Abstract
Nuclei in dry Arabidopsis thaliana seeds are very small and have highly condensed chromatin. Nuclear shrinkage and chromatin compaction occur during seed maturation and have been shown to be independent, developmentally controlled processes. To confirm this genetically, we studied chromatin compaction in a mutant of the seed developmental regulator ABA INSENSITIVE 3, and in a double mutant of the nuclear matrix proteins LITTLE NUCLEI 1 and 2. Our results indicated that the nuclear shrinking and chromatin condensation during seed maturation can be genetically uncoupled, confirming that these are independent processes. In addition, we demonstrated that transcript levels of siliques toward the end of seed maturation are comparable to those in vegetative tissues, despite the highly compacted chromatin, small nuclear volume and low hydration status of seeds.
Keywords: ABA INSENSITIVE 3, LITTLE NUCLEI, chromatin organization, seed development, seed maturation, transcription
Introduction
Plant embryos are protected within the seed, which allows their dispersal. In addition, the extremely low metabolic activities and a moisture level below 10% contribute to seed survival under hostile environmental conditions for extended periods.
The seed maturation phase in the model plant Arabidopsis thaliana is initiated after the embryo has been fully developed, starting at ~10 days after pollination (DAP) and ends when the seed is mature and desiccated (~20 DAP). The transcription factors ABSCISIC ACID INSENSITIVE 3 (ABI3), LEAFY COTYLEDON 1 (LEC1), LEC2 and FUSCA 3 (FUS3) control seed maturation in Arabidopsis.1 Mutants in these central regulators accumulate less storage compounds, show reduced desiccation tolerance and reduced seed dormancy.1-3
Studies on chromatin organization in seeds focused on seedling establishment and on the early stages of embryo development. It has for instance been shown that germinating seeds lack conspicuous heterochromatic DNA-domains, which re-appear during seedling establishment.4 In addition, diverse chromatin remodeling factors are required to repress embryonic properties in adult plants.5,6 We have recently analyzed nuclear morphology and chromatin organization in maturing and dry seeds at the microscopic level, using 4',6-diamidino-2-phenylindole (DAPI) staining. Interestingly, this revealed a major decrease in nuclear size between 8 and 12 DAP, before major dehydration of the maturing seed.7 Nuclei increased again in size during imbibition/germination. Although seeds are saturated with water at 1–2 h after imbibition, a strong increase in nuclear size was observed only after 24 h of imbibition, indicating that germination (and not rehydration per se) is required for the increase in nuclear size. In conclusion, it was shown that the dynamics in nuclear size is developmentally controlled, independently from changes in moisture content. Accordingly, a similar reduction in nuclear size was observed in desiccated leaves of the resurrection plant Craterostigma plantagineum.7 This suggests that reduced nuclear size is likely to be part of a general mechanism to establish desiccation tolerance in plants.
Analysis of heterochromatic sequences in nuclei by fluorescent In situ hybridization (FISH) and calculation of the relative heterochromatic fraction (RHF) showed that the chromatin of embryonic cotyledon nuclei from mature seeds is highly condensed. Kinetic analysis revealed that the changes in chromatin condensation can be uncoupled from the changes in nuclear size and hydration status of the seed, indicating that the increase in chromatin compaction is an independent process.7
In this short communication, we follow up on these observations and study the role of the genes ABI3, LITTLE NUCLEI 1 (LINC1) and LINC2 in the genetic uncoupling of nuclear size and chromatin condensation. In addition, we show that transcript levels in mature siliques at the end of seed maturation are similar to those in other tissues, despite the highly compact chromatin, the small nuclear volume and the low moisture content of seeds.
Results and Discussion
Genetic uncoupling of nuclear size reduction and chromatin compaction in seeds
The transcription factor ABI3 is involved in the reduction of nuclear size during seed maturation, whereas the nuclear matrix proteins LINC1 and LINC2 are required for the increase in size during imbibition/germination.7 The linc1-1 linc2-1 double mutant has constitutive small nuclei,8 which only slightly reduce in size during seed maturation. We were interested whether the abi3-5 and linc1-1 linc2-1 mutants also affect chromatin condensation during seed maturation. Therefore, we calculated the relative heterochromatin fraction (RHF), an indicator of chromatin compaction,9 at the beginning and the end of seed maturation.
The abi3-5 mutant shows similar RHF values at 10 DAP and 20 DAP (Fig. 1). These values are comparable to those of wild-type Landsberg erecta nuclei at the beginning of seed maturation. ABI3 is a central regulator of seed maturation and many aspects of seed maturation are affected in the abi3-5 mutant, including the decrease in nuclear size.7 Therefore, ABI3 is required for the regulation of both the decrease in nuclear size and the increase in chromatin compaction during seed maturation. In contrast, the linc1-1 linc2-1 double mutant shows an increase in RHF similar to wild-type, despite the absence of a strong reduction in nuclear size.7 This underlines that the decrease in nuclear size and the increase in chromatin compaction can be uncoupled and confirms the independent regulation of these two processes.
Figure 1.
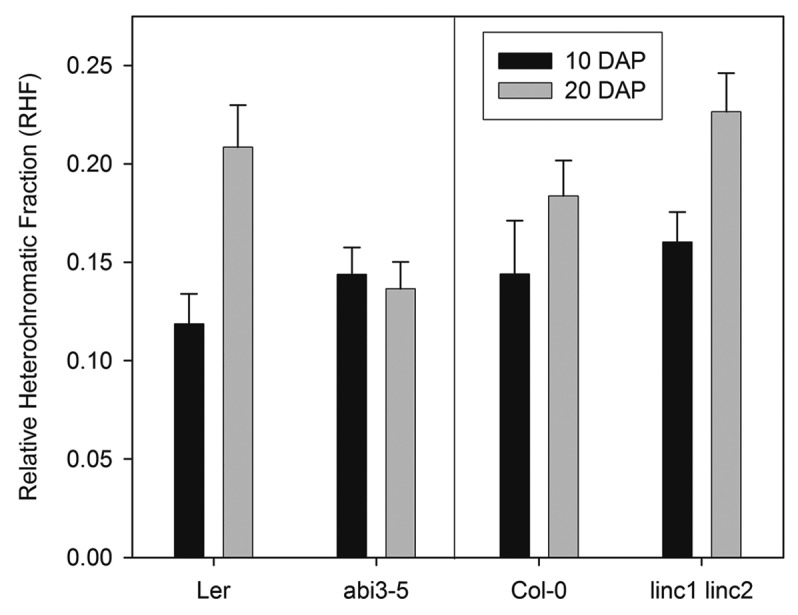
Chromatin compaction in embryonic cotyledon nuclei during seed maturation of abi3-5 and linc1-2 linc2-1 mutants. Relative heterochromatin fraction (RHF) at 10 DAP (black bars) and 20 DAP (gray bars) in abi3-5 and linc1-2 linc2-1 and the corresponding wild types Landsberg erecta (Ler) and Columbia-0 (Col-0). Error bars represent SE, n ≥ 99.
Maintenance of transcription levels in mature siliques
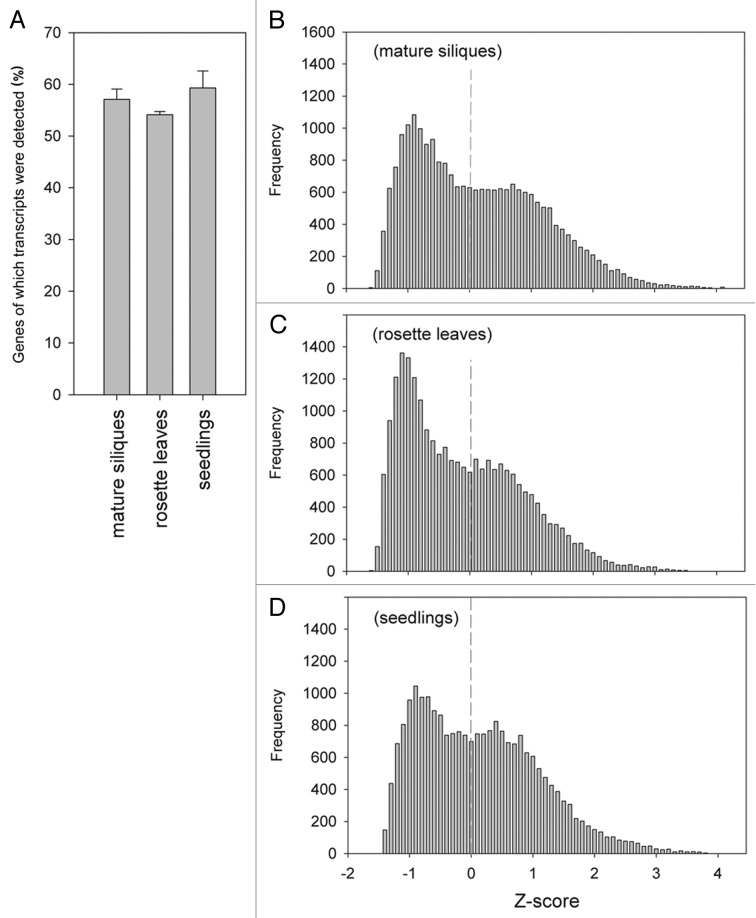
Increased chromatin compaction levels are generally associated with decreased transcriptional activity.10,11 Interestingly, the decrease in nuclear volume and increase in chromatin condensation of cotyledons during seed maturation is probably not associated with a general reduction in transcription levels, as the fraction of genes for which gene expression is detected in mature siliques is comparable to that of leaves and seedlings (Fig. 2A). In addition, a comparison of the transcription dynamics in these tissues using Z-scores12 showed similar distributions of genes with expression below or above the average expression level. Mature siliques even showed a higher fraction of genes expressed above average than rosette leaves (Fig. 2B–D). Mature siliques consist of several tissues that could all contribute to the observed high expression levels. However, siliques and testa are dead tissues and it is plausible that the cotyledons indeed contain a combination of high expression levels and high chromatin compaction.
Figure 2.
Comparison of transcriptomics data between diverse plant tissues. (A) Fraction of genes for which transcripts were detected (% of probe sets with detection p value < 0.05) in different tissues of Ler wild-type. Siliques were harvested at 18–19 DAP13 and rosette leaves and seedling data were obtained from publicly available microarray data NASCArray ID: affymetrix.arabidopsis.info/narrays/experimentpage.pl?experimentid = 327 and GEO (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE911), respectively. Error bars represent SE (B–D) Histograms of relative expression levels (Z-scores) of (B) siliques at 18–19 DAP, (C) rosette leaves and (D) seedlings. Note that the average expression level is X = 0 and is shown as a dashed line.
Possibly, this apparent discrepancy could be explained by the observation that several components of the RNA polymerase II associated factor 1 complex (PAF1C) are upregulated toward the end of seed maturation to maintain transcription in small nuclei with highly condensed chromatin.13 PAF1C facilitates transcription by providing a platform for the association of complexes that modulate the structure of local chromatin during transcription elongation.14 Another likely explanation is that these experiments do not only reveal actively transcribed mRNA, but show the total amount of RNA that is present in seeds including stored mRNA transcribed during earlier phases of seed maturation. This would imply that mRNA in seeds is very stable.
It is not possible to measure active transcription in dry seeds, but run-on experiments can determine the genes that are in a transcriptionally competent state. Comai and Harada15 analyzed several genes in Brassica napus seeds and concluded that their transcriptional competence was reduced but not absent in dry seeds compared with maturing seeds. Although these genes still have the ability to be transcribed in vitro, they are probably not actively transcribed in vivo in the dry seed due to the extremely low moisture content and the highly compacted chromatin.
Materials and Methods
Tissues were fixed in Carnoys fixative. Spread preparations of nuclei were made as described previously.16 RHF (fluorescence intensity of intensely DAPI-stained chromocenters, relative to the fluorescence of the entire nucleus) measurements were performed using a macro17 in ImagePro-Plus (Media Cybernetics) as described previously.7,9
The Z score transformation is a common approach usually applied for analyzing expression behavior within multiple experiments.12 It has been applied to depict expression dynamics of microarray data.18,19 The Z score transformation adjusts the logarithmic gene expression values such that each experiment has zero mean and unit variance. Z scores are calculated by subtracting the global average to the gene expression value and dividing that result by the global standard deviation (SD): Z score = (intensityG - mean intensityG1. . .Gn)/SDG1. . .Gn.
In order to compare the average expression level in mature siliques to that in other tissues, we downloaded publicly available microarray data of rosette leaves and seedlings from NASCArrays (affymetrix.arabidopsis.info/narrays/experimentpage.pl?experimentid=327) and GEO (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE911), with three and two replicates, respectively. These data are all from the Ler accession and generated using the Affymetrix Arabidopsis ATH1 Genome Array. The processing of the microarray data was done as previously18 and the Z scores were then calculated as above.
Acknowledgments
This work was supported by an EMBO Long-term Fellowship (ALTF 700-2010) to M.vZ. and by the Deutsche Forschungsgemeinschaft (SO 691/3-1) and the Max Planck Society to W.J.J.S.
Glossary
Abbreviations:
- DAP
days after pollination
- DAPI
4',6-diamidino-2-phenylindole
- FISH
fluorescent in situ hybridization
- PAF1C
RNA polymerase II associated factor 1 complex
- RHF
relative heterochromatin fraction
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/19281
References
- 1.To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, et al. A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell. 2006;18:1642–51. doi: 10.1105/tpc.105.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holdsworth MJ, Bentsink L, Soppe WJJ. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 3.Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008;54:608–20. doi: 10.1111/j.1365-313X.2008.03461.x. [DOI] [PubMed] [Google Scholar]
- 4.Mathieu O, Jasencakova Z, Vaillant I, Gendrel AV, Colot V, Schubert I, et al. Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell. 2003;15:2929–39. doi: 10.1105/tpc.017467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:13839–44. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka M, Kikuchi A, Kamada H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008;146:149–61. doi: 10.1104/pp.107.111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Zanten M, Koini MA, Geyer R, Liu Y, Brambilla V, Bartels D, et al. Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc Natl Acad Sci U S A. 2011;108:20219–24. doi: 10.1073/pnas.1117726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell. 2007;19:2793–803. doi: 10.1105/tpc.107.053231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tessadori F, van Zanten M, Pavlova P, Clifton R, Pontvianne F, Snoek LB, et al. Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000638. doi: 10.1371/journal.pgen.1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fransz P, de Jong H. From nucleosome to chromosome: a dynamic organization of genetic information. Plant J. 2011;66:4–17. doi: 10.1111/j.1365-313X.2011.04526.x. [DOI] [PubMed] [Google Scholar]
- 11.Tessadori F, van Driel R, Fransz P. Cytogenetics as a tool to study gene regulation. Trends Plant Sci. 2004;9:147–53. doi: 10.1016/j.tplants.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Geyer R, van Zanten M, Carles A, Li Y, Hörold A, et al. Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS One. 2011;6:e22241. doi: 10.1371/journal.pone.0022241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–67. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 15.Comai L, Harada JJ. Transcriptional activities in dry seed nuclei indicate the timing of the transition from embryogeny to germination. Proc Natl Acad Sci U S A. 1990;87:2671–4. doi: 10.1073/pnas.87.7.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schubert I, Fransz PF, Fuchs J, de Jong JH. Chromosome painting in plants. Methods Cell Sci. 2001;23:57–69. doi: 10.1023/A:1013137415093. [DOI] [PubMed] [Google Scholar]
- 17.Pavlova P, Tessadori F, de Jong HJ, Fransz P. Immunocytological analysis of chromatin in isolated nuclei. Methods Mol Biol. 2010;655:413–32. doi: 10.1007/978-1-60761-765-5_28. [DOI] [PubMed] [Google Scholar]
- 18.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–6. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Xie W, Chen Y, Tang W, Yang J, Ye R, et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010;61:752–66. doi: 10.1111/j.1365-313X.2009.04100.x. [DOI] [PubMed] [Google Scholar]