-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olson S, et al. A regulator of Dscam mutually exclusive splicing fidelity. Nature Struct. Mol. Biol. 2007;14:1134–1140. doi: 10.1038/nsmb1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. This paper shows that the neural-specific splicing regulator NOVA functions as either an activator or a repressor depending on the context.
- 54.Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nature Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 55.Sorek R, Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugnet CW, Kent WJ, Ares MJ, Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 2004;2004:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- 57. Ohler U, Shomron N, Burge CB. Recognition of unknown conserved alternatively spliced exons. PLoS Comput. Biol. 2005;1:113–122. doi: 10.1371/journal.pcbi.0010015. This paper shows that previously unannotated alternatively spliced exons can be identified accurately by using DNA sequence alone.
- 58. Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. This paper describes one of the first genome-wide predictions of splicing regulatory sequences.
- 59.Goren A, et al. Comparative analysis identifies exonic splicing regulatory sequences the complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 60.Shepard PJ, Hertel KJ. Conserved RNA secondary structures promote alternative splicing. RNA. 2008;14:1463–1469. doi: 10.1261/rna.1069408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hasselmann M, et al. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature. 2008;454:519–522. doi: 10.1038/nature07052. [DOI] [PubMed] [Google Scholar]
- 62.Beye M, Hasselmann M, Fondrk MK, Page RE, Omholt SW. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell. 2003;114:419–429. doi: 10.1016/s0092-8674(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 63.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 64.Xing Y, Lee C. Alternative splicing and RNA selection pressure — evolutionary consequences for eukaryotic genomes. Nature Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- 65.Zhang XH, Chasin LA. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc. Natl Acad. Sci. USA. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calarco JA, et al. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Graveley BR. The haplo-spliceo-transcriptome: common variations in alternative splicing in the human population. Trends Genet. 2008;24:5–7. doi: 10.1016/j.tig.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 69.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 70.Clamp M, et al. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl Acad. Sci. USA. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hillier LW, et al. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res. 2009;19:657–666. doi: 10.1101/gr.088112.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stolc V, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- 73.Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nern A, et al. An isoform-specific allele of Drosophila N-cadherin disrupts a late step of R7 targeting. Proc. Natl Acad. Sci. USA. 2005;102:12944–12949. doi: 10.1073/pnas.0502888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wojtowicz WM, et al. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Demir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121:785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 78.Kwon SY, Xiao H, Wu C, Badenhorst P. Alternative splicing of NURF301 generates distinct NURF chromatin remodeling complexes with altered modified histone binding specificities. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000574. e1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goodman SJ, Branda CS, Robinson MK, Burdine RD, Stern MJ. Alternative splicing affecting a novel domain in the C. elegans EGL-15 FGF receptor confers functional specificity. Development. 2003;130:3757–3766. doi: 10.1242/dev.00604. [DOI] [PubMed] [Google Scholar]
- 80.Muriel JM, Dong C, Hutter H, Vogel BE. Fibulin-1C and Fibulin-1D splice variants have distinct functions and assemble in a hemicentin-dependent manner. Development. 2005;132:4223–4234. doi: 10.1242/dev.02007. [DOI] [PubMed] [Google Scholar]
- 81.Ono K, Yamashiro S, Ono S. Essential role of ADF/cofilin for assembly of contractile actin networks in the C. elegans somatic gonad. J. Cell Sci. 2008;121:2662–2670. doi: 10.1242/jcs.034215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to α- and β-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 83.Chen H, et al. The N-terminal domain of Slack determines the formation and trafficking of Slick/Slack heteromeric sodium-activated potassium channels. J. Neurosci. 2009;29:5654–5665. doi: 10.1523/JNEUROSCI.5978-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sorensen JB, et al. Differential control of the releasable vesicle pools by SNAP-25 splice variants and SNAP-23. Cell. 2003;114:75–86. doi: 10.1016/s0092-8674(03)00477-x. [DOI] [PubMed] [Google Scholar]