Abstract
The homeodomain transcription factor KNOTTED1 (KN1) functions in shoot meristem maintenance and is thought to move from cell to cell in a similar fashion as viral movement proteins. Both types of transported proteins bind to RNA, and associate with intercellular bridges formed by plasmodesmata. In a mutant screen for KN1 transport deficiency, a component of a type II chaperonin complex, CCT8, was identified, and found to interact with non-cell-autonomous proteins. The cct8 mutants are characterized by limited functionality of non-cell-autonomous proteins after their movement, and a phenotype resembling lack of homeodomain protein activity. Evidence suggests that CCT8 functions in post-translocational refolding of transported proteins. Here we show that spread of tobamovirus infection is reduced in a cct8 mutant. This suggests that similar to KN1, viral movement proteins are unfolded and refolded during transport to gain functionality in the receiving cells.
Keywords: chaperonins, homeodomain, intercellular transport, plasmodesmata
Introduction and Results
The stem cell identity homeodomain (HD) transcription factor KNOTTED1 (KN1) is suggested to move from cell to cell via intercellular channels named plasmodesmata.1-7 The translocated protein is active in neighboring cells, and its transport is most likely mediated by factors interacting specifically with a HD motif present in a subclass of KNOTTED1-related proteins.4 This HD motif is also recognized by MOVEMENT PROTEIN BINDING PROTEIN 2C (MPB2C), a microtubule-associated protein that functions as a negative regulator of movement.3 Microinjection of structurally altered KN1 protein revealed that intercellular transport is blocked by fixing its tertiary protein structure, and that transport might occur without opening (gating) of the plasmodesmata.1,2 These observations are consistent with a model suggesting that mobile proteins are at least partially unfolded during the translocation process.8
This previous viewpoint found support in a mutant identified in a genetic screen for factors interfering with transport of KN1 fusion proteins. A gene named CHAPERONIN CONTAINING TCP1 (CCT) 8 was uncovered to be necessary for intercellular transfer of functional HD protein fusions.9 The mutant screen used a so-called trichome rescue line (TR line).4 TR plants express GL1-GFP-KN1C fusion protein sub-epidermally in trichome-less gl1 mutants. After intercellular transport of the fusion protein, GL1 induces the formation of trichomes in the epidermis of the gl1 mutants. KN1C is a N-terminally truncated version of KN1 lacking the MEINOX domain, with the C-terminal trafficking domain of KN1 intact. Such transgenic plants, which exhibit trichomes due to transport of GL1 mediated by the KN1 HD, were EMS mutagenized and a mutant cct8-1 line was identified.9
The CCT8 gene encodes a component of a type II chaperonin, which forms a large cytosolic oligomeric double-ring complex that assists in protein folding.10 CCT8 binds to a number of non-cell-autonomous proteins such as KN1, SHOOT MERISTEMLESS (STM), and TRANSPARENT TESTA GLABROUS 1 (TTG1), but not to the movement protein of Cucumber mosaic virus (CMV-MP). In cct8 mutants, KN1 and STM exhibit limited functional activity after cell-to-cell movement.9 Most likely a loss of post-translocational refolding of the fusion protein in the epidermis, due to the ctt8-1 mutation, interferes with the proper function of the homeodomain transcription factors in the targeted cells. This observation is consistent with the results obtained in microinjection experiments performed with cross-linked KN1 proteins.1 Such a structurally fixed KN1 molecule, although interacting with the plasmodesmatal transport system, was not transported into neighboring cells.
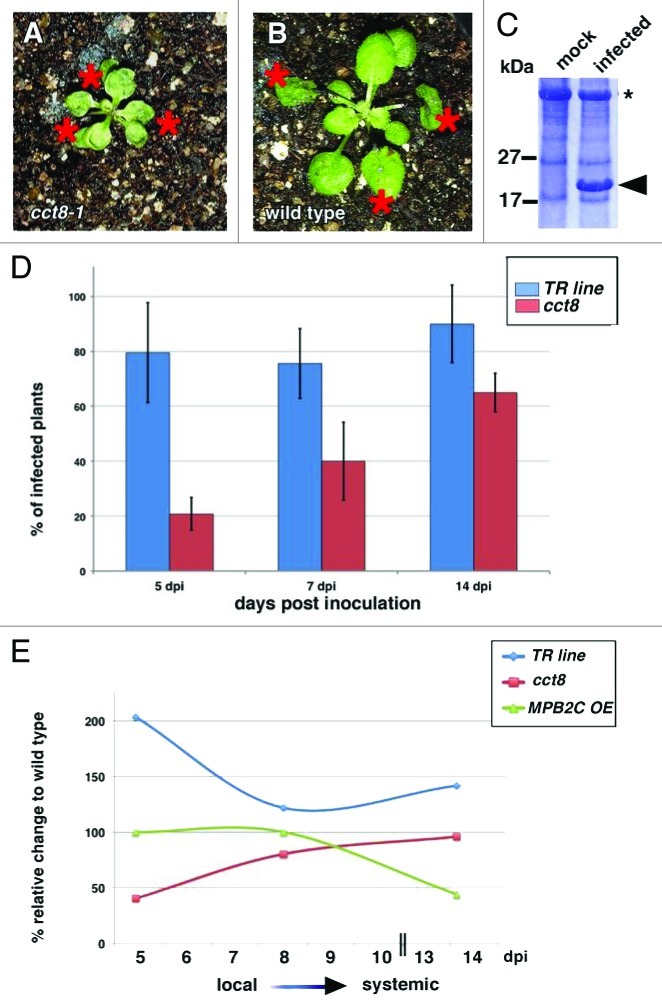
Interestingly, the protein binding characteristics of CCT8 show some parallels to MPB2C, which binds to the tobamoviral movement protein TMV-MP11 as well as KN1 and STM.3 However, like MPB2C, CCT8 does not interact with the Cucumber mosaic virus MP.9,11 Elevated MPB2C levels reduce the infection efficiency of TMV and the related Oilseed rape mosaic virus (ORMV).12 This inhibition is most likely due to MPB2C binding to the MP.11 As MPB2C and CCT8 interact with the same mobile HD proteins, we tested whether cct8 mutants also reduce systemic spread of ORMV. We infected ctt8-1 plants with ORMV particles (Fig. 1A), and as a control we infected the progenitor trichome rescue (TR)-line plants, plants harboring a 35S::MPB2C construct, and wild-type plants (Fig. 1B). We compared the infection rate over 14 d, by determining the presence of the ORMV coat protein in at least 30 plants for each time point (Fig. 1C).
Figure 1.
ORMV infection experiments. (A) and (B) inoculated wild-type (A) and cct8–1 mutant plant (B). Inoculated leaves are marked with an asterisk. (C) Representative Coomassie stained 10% polyacrylamide gel showing the presence of the ORMV coat protein in infected plants. Mock: buffer inoculated plant. Infected: viral coat protein band is marked with an arrowhead. (D) Percentage of infected TR parent and cct8-1 mutant plants harvested at different time points after infection. (E) Infection rates of TR parent, cct8-1 mutant, and MPB2C overexpression (OE) plants in comparison to the wild-type infection levels.
Five days after inoculation, a striking difference could be observed between the number of ORMV infected cct8-1 plants and the TR parent plants (Fig. 1D and E). ORMV infection was significantly reduced in the cct8-1 mutant compared with the parent TR line. As the inoculated leaves senesce ~6 d post infection, the remaining leaves are an indicator for systemic infection (Fig. 1E). At a later stage of infection (7 and 14 d after inoculation) the infection levels in cct8-1 line remained well below the percentage observed in infected TR control plants. Also in comparison to the wild type, the cct8-1 mutant showed a decreased infection rate. The percentage of infected plants remained below wild-type levels until 14 d after inoculation (Fig. 1E). Interestingly, the TR parent line, expressing the GL1-GFP-KN1C fusion protein, was nearly 2-fold (indicated as ~200% in Fig. 1E) more susceptible than wild type to ORMV. This suggests that the presence of the mobile KN1 HD enhances transport activity of the virus. At later time points, the infection rate of cct8 mutants slowly approaches wild-type levels (Fig. 1E). This might be due to host factors, distinct from chaperonins, involved in phloem-mediated systemic transport of viral complexes. In any case the reduced early infection suggests that CCT8 facilitates efficient tobamoviral infections, perhaps by promoting cell-to-cell movement. By analogy to KN1, this could occur through refolding of the viral MP after cell-to-cell transport through plasmodesmata, and suggests an interaction of the tobamoviral MP with CCT8.
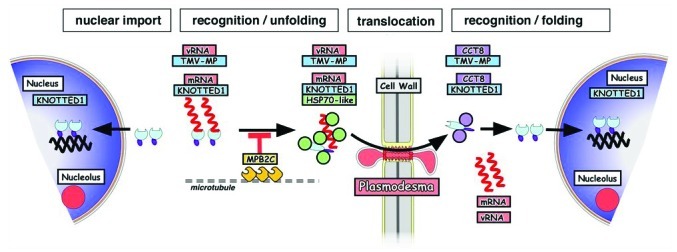
In a functional transport model (Fig. 2), endogenous non-cell-autonomous proteins, such as KN1 and tobamoviral MPs, are RNA binding proteins moving from cell to cell via plasmodesmata. Intercellular transport of the mobile protein-RNA complexes is negatively regulated by microtubule-associated factors such as MPB2C.3 If they are available for transport, the complexes might bind to HSP70-like chaperones found at plasmodesmata.13 We hypothesize that chaperone activity initiates partial unfolding of the proteins and associated RNAs prior or during transport through plasmodesmata.1 After transport to a neighboring cell, the chaperonin complex, containing CCT8, enables refolding of the transported polypeptides, to ensure their functionality.9
Figure 2.
Model showing the factors facilitating viral (ORMV-MP, TMV-MP) and plant endogenous protein (KN1, STM) cell-to-cell transport (see text for details).
Materials and Methods
Viral particles were isolated from infected material, and three weeks old plants were inoculated mechanically with a total of approx. 600 ng viral particles spread on three leaves using a brush.12 For infection experiments, plants were grown on soil at 16 h light, 22°C day/18°C night in a Percival growing chamber. Material from individuals was harvested at different time points after inoculation, and analyzed for the presence of the coat protein. For this, total protein extracts were prepared in a denaturing buffer (30% glycerol, 2% SDS, 50 mM TRIS-HCl pH 6,8, 36% urea, 0,1 M DTT), separated by SDS-PAGE, and visualized by Coomassie blue staining (Fig. 1C). Plants were scored as infected as previously described.12 In general, at 5 dpi plants were scored as infected when the coat protein appeared at an relative intensity of at least ~20% of the RuBisCo protein. Protein samples of mock-infected plants were used in each infection round to ensure correct identification of the coat protein band. On later time points plants were scored as infected when the coat protein band was detected in a similar intensity as the RuBisCo protein band appearing on Coomassie blue stained gels.
Acknowledgments
We thank Fernando Ponz and Carmen Mansilla for providing ORMV- infected material. D.F. and F.K. were supported by the Austrian Science Funds (FWF Project # P19682-Kragler).
Glossary
Abbreviations:
- KN1
KNOTTED1
- CCT8
CHAPERONIN CONTAINING TCP1 8
- HSC70
heatshock cognate protein 70
- MP
viral movement protein
- ORMV
Oilseed rape mosaic virus
- MPB2C
movement protein binding protein 2C
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/19152
References
- 1.Kragler F, Monzer J, Shash K, Xoconostle-Cazares B, Lucas WJ. Cell-to-cell transport of proteins: requirement for unfolding and characterization of binding to a putative plasmodesmal receptor. Plant J. 1998;15:367–81. doi: 10.1046/j.1365-313X.1998.00219.x. [DOI] [Google Scholar]
- 2.Kragler F, Monzer J, Xoconostle-Cázares B, Lucas WJ. Peptide antagonists of the plasmodesmal macromolecular trafficking pathway. EMBO J. 2000;19:2856–68. doi: 10.1093/emboj/19.12.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winter N, Kollwig G, Zhang S, Kragler F. MPB2C, a microtubule-associated protein, regulates non-cell-autonomy of the homeodomain protein KNOTTED1. Plant Cell. 2007;19:3001–18. doi: 10.1105/tpc.107.044354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JY, Rim Y, Wang J, Jackson D. A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 2005;19:788–93. doi: 10.1101/gad.332805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JY, Yuan Z, Cilia M, Khalfan-Jagani Z, Jackson D. Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc Natl Acad Sci U S A. 2002;99:4103–8. doi: 10.1073/pnas.052484099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JY, Yuan Z, Jackson D. Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development. 2003;130:4351–62. doi: 10.1242/dev.00618. [DOI] [PubMed] [Google Scholar]
- 7.Lucas WJ, Bouché-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, et al. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science. 1995;270:1980–3. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz-Medrano R, Xoconostle-Cazares B, Kragler F. The plasmodesmatal transport pathway for homeotic proteins, silencing signals and viruses. Curr Opin Plant Biol. 2004;7:641–50. doi: 10.1016/j.pbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Xu XM, Wang J, Xuan Z, Goldshmidt A, Borrill PG, Hariharan N, et al. Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science. 2011;333:1141–4. doi: 10.1126/science.1205727. [DOI] [PubMed] [Google Scholar]
- 10.Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115–45. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 11.Kragler F, Curin M, Trutnyeva K, Gansch A, Waigmann E. MPB2C, a microtubule-associated plant protein binds to and interferes with cell-to-cell transport of tobacco mosaic virus movement protein. Plant Physiol. 2003;132:1870–83. doi: 10.1104/pp.103.022269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruggenthaler P, Fichtenbauer D, Krasensky J, Jonak C, Waigmann E. Microtubule-associated protein AtMPB2C plays a role in organization of cortical microtubules, stomata patterning, and tobamovirus infectivity. Plant Physiol. 2009;149:1354–65. doi: 10.1104/pp.108.130450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoki K, Kragler F, Xoconostle-Cazares B, Lucas WJ. A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proc Natl Acad Sci U S A. 2002;99:16342–7. doi: 10.1073/pnas.252427999. [DOI] [PMC free article] [PubMed] [Google Scholar]