Abstract
Copper-zinc superoxide dismutase (CuZnSOD; CSD) is an important antioxidant enzyme for oxidative stress protection. To date, two activation pathways have been identified in many species. One requiring the CCS, Cu chaperone for SOD, to insert Cu and activate CSD (referred to as CCS-dependent pathway), and the other works independently of CCS (referred to as CCS-independent pathway). In our previous study, we suggest an unidentified factor will work with glutathione (GSH) for CSD activation in the absence of the CCS. Here, two models of the CCS-independent mechanism are proposed. The role of the unidentified factor may work as a scaffold protein, which provides a platform for the CSD protein and Cu-GSH to interact, or as a Cu carrier, which itself can bind Cu and interact with CSD proteins. We also suggest that the CSD protein conformation at C-terminal is important in providing a docking site for unidentified factor to access.
Keywords: Arabidopsis, CCS, CuZnSOD, GSH, SOD activation
Superoxide dismutases (SODs) can defend against free radical species by disproportionation O2- into H2O2 and O2 molecules,1,2 which are classified as Cu-Zn SOD (CuZnSOD), Fe SOD (FeSOD), Mn SOD (MnSOD) or Ni SOD (NiSOD).3,4 In Arabidopsis thaliana, there are three CuZnSOD (CSD) isoforms: CSD1 in the cytoplasm, CSD2 in chloroplasts, and CSD3 in peroxisomes.3,5 It is known that CSD activation involves two pathways.6,7 The CCS-dependent pathway has been well studied, which requires the CCS (Cu chaperone of SOD).8 An alternative pathway, which acts in the absence of CCS, is observed in mice, nematode and Arabidopsis,7,9-12 with its mechanism unclear. Depending on an organism’s lifestyle and complexity, it is very likely that different eukaryotes may have evolved to favor one pathway over the other.
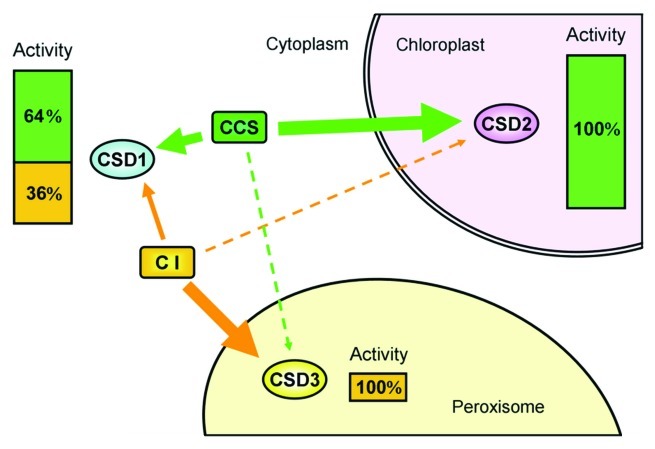
In our previous study,7 the three Arabidopsis CSD isomers show different favor for the two activation pathways, which is due to cellular environments they localized but not their protein structures (Fig. 1): cytoplasmic CSD1 is activated mainly depending on CCS (64%) and partially by the CCS-independent pathway (36%); chloroplastic CSD2 activation depends completely on CCS (100%); and peroxisomal CSD3 activated mainly by the CCS-independent pathway (100%). The extremely different preference of three CSD present in a single cell makes Arabidopsis a good species for the investigation of the CuZnSOD activation preference.
Figure 1. Variable levels of CCS-dependent and -independent activation of three Arabidopsis CSDs localized in different cellular compartments. Assuming that the total activity of each CSD in the WT is 100%, the fraction activated by AtCCS (CCS) is in green and that activated through the CCS-independent (CI) pathway is in orange. Solid arrows represent the activation that was demonstrated experimentally, and the dashed arrows indicate activation levels that were undetectable in our system.
Two Models of How the Unidentified Factor Involves In the CCS-Independent Pathway
Based on our previous study, an unidentified factor is participated in the CCS-independent pathway and may cooperate with glutathione (GSH) to activate CSD.7 Here, we propose two models for CSD activation in the absence of the CCS (Fig. 2). In the first model, the Cu cofactor is transferred from Cu-GSH complexes to CSD, but interaction between the Cu-GSH and CSD proteins requires the assistance of an unidentified factor (model A). The role of the unidentified factor is a scaffold protein, which provides a platform for the CSD protein and Cu-GSH to interact. In the second model, Cu is first transferred from Cu-GSH to the unidentified factor and then transferred to a CSD protein (model B). Here, the role of the unidentified factor could be as a Cu carrier, which itself can bind Cu and interact with CSD proteins. Nevertheless, the interaction between CSD and the unidentified factor is essential in either model. Of note, the importance of proline 144 on the carboxyl terminus of human CSD (hSOD1) has been noted as a potential Cu-GSH docking site of CSD protein,10 which implies the approach of Cu-GSH to the CSD protein (model A), for a more convincing model.
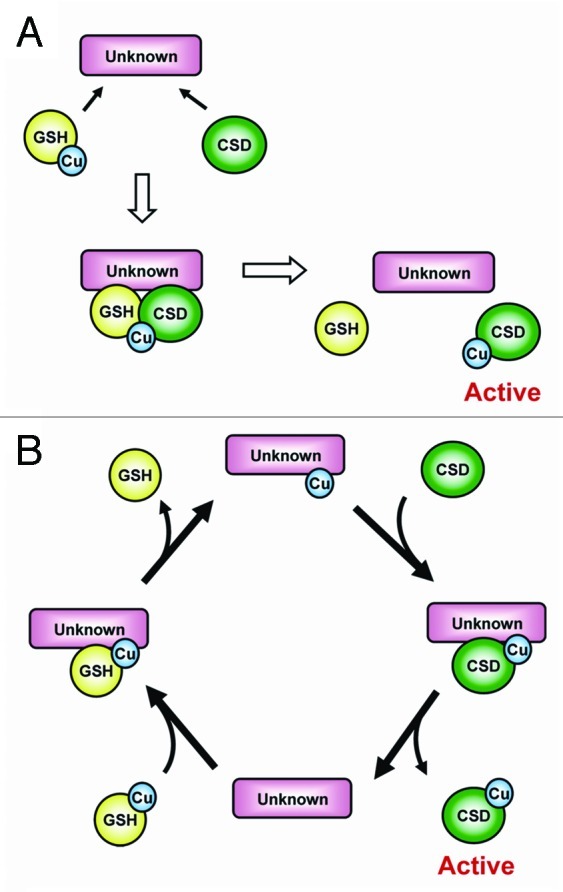
Figure 2. Two possible models for CCS-independent activation of CSD. (A) The unidentified factor (unknown), acting as a scaffold protein, first interacts with both GSH and the CSD protein, and the GSH-bound Cu cofactor is then transferred to CSD. (B) The Cu cofactor is first transferred from GSH to the unidentified factor and then transferred to a CSD protein. In this model, the unidentified factor functions as a Cu carrier.
C-Terminal of CSD Protein May Determines Its Activation by CCS or the Unidentified Factor
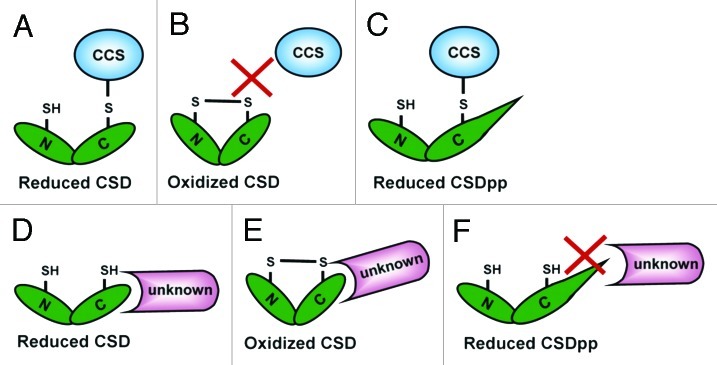
An important difference between CCS-dependent and -independent pathways concerns the status of the CSD disulfide bond.6 CCS can activate only disulfide-reduced CSD proteins, because the interaction between CCS and CSD involves formation of an intermolecular disulfide bond (Fig. 3A).13 Thus, intramolecular disulfide-oxidized CSD would preclude the interaction (Fig. 3B). In contrast, both disulfide-reduced and -oxidized CSD can be activated by the CCS-independent pathway.6 This observation led us to suggest that the interaction of CSD and the unidentified factor differs from that of CSD and CCS, which does not require the formation of the intermolecular disulfide bond (Fig. 3D), so the disulfide-oxidized CSD can still interact with the unidentified factor (Fig. 3E).
Figure 3. CSD interacts with the CCS vs. the unidentified factor. (A–C) The CCS interacts with disulfide-reduced CSD and CSDPP but not disulfide-oxidized CSD. (D–F), The unidentified factor (unknown) interacts with disulfide-reduced and -oxidized CSD but not CSDPP.
As mentioned previously, the presence of proline at position 144 of yeast CSD (ySOD1) prevents activation by the CCS-independent pathway.10,12 The most recent model indicates that this proline results in a conformational restriction that prevents disulfide bond formation within CSD in the absence of the CCS.6 This restriction on disulfide bond formation can be overcome by the action of CCS but not the CCS-independent mechanism (Fig. 3C and F, respectively). Here, we suggest another reason for an essential unidentified factor in CCS-independent pathway interacting with the disulfide-oxidized CSD. In this model, the molecular conformation adopted by CSD containing proline 144 may prevent interaction of the unidentified factor with the CSD protein. This situation would explain why proline 144 blocked only CCS-independent activity but not that conferred by CCS. Further investigation is required to characterize the unidentified factor and elucidate the complete mechanism.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/19192
References
- 1.Beyer W, Imlay J, Fridovich I. Superoxide dismutases. Prog Nucleic Acid Res Mol Biol. 1991;40:221–53. doi: 10.1016/S0079-6603(08)60843-0. [DOI] [PubMed] [Google Scholar]
- 2.Bowler C, Van Montagu MV, Inzé D. Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:83–116. doi: 10.1146/annurev.pp.43.060192.000503. [DOI] [Google Scholar]
- 3.Alscher RG, Erturk N, Heath LS. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot. 2002;53:1331–41. doi: 10.1093/jexbot/53.372.1331. [DOI] [PubMed] [Google Scholar]
- 4.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337–49. doi: 10.1016/S0891-5849(02)00905-X. [DOI] [PubMed] [Google Scholar]
- 5.Kliebenstein DJ, Monde RA, Last RL. Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998;118:637–50. doi: 10.1104/pp.118.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leitch JM, Yick PJ, Culotta VC. The right to choose: multiple pathways for activating copper,zinc superoxide dismutase. J Biol Chem. 2009;284:24679–83. doi: 10.1074/jbc.R109.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang CH, Kuo WY, Weiss C, Jinn TL. Copper chaperone-dependent and -independent activation of three copper-zinc superoxide dismutase homologs localized in different cellular compartments in Arabidopsis. Plant Physiol. 2011;158:737–46. doi: 10.1104/pp.111.190223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culotta VC, Klomp LWJ, Strain J, Casareno RLB, Krems B, Gitlin JD. The copper chaperone for superoxide dismutase. J Biol Chem. 1997;272:23469–72. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 9.Wong PC, Waggoner D, Subramaniam JR, Tessarollo L, Bartnikas TB, Culotta VC, et al. Copper chaperone for superoxide dismutase is essential to activate mammalian Cu/Zn superoxide dismutase. Proc Natl Acad Sci U S A. 2000;97:2886–91. doi: 10.1073/pnas.040461197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll MC, Girouard JB, Ulloa JL, Subramaniam JR, Wong PC, Valentine JS, et al. Mechanisms for activating Cu- and Zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc Natl Acad Sci U S A. 2004;101:5964–9. doi: 10.1073/pnas.0308298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu CC, Lee WC, Guo WY, Pan SM, Chen LJ, Li HM, et al. A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol. 2005;139:425–36. doi: 10.1104/pp.105.065284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen LT, Culotta VC. Activation of CuZn superoxide dismutases from Caenorhabditis elegans does not require the copper chaperone CCS. J Biol Chem. 2005;280:41373–9. doi: 10.1074/jbc.M509142200. [DOI] [PubMed] [Google Scholar]
- 13.Culotta VC, Yang M, O’Halloran TV. Activation of superoxide dismutases: putting the metal to the pedal. Biochim Biophys Acta. 2006;1763:747–58. doi: 10.1016/j.bbamcr.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]