Abstract
Two Bacillus subtilis genes encoding two proteins (currently annotated ThiD and YjbV) were overexpressed and characterized. YjbV has 4-amino-5-hydroxymethyl-2-methylpyrimidine and 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate kinase activity and should be reannotated ThiD, and B. subtilis ThiD has pyridoxine, pyridoxal, and pyridoxamine kinase activity and should be reannotated PdxK.
The biosynthesis of thiamine pyrophosphate (TPP) involves the coupling of 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate (HMP-PP) and 4-methyl-5-β-hydroxyethylthiazole phosphate (Thz-P) to form thiamine phosphate followed by a final phosphorylation (1). In addition to the de novo biosynthesis, microorganisms have developed several salvage pathways for the biosynthesis of TPP (Table 1). Thiamine from the growth medium is either phosphorylated by thiamine kinase or pyrophosphorylated by thiamine pyrophosphokinase (J. Melnick, E. Lis, J.-H. Park, H. Mori, C. Kinsland, J. Perkins, G. Schyns, A. Osterman, and T. P. Begley, submitted for publication). The pyrimidine and thiazole components can also be salvaged: thiazole is phosphorylated by thiazole kinase (2, 4, 6), HMP is phosphorylated to HMP-P by both ThiD and PdxK (3, 7, 10), and the phosphorylation of HMP-P is catalyzed by ThiD (5, 6, 7). Thus, ThiD has both a biosynthetic and a salvage function in thiamine biosynthesis. PdxK is able to phosphorylate a broad range of substrates, including HMP, pyridoxal (PL), pyridoxamine (PM), and pyridoxine (PN), and is a salvage enzyme in the biosynthesis of thiamine as well as that of PL phosphate (PLP).
TABLE 1.
Microbial thiamine salvage enzymes
| Enzyme | Microorganism | Substrate | Product | Reference(s) |
|---|---|---|---|---|
| ThiM | Bacillus subtilis | Thz | Thz-P | 2, 4, 6 |
| Escherichia coli | ||||
| Salmonella typhimurium | ||||
| PdxK | Escherichia coli | HMP | HMP-P | 10 |
| ThiD | Escherichia coli | HMP | HMP-P | 7 |
| ThiK | Escherichia coli | Thiamine | Thiamine phosphate | Melnick et al., submitted |
| ThiN | Bacillus subtilis | Thiamine | TPP | Melnick et al., submitted |
A search of the Bacillus subtilis genomic database (http://genolist.pasteur.fr/SubtiList/index.html) shows homologues of Escherichia coli ThiD and PdxK named YjbV (1246149-1246961) and ThiD (3899983-3900795). They are both 271-amino-acid proteins. yjbV is located immediately downstream of the thiOSGF operon that is involved in Thz-P biosynthesis, while thiD is not clustered with any of the thiamine or PLP biosynthetic genes. E. coli PdxK shows 24 and 25% sequence identity with B. subtilis YjbV and ThiD, respectively, and E. coli ThiD shows 41 and 35% identity with B. subtilis YjbV and ThiD, respectively. The level of sequence homology between these two proteins is too high to allow the preferred substrate to be predicted for either protein. However, the occurrence of yjbV in the thiazole biosynthetic operon suggests that these proteins are incorrectly annotated and that YjbV might function as the B. subtilis HMP/HMP-P kinase. Here we report the overexpression of YjbV and ThiD from B. subtilis and the identification of the substrate preferences of the two proteins.
The amino acid sequences of E. coli ThiD and PdxK were obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and used with the SubtiList World Wide Web server for a BLAST search. For cloning B. subtilis thiD and yjbV, standard DNA restriction endonuclease digestion, ligation, and transformation methods were used (9). Genomic DNA and plasmid DNA were purified with a Wizard Plus SV genomic DNA kit and a DNA Miniprep kit, respectively (Promega). DNA fragments were separated by agarose gel electrophoresis, excised, and purified with a QIAquick gel extraction kit (Qiagen). pET-16b plasmid was obtained from Novagen. E. coli strain DH5α was used as a recipient for transformation during plasmid construction and for plasmid propagation and storage. E. coli BL21(DE3) was purchased from Novagen and used as a host strain for the overexpression of the proteins. A Perkin Elmer GeneAmp PCR System 2400 apparatus and Platinum Pfx DNA polymerase (Gibco Life Technologies) were used for PCR. B. subtilis CU1065 genomic DNA was used as a template for PCR. Primer synthesis and DNA sequencing were performed by the Bioresource Center at Cornell University. Primers introduced NdeI and XhoI restriction enzyme sites at the 5′ and 3′ ends, respectively.
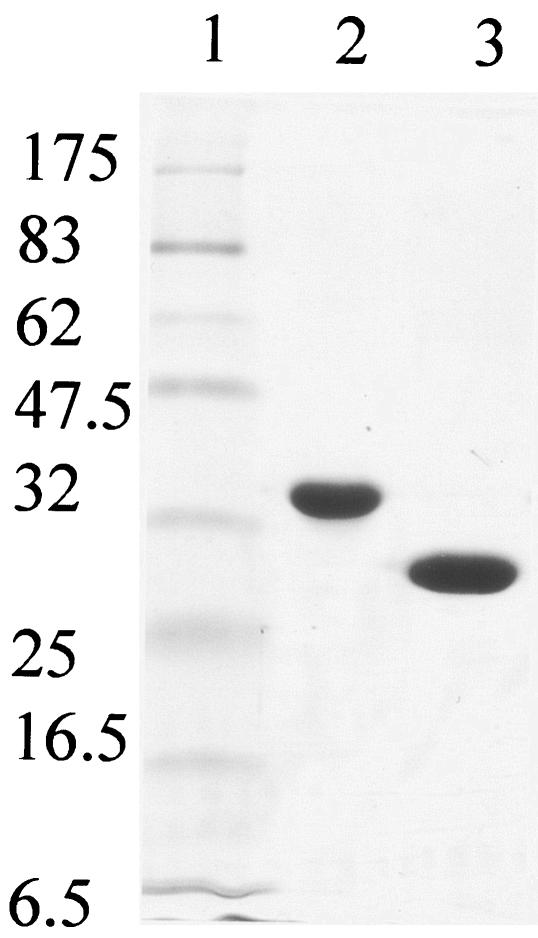
For the overexpression and purification of ThiD and YjbV, their corresponding overexpression plasmids were transformed into competent E. coli BL21(DE3) cells and the transformed cells were grown at 37°C in Luria-Bertani medium containing 50 mg of ampicillin/liter. To induce the overexpression of proteins, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture (when the optical density at 595 nm reached 0.6) to achieve a final concentration of 1 mM. Culture growth was continued for 8 h at 28°C, after which the cells were harvested and stored at −80°C until further use. The proteins were purified according to a Qiagen protocol for the purification of His-tagged proteins. The eluted proteins were rapidly desalted using a PD-10 column (Amersham Pharmacia) because of instability under high-salt concentrations and stored in 5% glycerol at −80°C. ThiD was soluble and stable in 50 mM Tris buffer (pH 8), but YjbV solutions rapidly became turbid. The results of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with the purified proteins are shown in Fig. 1. Although the migration characteristics of the purified proteins were different, their molecular weights were confirmed by mass spectrometry (data not shown).
FIG. 1.
SDS-PAGE (12%) analysis of purified B. subtilis ThiD and YjbV. Lane 1, molecular mass markers (in kilodaltons); lane 2, His-tagged ThiD; lane 3, His-tagged YjbV. Although ThiD and YjbV are predicted to have the same molecular mass, they migrate differently on the gel.
The reaction mixtures for B. subtilis ThiD and YjbV enzymatic assays contained 1 mM ATP, 1 mM HMP, 2 mM MgCl2, and 40 μg of enzyme in 100 μl of 50 mM Tris-HCl (pH 8). After incubation at 37°C for 10 min, the reaction was quenched by the addition of 100 μl of 10% trichloroacetic acid and centrifuged to remove proteins. A total of 20 μl of the reaction mixture was analyzed by high-pressure liquid chromatography (HPLC) (Supelcosil LC-18-T) (15- by 4.6-mm column). The elution conditions were as follows: flow rate, 1 ml/min; elution time, 0 to 20 min; elution buffer, 100% of 0.1 M potassium phosphate (pH 6.6). To conduct a competition assay, ThiD was incubated with all four substrates (0.3 mM concentrations each of HMP, PL, PM, and PN) for 30 min under the conditions described above (except that 2 mM ATP was used and the reaction mixture was analyzed by HPLC).
For kinetic studies, ADP produced by the kinase activity of ThiD or YjbV was assayed using a pyruvate kinase-lactate dehydrogenase-coupled system (which uses ADP and NADH as substrates). The consumption of NADH by this coupled system can be measured by monitoring the decrease in absorbance at 340 nm (7). Pyruvate kinase, lactate dehydrogenase, phosphoenolpyruvate, NADH, and PL were purchased from Sigma. HMP was synthesized as previously described (8). The assay mixture for the kinetic analysis of ThiD in the presence of HMP or PL contained saturating concentrations of ATP (5 mM), 30 to 400 μM HMP (or 30 to 300 μM PL), 10 mM MgCl2, 50 mM KCl, 0.2 mM NADH, 1 mM phosphoenolpyruvate, 8 units of pyruvate kinase/ml, and 10 units of lactate dehydrogenase/ml in 0.6 ml of 50 mM Tris-HCl (pH 8). Addition of ThiD to achieve a final concentration of 6.7 μM initiated the reactions, which were then monitored over 5 min for NADH consumption at 340 nm.
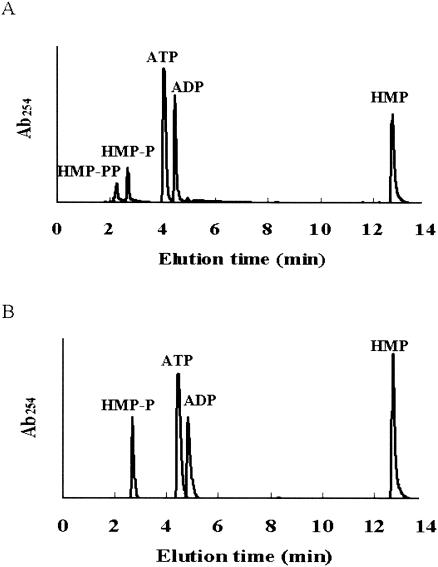
HPLC analysis of the reaction mixture containing B. subtilis YjbV showed the appearance of two new peaks corresponding to HMP-P and HMP-PP (Fig. 2A). The reaction mixture containing B. subtilis ThiD showed only one pyrimidine product peak, which corresponds to HMP-P (Fig. 2B). In addition to the phosphorylation of HMP, B. subtilis ThiD was able to phosphorylate PL, PM, and PN, producing PLP, PMP, and PNP, respectively. Under similar conditions, YjbV did not catalyze the phosphorylation of these compounds (data not shown). A competition assay using the substrates of ThiD revealed a preference for PL followed by HMP, PN, and PM (8:2.4:1.1:1 product ratios). The kinetic parameters for B. subtilis ThiD are shown in Table 2. The kinetic parameters of B. subtilis YjbV could not be determined, because the reaction mixture became turbid immediately after the reaction began.
FIG. 2.
HPLC analysis of the ThiD- and YjbV-catalyzed reactions. (A) YjbV catalyzed phosphorylation of HMP and HMP-P. (B) ThiD catalyzed phosphorylation of HMP.
TABLE 2.
Kinetic parameters for substrate phosphorylation by B. subtilis ThiD
| Substrate | Km (μM) | kcat (s-1) | kcat/Km (s-1 μM-1) |
|---|---|---|---|
| HMP | 2,030 | 0.36 | 1.8 × 10−4 |
| PL | 46.6 | 0.032 | 6.9 × 10−4 |
Overall our results indicate that B. subtilis YjbV has HMP/HMP-P kinase activity and should be reannotated ThiD (i.e., the name should be changed from YjbV to ThiD) and that B. subtilis ThiD has PN/PL/PM/HMP kinase activity and should be reannotated PdxK (i.e., from ThiD to PdxK).
Acknowledgments
This research was supported by a grant from NIH (DDK44083).
REFERENCES
- 1.Begley, T. P., D. M. Downs, S. E. Ealick, F. W. McLafferty, A. P. G. M. Van Loon, S. Taylor, N. Campobasso, H.-J. Chiu, C. Kinsland, J. J. Reddick, and J. Xi. 1999. Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 171:293-300. [DOI] [PubMed] [Google Scholar]
- 2.Campobasso, N., I. I. Mathews, T. P. Begley, and S. E. Ealick. 2000. Crystal structure of 4-methyl-5-β-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 Å resolution. Biochemistry 39:7868-7877. [DOI] [PubMed] [Google Scholar]
- 3.Mizote, T., and H. Nakayama. 1989. Purification and properties of hydroxymethylpyrimidine kinase from Escherichia coli. Biochim. Biophys. Acta 991:109-113. [DOI] [PubMed] [Google Scholar]
- 4.Mizote, T., and H. Nakayama. 1989. The thiM locus and its relation to phosphorylation of hydroxyethylthiazole in Escherichia coli. J. Bacteriol. 171:3228-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizote, T., M. Tsuda, D. D. S. Smith, H. Nakayama, and T. Nakazawa. 1999. Cloning and characterization of the thiD/J gene of Escherichia coli encoding a thiamin-synthesizing bifunctional enzyme, hydroxymethylpyrimidine kinase/phosphomethylpyrimidine kinase. Microbiology 145:495-501. [DOI] [PubMed] [Google Scholar]
- 6.Petersen, L. A., and D. M. Downs. 1997. Identification and characterization of an operon in Salmonella typhimurium involved in thiamine biosynthesis. J. Bacteriol. 179:4894-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddick, J. J., C. Kinsland, R. Nicewonger, T. Christian, D. M. Downs, M. E. Winkler, and T. P. Begley. 1998. Overexpression, purification and characterization of two pyrimidine kinases involved in the biosynthesis of thiamin: 4-amino-5-hydroxymethyl-2-methylpyrimidine kinase and 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase. Tetrahedron 54:15983-15991. [Google Scholar]
- 8.Reddick, J. J., R. Nicewonger, and T. P. Begley. 2001. Mechanistic studies on thiamin phosphate synthase: evidence for a dissociative mechanism. Biochemistry. 40:10095-10102. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Yang, Y., G. Zhao, and M. E. Winkler. 1996. Identification of the pdxK gene that encodes pyridoxine (vitamin B6) kinase in Escherichia coli K-12. FEMS Microbiol. Lett. 141:89-95. [DOI] [PubMed] [Google Scholar]