Abstract
Poly-ADP ribose polymerase (PARP) inhibitors have shown promise in the treatment of human malignancies characterized by deficiencies in the DNA damage repair proteins BRCA1 and BRCA2 and preclinical studies have demonstrated the potential effectiveness of PARP inhibitors in targeting ataxia-telangiectasia mutated (ATM)-deficient tumours. Here, we show that mantle cell lymphoma (MCL) cells deficient in both ATM and p53 are more sensitive to the PARP inhibitor olaparib than cells lacking ATM function alone. In ATM-deficient MCL cells, olaparib induced DNA-PK-dependent phosphorylation and stabilization of p53 as well as expression of p53-responsive cell cycle checkpoint regulators, and inhibition of DNA-PK reduced the toxicity of olaparib in ATM-deficient MCL cells. Thus, both DNA-PK and p53 regulate the response of ATM-deficient MCL cells to olaparib. In addition, small molecule inhibition of both ATM and PARP was cytotoxic in normal human fibroblasts with disruption of p53, implying that the combination of ATM and PARP inhibitors may have utility in targeting p53-deficient malignancies.
Keywords: ATM, DNA-PK, olaparib, p53, PARP
INTRODUCTION
Synthetic lethality using poly-ADP ribose polymerase (PARP) inhibitors has emerged as a new potential therapeutic strategy to exploit tumour-specific genetic alterations. Synthetic lethality is defined as the premise, whereby, deletion of one of two genes independently has no effect on cellular viability, whereas, simultaneous loss of both genes is lethal (Kaelin, 2005). The utility of this approach was first demonstrated when cells with mutations in the breast and ovarian susceptibility genes BRCA1 and BRCA2 were shown to be extremely sensitive to small molecule inhibitors of the DNA single strand break (SSB) sensing protein PARP-1 (Bryant et al, 2005; Farmer et al, 2005). PARP inhibitors have subsequently shown promise in Phase I and Phase II clinical trials for the treatment of BRCA1/2-deficient breast, ovarian and prostate tumours (Audeh et al, 2010; Fong et al, 2009; Tutt et al, 2010). However, a recently completed Phase III study combining PARP inhibition with chemotherapy did not generate the anticipated survival gains (Guha, 2011; O'Shaughnessy et al, 2011) suggesting that additional, as yet unidentified, molecular factors may influence the in vivo anti-tumour effectiveness of this class of drugs.
BRCA1 and BRCA2 both function in the homologous recombination (HR) pathway for the repair of DNA double strand breaks (DSBs; San Filippo et al, 2008). Therefore, one proposed explanation for synthetic lethality in this context is that inhibition of PARP leads to accumulation of DNA damage, in particular SSBs, which are converted to dsDNA ends during DNA replication and are repaired by the HR pathway. Consequently, cells that are deficient in BRCA1 or BRCA2 have defective HR and are more sensitive to PARP inhibitors than their normal counterparts (Evers et al, 2010; McCabe et al, 2006). Subsequent experiments revealed that additional proteins share a synthetic lethal interaction with PARP, including the DNA damage activated protein kinase ataxia-telangiectasia mutated (ATM; Lord et al, 2008; Turner et al, 2008).
ATM, like ATM- and Rad3-related (ATR) and the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), is a member of the phosphatidyl-inositol kinase-like kinase (PIKK) family of protein kinases and a key regulator of the cellular response to genotoxic stress, in particular DNA DSBs (Lavin, 2008). Following DNA damage, ATM phosphorylates multiple downstream effector proteins including the checkpoint kinase Chk2, DNA repair factors including BRCA1 and critical transcriptional regulators such as p53 (Jackson & Bartek, 2009; Lavin, 2008). Through these phosphorylation events, the cell initiates cell cycle checkpoint arrest, DNA repair and/or apoptosis (Lavin, 2008). In contrast, the related protein DNA-PKcs primarily acts to repair DNA DSBs via the non-homologous end-joining (NHEJ) pathway (Mahaney et al, 2009). However, in the absence of ATM, DNA-PKcs can phosphorylate many of the same effector proteins as ATM, including H2AX and p53 (Callén et al, 2009; Stiff et al, 2004). Interestingly, a recent study found that PARP inhibitor sensitivity of BRCA1-, BRCA2- or ATM-deficient cells was alleviated by inhibition of DNA-PK, suggesting that functional DNA-PK, and consequently NHEJ, is required for PARP inhibitor-induced cytotoxicity (Patel et al, 2011).
ATM is frequently altered or deleted in breast cancer (Fang et al, 2010), in particular the triple negative subtype (Tommiska et al, 2008), as well as in gastric (Kang et al, 2008) and lung cancer (Ding et al, 2008). However, the highest rate of ATM alteration is seen in haematological cancers where B-cell lymphocytic leukaemia (B-CLL), follicular lymphoma (FL) and mantle cell lymphoma (MCL) are all characterized by high rates of inactivating mutations and/or deletion of ATM (Boultwood, 2001; Schaffner et al, 1999). Amongst these lymphoma subtypes, ATM alteration is most common in MCL, where mutations or deletions are seen in up to half of all cases (Fang et al, 2003). Cells in which ATM activity was disrupted either through genetic mutation, RNA interference or small molecule inhibition displayed increased sensitivity to PARP inhibitors (Bryant & Helleday, 2006; Turner et al, 2008; Williamson et al, 2010). Moreover, we recently demonstrated that MCL cell lines with loss of ATM function have increased sensitivity to PARP inhibitors in vitro and in vivo (Williamson et al, 2010). Subsequently, ATM-deficient B-CLL cells were also shown to be sensitive to PARP inhibitors (Weston et al, 2010). Together, these studies confirm that ATM and PARP have a synthetic lethal relationship and suggest that PARP inhibitors may have utility against ATM-deficient malignancies. However, whether additional genetic alterations impact upon the synthetic lethal interaction between PARP and ATM has not been addressed.
Genetic alterations of TP53 can be found in every major solid and haematological form of cancer and it has been estimated that up to 50% of all cancers have either mutation or deletion of TP53 (Hoeijmakers, 2001; Hollstein et al, 1991). Indeed, in MCL, 26% of cases contain TP53 mutation/deletion, 56% ATM alteration and 10% contain changes in both genes (Greiner et al, 2006). The p53 protein is phosphorylated on multiple sites in response to a large number of cellular stresses, including DNA DSBs, blocking its proteosomal degradation and promoting stabilization (Lavin, 2008; Maclaine & Hupp, 2011). Once stabilized, p53 transcriptional activity leads to expression of a variety of genes, including those involved in cell cycle checkpoint arrest like p21 and GADD45α, as well as NOXA and Puma, which promote apoptosis (Maclaine & Hupp, 2011). DNA damage-induced phosphorylation and stabilization of p53 is critical for maintaining genomic stability and acts as a barrier to tumourigenesis (Bartkova et al, 2006).
Several recent studies have found that concurrent alteration of ATM and p53 within the same cell causes sensitivity to both endogenous and exogenous genotoxic stresses. For example, combined loss of ATM and p53 in mouse embryonic fibroblasts (MEFs) leads to increased sensitivity to DNA damaging agents such as doxorubicin in vitro (Jiang et al, 2009). Clinically, cases of ATM/p53 co-mutation in breast cancer have improved response to therapy and clinical outcome compared to cases in which either protein is altered individually (Jiang et al, 2009). Also, there is increasing evidence from detailed genomic sequencing of different stages of lung (Ding et al, 2008) and breast (Jiang et al, 2009) cancers that early precursor lesions require disruption of either ATM or p53 to progress to malignancy. However, what effect, if any, p53 has on PARP inhibitor mediated synthetic lethality in ATM-deficient cells has not been examined.
Here, we demonstrate that MCL cells with genetic disruptions in both ATM and p53 display increased sensitivity to olaparib, in vitro and in vivo, compared to those deficient for only ATM. Direct disruption of p53 transcriptional activity, either by expression of dominant-negative (DN) p53 or by small molecule inhibition (pifithrin-α; Komarov et al, 1999), heightened the sensitivity of ATM-deficient cells to olaparib. Furthermore, the combined inhibition of ATM and PARP induced greater cytotoxicity in MCL cell lines and normal human fibroblasts with disruption of p53 than in cells with wild-type (WT) p53. Inhibition of PARP in ATM-deficient cells induced phosphorylation and stabilization of p53, which was largely dependent upon DNA-PK protein kinase activity, while direct inhibition of DNA-PK reduced the toxicity of olaparib in ATM-deficient cells, suggesting that in the absence of ATM, inappropriate NHEJ directs PARP inhibitor-mediated cell death. Together, these results indicate that the functional status of both DNA-PK and p53 may impact the effectiveness of PARP inhibitors in ATM-deficient malignancy and suggest that a more detailed molecular-based tumour characterization may be required to better target these agents. Moreover, our findings raise the prospect of targeting p53-deficient cancers with the combined use of ATM and PARP inhibitors.
RESULTS
UPN2 cells are more sensitive to olaparib in vitro than Granta-519 and this difference is not due to residual ATM activity
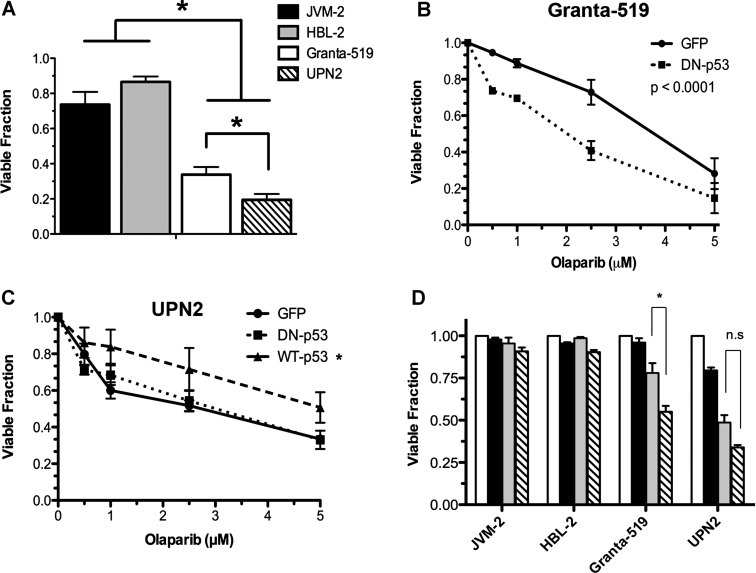
We previously characterized a panel of MCL cell lines for ATM function (Table 1 of Supporting Information and Williamson et al, 2010). Both Granta-519 and UPN2 cells displayed low ATM protein levels, reduced ATM-dependent signalling, radiation sensitivity and cell cycle checkpoint defects; all characteristics of cells lacking ATM function (Williamson et al, 2010). Moreover, these ATM-deficient MCL cell lines showed increased sensitivity towards small molecule inhibitors of PARP compared to their ATM-proficient counterparts in both in vitro (cell line) and in vivo (animal) models (Williamson et al, 2010). While comparing the level of PARP inhibitor sensitivity between these cell lines, we noted that, in vitro, UPN2 were more sensitive to the clinically relevant compound olaparib (AZD2881) than were Granta-519 (Fig 1A). We considered that one potential explanation for these findings might be a gene dose effect as Granta-519 cells retain a small amount of ATM protein expression, while UPN2 cells express undetectable levels of ATM (Fig 1A of Supporting Information).
Figure 1. Loss of p53 transcriptional activity increases the sensitivity of ATM-deficient MCL cells to PARP inhibition.
- MCL cell lines JVM-2 (black), HBL-2 (grey), Granta-519 (white) and UPN2 (hatched) cells were exposed to 5 µM olaparib for 96 h prior to determining cellular viability by trypan blue exclusion. Viability was normalized to DMSO treated controls for each individual cell line. Results were analysed by Student's t-test, where n = 6 for JVM-2, 5 for Granta-519, 6 for HBL-2 and 4 for UPN2. Error bars represent SEM. p-Values were 0.0408 for Granta-519 compared to UPN2, 0.0016 for JVM-2 compared to Granta-519 and <0.0001 for JVM-2 compared to UPN2 and HBL-2 compared to UPN2. Asterisks indicate p < 0.05.
- Granta-519 cells were transfected with either DN-p53 (dotted line) or GFP vector (solid line). Olaparib was added 24 h post transfection and cellular viability was determined by trypan blue exclusion following 96 h of exposure to increasing concentrations of olaparib. Results were analysed by two-way ANOVA, n = 3, error bars represent SEM, p < 0.0001.
- UPN2 cells were transfected with DN-p53 (dotted line), WT p53 (dashed line) or GFP vector (solid line), prior to exposure to increasing concentrations of olaparib and assayed as in panel B. Results were analysed by 2-way ANOVA, n = 3, error bars represent SEM, the p-value for GFP-vector compared to WT p53 was 0.0006, and for DN-p53 compared to wt-p53, 0.0023. p-Values <0.05 were considered statistically significant and marked by an asterisk.
-
JVM-2, HBL-2, Granta-519 and UPN2 cells were exposed to either DMSO (white bars), pifithrin-α (black bars; 50 µM), olaparib (grey bars; 2.5 µM) or both pifithrin-α and olaparib (hatched bars; at 50 and 2.5 µM, respectively), for 96 h prior to determining cell viability as in panel A. Viability was normalized to DMSO treated controls for each cell line. Results were analysed by Student's t-test, n = 3, p-values were 0.0145 for Granta-519 and 0.0831 for UPN2.Comparisons were considered statistically significant when p < 0.05 and are marked by an asterisk. Results that were not considered statistically significant are indicated by n.s.
To determine whether residual low levels of ATM activity could account for the decreased sensitivity of Granta-519 compared to UPN2, we treated both cell lines with either olaparib alone or olaparib in combination with the ATM kinase inhibitor KU55933, which has been shown to increase the sensitivity of ATM-proficient cells to PARP inhibitors (Bryant & Helleday, 2006). No increase in olaparib toxicity was seen in either Granta-519 or UPN2 cells (Fig 1B of Supporting Information), indicating that the increased resistance of Granta-519 cells is not due to the presence of low levels of ATM activity.
Disruption of p53 transcriptional activity increases the sensitivity of ATM-deficient cells to olaparib
Another potential difference between these two cell lines is the status of the tumour suppressor p53 (Table 1 of Supporting Information). Granta-519 retain one WT TP53 allele, while UPN2 contain only a single TP53 allele with a mutation (R175H) in the DNA binding domain, previously demonstrated to disrupt transcriptional activity (M'kacher et al, 2003). To investigate whether p53 status impacts the sensitivity of ATM-deficient MCL cells to olaparib, we transfected both Granta-519 and UPN2 cells with a vector expressing DN-p53. Transfection of DN-p53 reduced both IR-induced stabilization of p53 and induction of p21, indicating that p53 function had been suppressed (Fig 2 of Supporting Information). Following transfection, cellular viability was determined in response to increasing concentrations of olaparib (Fig 1B). Expression of DN-p53 sensitized Granta-519 cells to olaparib compared to transfection with control (GFP) vector, as the LD50 (lethal dose required to reduce viability by 50%) decreased by approximately 50% (Fig 1B). In contrast, expression of DN-p53 in UPN2 cells, which lack endogenous p53 function, had no effect on olaparib sensitivity, while expression of functional WT p53 reduced the toxicity of olaparib (Fig 1C). Together, these studies suggest that inactivation of p53 increases the toxicity of olaparib in ATM-deficient MCL cell lines.
We also examined the effects of the small molecule inhibitor, pifithrin-α, which blocks p53 transcriptional activity (Komarov et al, 1999), on the sensitivity of MCL cell lines to olaparib. Combination of olaparib with pifithrin-α, at a concentration that reduced p53 transcriptional activity (Fig 3 of Supporting Information), significantly reduced the viability of Granta-519 cells compared to either treatment alone (Fig 1D). In contrast, pifithrin-α did not enhance sensitivity to olaparib in UPN2 cells (Fig 1D), and co-treatment of Granta-519 cells with olaparib and pifithrin-α reduced cellular viability to a similar level as treatment of UPN2 cells with olaparib alone (Fig 1D). Thus, disruption of p53 transcriptional activity increases the sensitivity of ATM-deficient MCL cells to olaparib in vitro.
Olaparib is more effective in vivo against UPN2 xenografts than Granta-519
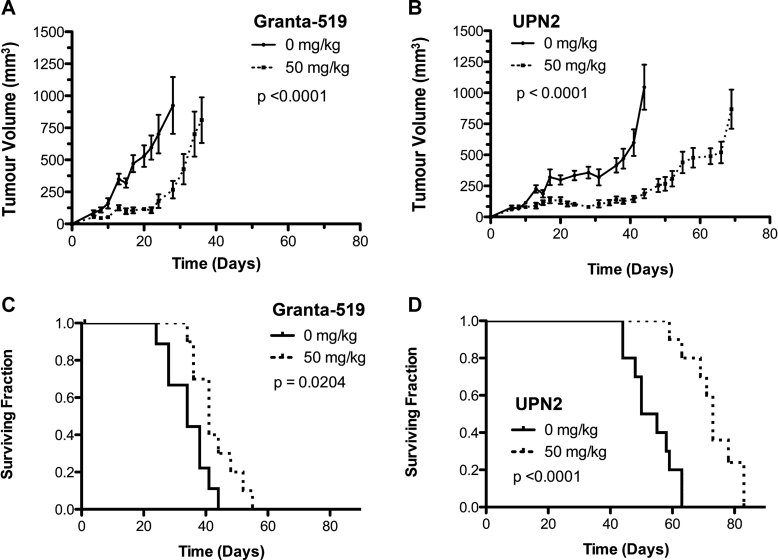
We next examined the combined impact of p53 and ATM status on olaparib sensitivity in vivo using ATM-deficient MCL cell lines Granta-519 and UPN2 as well as the ATM-proficient MCL cell lines JVM-2 (p53 WT) and HBL-2 (p53 mutated; Fig 2). Subcutaneous xenografts were generated in RAG2−/− immune-compromised mice as described previously (Williamson et al, 2010). After xenograft implantation, mice were divided into two groups and treated daily with either 50 mg/kg olaparib or vehicle control in accordance with previous in vivo experiments (Rottenberg et al, 2008; Williamson et al, 2010). As expected, olaparib treatment had no impact on tumour volume in ATM-proficient xenografts of JVM-2 and HBL-2 cells (Fig 4A and B and Table 1 of Supporting Information). Similarly, no significant differences were seen in the median survival time between olaparib and vehicle treated mice in either of these groups (Fig 4C and D and Table 1 of Supporting Information). In contrast, olaparib treatment significantly delayed tumour growth and increased median overall survival in mice bearing Granta-519 xenografts (Fig 2A and C). Consistent with in vitro observations, the effect of olaparib was even more pronounced in mice bearing UPN2 xenografts (Fig 2B and D). The time required for the average olaparib treated Granta-519 xenograft to reach a tumour volume of 750 mm3 was delayed by 40% over vehicle treated animals, while in UPN2 xenografts the same end point was delayed by 58% (Table 1). Moreover, while median overall survival of olaparib treated Granta-519 animals increased by 7 days (21%) over controls, olaparib treatment increased overall survival of UPN2 xenograft mice nearly 24 days (45%) over vehicle treated animals. These results indicate that olaparib is effective in vivo against ATM-deficient MCL xenografts, however, its efficacy is even greater against ATM-deficient xenografts of UPN2 cells that also lack p53 function. The effect of p53 appears to be limited to ATM-deficient xenografts as olaparib had little effect against either HBL-2 (ATM+/p53−) or JVM-2 (ATM+/p53+) xenografts (Fig 4 of Supporting Information).
Figure 2. Olaparib is more effective in vivo against MCL xenografts, which lack both ATM and p53 activity.
- A,B. MCL xenografts were established in RAG2−/− mice using Granta-519 (A) or UPN2 cells (B). Beginning 5 days following xenograft implantation, mice were injected daily with either 50 mg/kg olaparib (dotted line) or vehicle control (solid line). Tumour dimensions were measured as described in Materials and Methods Section. The average tumour volume of each treatment groups is shown. Panel A: Results were analysed by two-way ANOVA, n = 9 for mice treated at 0 mg/kg, and n = 10 for mice treated at 50 mg/kg. The p-value was <0.0001. Panel B: Results were analysed by two-way ANOVA, n = 10 for mice treated at 0 mg/kg, and n = 9 for mice treated at 50 mg/kg. The p-value was <0.0001.
- C,D. Kaplan–Meier survival curves of RAG2−/− mice bearing MCL xenografts generated using Granta-519 (C) and UPN2 (D) cells. Solid lines represent mice treated with vehicle solution, while dotted lines represent mice treated with 50 mg/kg olaparib. p-Values were determined using the Log-rank (Mantel–Cox) test. Panel C: n = 9 for mice treated with 0 mg/kg, and n = 10 for mice treated with 50 mg/kg, p = 0.0204. For panel D: n = 10 for 0 mg/kg, and n = 9 for 50 mg/kg, p < 0.0001. In all panels, p-values of < 0.05 were considered statistically significant.
Table 1.
Summary of in vivo experiment
| Cell line xenograft | ATM status | p53 status | Drug treatment (mg/kg) | Time (days) for tumour volume to reach 750 mm3 | % Delay in tumour growth (olaparib vs. saline) | Median Survival (days) | % Survival increase (olaparib vs. saline) | Survival range (days) | # Mice |
|---|---|---|---|---|---|---|---|---|---|
| JVM-2 | WT/WT | WT/WT | 0 | 21 | 0(ns) | 30 | + 3(ns) | 24–36 | 10 |
| 50 | 21 | 31 | 24–38 | 9 | |||||
| HBL-2 | WT/WT | Del/Mut | 0 | 30 | −13(ns) | 34 | −9(ns) | 27–41 | 10 |
| 50 | 26 | 31 | 27–45 | 9 | |||||
| Granta-519 | Del/Mut | WT/Del | 0 | 25 | +40* | 34 | 21** | 24–44 | 9 |
| 50 | 35 | 41 | 34–55 | 10 | |||||
| UPN2 | Del/Mut | Del/Mut | 0 | 43 | +58** | 52.5 | 39** | 44–63 | 10 |
| 50 | 68 | 73** | 59–83 | 9 |
RAG2−/− mice (10/group) were implanted with xenografts of one of four MCL cell lines with varying ATM and p53 status. Mice were treated with either 50 mg/kg olaparib or an equal volume of vehicle control, every day, beginning 5 days after cell implantation and continuing until mice were sacrificed. Percent delay in tumour growth and % increase in survival were determined by dividing the appropriate values for the drug treated group by the saline treated group. (ns) indicates not statistically significant, * indicates statistical significance with a p-value <0.05, ** indicates statistical significance with a p-value <0.0001. Actual p-values are provided in the legend to Fig 2. Survival range indicates the range between the day of sacrifice of the first and last mouse within the same group.
Inhibition of ATM activity preferentially sensitizes p53 mutant cells to olaparib
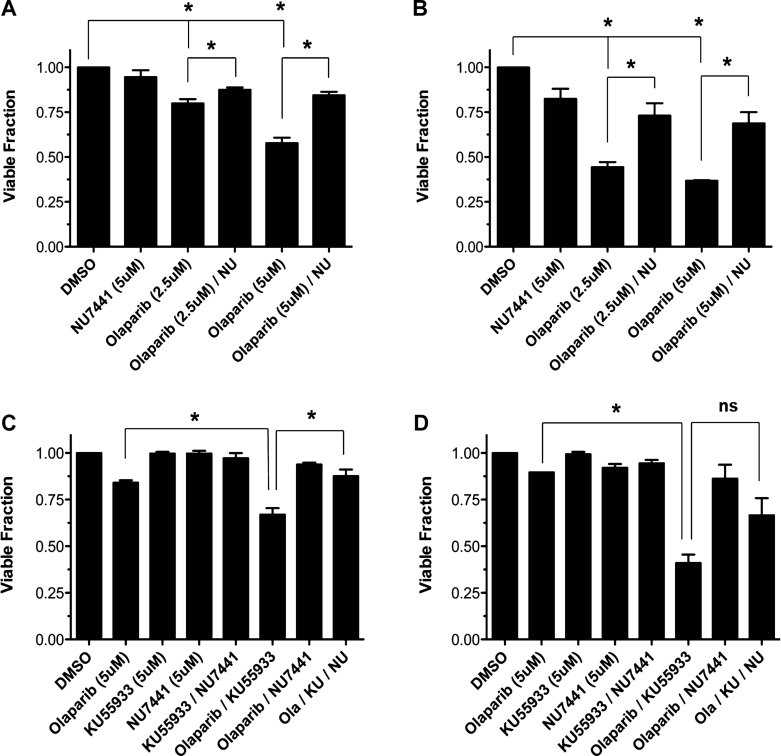
Our data suggest that cells in which both ATM and p53 are inactivated are more sensitive to PARP inhibition than cells with inactivation of ATM alone. MCL cells containing mutations in both ATM and p53 account for 10% of MCL cases, while a much larger percentage (26%) are predicted to be ATM positive but p53 defective (Greiner et al, 2006). Thus, we hypothesized that the combination of small molecule inhibitors of ATM and PARP in ATM-proficient MCL cells would be more toxic in cells lacking p53 function than in those that retained WT p53. To test this hypothesis, ATM-proficient MCL cell lines with either WT p53 (JVM-2, Z138) or mutant p53 (HBL-2, UPN1) were incubated with the ATM inhibitor KU55933 in combination with olaparib (Fig 3A). The combined inhibition of both ATM and PARP significantly reduced the cellular viability of UPN1 and HBL-2 cells, both of which contain inactivating p53 mutations, while producing only a modest decrease in the viability of p53-proficient JVM-2 and Z138 cells (Fig 3A).
Figure 3. Inhibition of ATM and PARP is more cytotoxic in cells in which p53 function has been disrupted.
- Z138 (grey bars), JVM-2 (black bars), HBL-2 (white bars) and UPN1 (hatched bars) were treated with DMSO (Untreated), KU55933 (5 µM), or olaparib (5 µM) or both KU55933 and olaparib (KU/Olaparib; both at 5 µM) for 96 h. Results were analysed by Student's t-test. n = 3, p-values were 0.013 for UPN1 versus Z138, 0.037 for UPN1 versus JVM-2, 0.0007 for Z138 versus HBL-2 and 0.0015 for Z138 versus JVM2. Results were considered statistically significant when p < 0.05, marked by *.
- Human BJ fibroblast cells and BJ cells expressing HPV genes E6/E7 were exposed to DMSO (grey bars), KU55933 (5 µM; black bars), olaparib (5 µM; while bars) or both KU55933 and olaparib (KU/Olaparib; both at 5 µM) for 96 h. In both panels, cell viability was determined by trypan blue exclusion and normalized to DMSO treated controls for each cell line. Results were analysed by Student's t-test, n = 3. p-Values were 0.00049 for KU55993/Olaparib versus DMSO, 0.00049 for KU55933/Olaparib versus KU55933 and 0.00174 for KU55933/Olaparib versus Olaparib. Results were considered statistically significant when p < 0.05, marked by an *.
As the above experiments were carried out using MCL cell lines derived from patient tumour samples, we also investigated whether p53 status affected susceptibility to olaparib in normal human cells. For these experiments, we utilized early passage, untransformed human foreskin fibroblasts (BJ), which contain WT p53 and BJ cells expressing the human papilloma virus 16 (HPV16) oncoproteins E6 and E7, which act to suppress the activities of p53 and Rb (Zou et al, 2009). BJ and BJ-E6/E7 cells were incubated with olaparib or KU55933 alone or in combination. Inhibition of both PARP and ATM produced a significant decrease in survival only in BJ-E6/E7 cells, in which the p53 axis is disrupted (Fig 3B), confirming that inactivation of ATM and p53 enhances susceptibility to olaparib in human cells. To ensure that this effect was not due to differences in ATM function between these two cell lines, IR-induced ATM autophosphorylation was examined 1 h following 2 Gy IR (Fig 5 of Supporting Information). Both BJ and BJ E6/E7 cells displayed IR-induced phosphorylation of ATM on serine 1981, indicating that the effect observed with olaparib was not due to differences in ATM activity.
Olaparib induces activation of p53 but apoptosis in ATM-deficient MCL cells is p53-independent
Our data indicates that in ATM-deficient cells, the presence of WT p53 reduces the toxicity of olaparib. We hypothesized that inactivation of PARP in ATM-deficient MCL would lead to DNA damage and activation of p53 by a related protein kinase, leading to upregulation of p53-responsive genes and enhanced cellular survival. To address this hypothesis, ATM-deficient MCL cells (Granta-519 and UPN2) were treated with olaparib and the effects on p53 phosphorylation at serine 15 and protein stabilization were examined. Olaparib induced serine 15 phosphorylation and concurrent stabilization of p53 protein, markers of p53 activation following cellular stress/DNA damage (Fig 4A). DNA damage-induced phosphorylation of p53 and other proteins in ATM-deficient B cells has been shown to be DNA-PK-dependent (Callén et al, 2009). Therefore, ATM-deficient MCL cells were incubated with the small molecule DNA-PK inhibitor NU7441 at a concentration shown to prevent DNA damage-induced activation of DNA-PK but not ATM (Fig 6 of Supporting Information). Inhibition of DNA-PK largely abrogated olaparib-induced p53 serine 15 phosphorylation (Fig 4A) implicating DNA-PK in the phosphorylation and stabilization of p53 in response to olaparib.
Figure 4. Olaparib induces DNA-PK-dependent phosphorylation as well as stabilization of p53 and upregulation of p53-responsive cell cycle checkpoint genes.
- Granta-519 and UPN2 cells were incubated for 1 h with either DMSO, KU55933 (5 µM) or NU7441 (10 µM). Following incubation cells were treated with olaparib (2.5 µM) or DMSO for 24 h. As a positive control for p53 cells were treated with IR (2 Gy) and incubated 2 h. Western blots of whole cell extracts were immuno-blotted with antibodies specific for P-S15 p53, p53 and Actin.
- Granta-519 and UPN2 cells were incubated in 2.5 µM olaparib for the indicated time and mRNA was analysed by qRT-PCR. Each value represents the mean fold change in expression (±SEM) determined from a minimum of three-independent experiments.
- Granta-519 and UPN2 cells were exposed to olaparib (2.5 µM) or DMSO (-) for the indicated time and then harvested and whole cell extractions prepared. Antibodies for Western blotting were as indicated.
- Relative p21 levels from immuno-histochemical staining of Granta-519 and UPN2 xenografts treated with vehicle or olaparib (50 mg/kg). p21 expression was measured by quantitative fluorescent IHC using HistoRX. Three areas of each tumour were used for analysis. Relative values were normalized to p21 levels in vehicle treated samples. Results were analysed using Student's t-test. For vehicle-treated Granta-519 xenografts, n = 25, and for olaparib-treated animals, n = 28. The p-value was 0.012 and is considered statistically significant, marked by an asterisk. For vehicle treated animals with UPN2 xenografts, n = 26 and for olaparib treated animals with UPN2 xenografts, n = 23. The p-value was 0.31 and is not considered statistically significant.
As p53 acts primarily as a transcription factor, we used quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) to ask whether olaparib induced the expression of p53-responsive genes. Consistent with activation of p53, in Granta-519 cells olaparib induced the expression of the cyclin-dependent kinase inhibitor CDKN1A (encoding p21) but unexpectedly, we did not detect any change in the expression of the pro-apoptotic genes Puma and NOXA (Fig 4B, left). In UPN2 cells, which lack functional p53, an increase in CDKN1A, Puma and NOXA mRNA levels was observed (Fig 4B, right). However, there was also a concomitant reduction in the expression of the control RNAs (Fig 7A of Supporting Information) so the apparent p53-independent increase in the expression of CDKN1A, Puma and NOXA is likely overestimated in these cells. Importantly, the anticipated pattern of p53-dependent gene expression was confirmed following exposure to IR, as several p53-regulated transcripts were induced more strongly in Granta-519 compared to UPN2 cells (Fig 7B of Supporting Information). Taken together, olaparib appears to be less effective than IR at inducing a p53 transcriptional response despite the DNA-PK-dependent upregulation of p53 (Fig 4A).
We also examined olaparib-induced protein expression by Western blot. Consistent with the modest p53 response in Granta-519 cells, olaparib led to increased expression of p21 and GADD45α but increased expression of NOXA and Puma proteins was not consistently observed (Fig 4C). A modest increase in p21 protein was also detected in UPN2 cells exposed to olaparib, consistent with qRT-PCR data (Fig 4B). We also used quantitative immunohistochemistry (IHC) to examine the level of p21 expression in vivo from Granta-519 and UPN2 xenografts treated with either vehicle only or olaparib (50 mg/kg). Consistent with subtle differences in vitro, mice bearing Granta-519 xenografts had a small but statistically significant increase in the level of p21 following treatment with olaparib (Fig 4D). However, there was no increase in p21 protein levels in UPN2 xenografts treated with olaparib (Fig 4D). Thus, olaparib treatment led to a small increase in p21 expression but the pro-apoptotic Puma or NOXA proteins were not induced in either cell line.
Despite the lack of Puma and NOXA expression in response to olaparib, both Granta-519 and UPN2 cells displayed markers of apoptosis, including cleavage of PARP-1 and Caspase-3 (Fig 4C). In addition, olaparib induced positive Annexin V and TdT-mediated dUTP nick end labelling (assay) (TUNEL) staining in both cell lines (Williamson et al, 2010; and Fig 8 of Supporting Information), indicating that olaparib-induced cell death occurs at least in part by apoptosis. Together, these data suggests that, in ATM-deficient cells, olaparib leads to the DNA-PK-dependent activation of p53 but that apoptosis in these cells is p53-independent.
Inhibition of DNA-PK reduces the toxicity of olaparib in ATM-deficient cells in a p53-independent manner
Our finding that olaparib induces DNA-PK-dependent phosphorylation of Ser-15 of p53 implicates DNA-PK in the pro-survival role of p53 in ATM-deficient MCL. We therefore hypothesized that inhibition of DNA-PK would increase the toxicity of olaparib in ATM-deficient MCL cells. However, to the contrary, inhibition of DNA-PK by NU7441 increased survival in olaparib-treated ATM-deficient MCL cell lines Granta-519 and UPN2 (Fig 5A and B). We then asked whether NU7441 could also reduce the toxicity of the combination of ATM and PARP inhibitors in ATM-proficient MCL cell lines (Fig 5C and D). While the combination of KU55933 and olaparib reduced the viability of both JVM-2 and HBL-2 cells, further treatment with NU7741 restored cellular viability in both cell lines (Fig 5C and D). The reduction of olaparib toxicity in ATM-deficient cells by NU7741 was observed in both p53-proficient and -deficient cell lines indicating that this effect is not dependent on p53 activity. Our results are in agreement with the findings of Patel et al, 2011, who showed that inhibition of DNA-PK rescued PARP inhibitor toxicity in ATM-deficient cells as well as in cells with mutations in either BRCA1 or BRCA2. Together, these findings support the notion that a functional NHEJ pathway is required for PARP inhibitor-induced cytotoxicity.
Figure 5. Inhibition of DNA-PK enhances survival in olaparib-treated MCL cells.
- A,B. Granta-519 (A) or UPN2 (B) cells were exposed to DMSO, olaparib (2.5 and 5 µM) and NU7441 (5 µM) alone or in combination continuously for 96 h. Cellular viability was determined as in Fig 1. Results were analysed using Students t-test. For panel A, n = 3, p-values were 0.001 for DMSO versus 2.5 µM olaparib, 0.0001 for DMSO versus 5 µM olaparib, 0.0486 for 2.5 µM olaparib versus 2.5 µM olaparib plus NU7441 and 0.0017 for 5 µM olaparib versus 5 µM olaparib plus NU7441. For panel B, n = 3, and p-values were <0.0001 for each set of samples (DMSO vs. 2.5 µM olaparib, DMSO vs. 5 µM olaparib, 2.5 µM olaparib vs. 2.5 µM olaparib plus NU7441 and 5 µM olaparib vs. 5 µM olaparib plus NU7441).
- C,D. JVM-2 (C) and HBL-2 (D) cells were exposed to either DMSO, olaparib (5 µM), KU55933 (5 µM), NU7441 (5 µM) alone or each combination thereof, continuously for 96 h. Viability was determined by trypan blue exclusion and results analysed by Student's t-test as above. For panel C, n = 3, p-values were 0.0148 for 5 µM olaparib versus 5 µM olaparib plus KU55933 and 0.0097 for olaparib plus KU55933 versus olaparib plus KU55933 and NU7441. For panel D, n = 3, and p-values were 0.0004 for 5 µM olaparib versus olaparib plus KU55933 and 0.066 for olaparib plus KU55933 versus olaparib plus KU55933 and NU7441. In all panels, comparisons marked by an * indicate statistical significance (p < 0.05), while the comparison marked by n.s. is not statistically significant.
In contrast, inactivation of DNA-PK has been reported to increase sensitivity to doxorubicin in ATM-depleted p53 positive cells (Jiang et al, 2009). We therefore asked whether or not inhibition of DNA-PK would protect the MCL cell lines from doxorubicin cytotoxicity. In MCL, NU7441 caused a modest increase in sensitivity to doxorubicin, regardless of ATM or p53 status (Fig 9 of Supporting Information).
DISCUSSION
MCL is an aggressive B-cell lymphoma that, despite recent advances, remains incurable with current treatment strategies (Williams et al, 2011). Since the ATM gene is frequently altered through mutation or deletion in MCL (Schaffner et al, 2000), the use of PARP inhibitors to induce synthetic lethality is an attractive proposition in this disease. We previously demonstrated that ATM deficiency increases the sensitivity of MCL cells to PARP inhibitors, including olaparib (Williamson et al, 2010). Like many other human malignancies, MCL is also frequently characterized by mutation of p53 (Greiner et al, 2006). In MCL, 26% of cases contain p53 mutation/deletion, 56% ATM alteration and 10% contain alterations in both genes (Greiner et al, 2006). Here, we show that MCL cell lines and normal human fibroblasts with inactivation or mutation of both ATM and p53 are more sensitive to the PARP inhibitor olaparib than the same cells with inactivation or mutation of ATM alone. In contrast, p53 mutation had little effect on olaparib-induced cytotoxicity in ATM-proficient cells. Our findings suggest that the clinical utility of PARP inhibitors may depend on the status of both ATM and p53. Our results also suggest that combining inhibitors of PARP and ATM may have utility in p53-deficient MCL and perhaps other cancers with p53 dysfunction, although additional in vivo studies are required to further investigate this possibility.
The results of this study have several implications for the potential treatment of ATM-deficient malignancies with PARP inhibitors. First, our work extends the understanding of factors influencing the susceptibility of MCL cells to synthetic lethality by demonstrating a role for p53 in the interaction between PARP and ATM. UPN2 cells, deficient for both ATM and p53 function, were more sensitive to olaparib – both in vitro and in vivo – than Granta-519 cells, which lack ATM function but retain WT p53. Blocking p53 transcriptional activity, either through expression of DN-p53 or by incubation with the small molecule p53 inhibitor pifithrin-α, increased the sensitivity of Granta-519 cells to olaparib. Conversely, the re-expression of WT p53 in UPN2 cells reduced the toxicity of olaparib. Together, this indicates that p53 status may impact the potential effectiveness of PARP inhibitors in ATM-deficient MCL, suggesting that future clinical trials should take note of both ATM and p53 status of tumours to be treated.
Secondly, our work suggests the possibility of combining targeted agents to induce a synthetic lethal response. We demonstrate that the combined inhibition of both ATM activity (by KU55933) and PARP activity (by olaparib) is particularly cytotoxic in MCL cells with p53 mutations as well as in human fibroblasts in which p53 has been disrupted by expression of E6/E7. These findings raise the interesting possibility that the combination of ATM and PARP inhibitors may have potential in the treatment of ATM-proficient MCL cases that carry mutations in TP53, a subpopulation, which may account for a quarter of all MCL patients (Greiner et al, 2006), as well as in other tumours in which p53 is mutated. However, we caution that potential concerns with this approach could be the induction of a synthetic lethal response in non-malignant, p53-proficient cells and enhanced cytotoxicity caused by combining both inhibitors, and further studies in animal models will be required to confirm the utility of this approach in an in vivo setting.
We also demonstrate that in cells lacking ATM function, olaparib induces DNA-PK-mediated p53 stabilization and phosphorylation. In cells retaining p53 function, an increase in both the mRNA and protein level of p53-responsive genes including p21 was observed (Fig 4). However, olaparib did not result in increased mRNA expression of the p53-responsive apoptotic genes Puma and NOXA. Since other apoptotic markers (PARP-1 and Caspase-3 cleavage, TUNEL staining) were observed, this suggests that olaparib induces apoptosis independently of p53 function and we suggest that the activation of the cell cycle checkpoint repertoire of p53-responsive genes may function to reduce the toxicity of olaparib in ATM-deficient cells that retain p53 function (Fig 6).
Figure 6. Model for the cellular effects of PARP inhibitors in MCL cells.
Olaparib prevents PARP-1 mediated repair of endogenous DNA SSBs generated by reactive oxygen species (ROS) and intermediates of base excision repair (BER), as well as one-ended DNA DSBs produced when replication forks collapse. In ATM-proficient cells, these DSBs activate ATM inducing transient cell cycle checkpoints and repair of the breaks by HR. In ATM-deficient cells, olaparib activates DNA-PK, and possibly ATR, which phosphorylate and stabilize p53 resulting in upregulation of cell cycle checkpoint genes GADD45α and p21, providing a survival advantage over cells with mutant p53. ATM-deficient cells initiate NHEJ to attempt to repair one-ended DNA DSBs regardless of p53 status, leading to cell death, via p53-independent apoptosis.
Our work also highlights the potential role of DNA-PK in the clinical exploitation of synthetic lethality. We show that DNA-PK activity is responsible, at least in part, for the phosphorylation/stabilization of p53, which we speculate could allow for a pro-survival response in cells with p53 function. However, paradoxically, DNA-PK activity was also required for the toxicity of olaparib, which occurs in a p53-independent fashion (Fig 5). Inhibition of DNA-PK kinase activity has been proposed as a therapeutic strategy to increase the effectiveness of genotoxic agents in ATM-deficient cancers (Jiang et al, 2009; Reinhardt et al, 2009), the rationale being that DNA-PK has the ability to act redundantly in the absence of ATM to activate p53 and cell cycle checkpoints, thus mitigating the cytotoxic effect of certain genotoxic agents. Our results support the idea that DNA-PK can phosphorylate p53 in the absence of ATM, however, they also suggest that the direct inhibition of DNA-PK reduces the toxicity of olaparib in this setting. Our findings are therefore in agreement with those of Patel et al, 2011, who reported that disruption of DNA-PK activity rescues the lethality of PARP inhibition in cell lines lacking BRCA2, BRCA1 or ATM.
In contrast, inhibition of DNA-PK has been shown to increase the cytotoxicity of doxorubicin in p53-proficient cells that lack functional ATM (Jiang et al, 2009; Reinhardt et al, 2009). Similarly, we found that inhibition of DNA-PK sensitized doxorubicin-treated MCL cells, regardless of ATM or p53 status (Fig 9 of Supporting Information). We speculate that the apparent discrepancy between the effects of DNA-PK inhibition on viability in response to olaparib compared to doxorubicin could be explained by olaparib inducing DNA damage in the form of replication-dependent one-ended DNA DSBs (Patel et al, 2011), while doxorubicin induces DNA DSBs by stabilization of topoisomerase II intermediates (Pommier et al, 2010). We propose that the action of DNA-PK in the NHEJ repair pathway would be unable to accurately repair the one-ended DSBs induced by PARP inhibition, but would be proficient to repair DNA DSBs induced by doxorubicin. Similarly, cell lines derived from Fanconi anaemia (FA) patients, that lack key proteins involved in the repair of DNA cross-links including BRCA1 (Adamo et al, 2010; Pace et al, 2010), are extremely sensitive to DNA cross-linking agents such as cisplatin, and inhibition of NHEJ by addition of the DNA-PK inhibitor NU7441 rescued this sensitivity (Adamo et al, 2010). Thus, inappropriate engagement of the error-prone NHEJ pathway appears to be responsible for the cytotoxicity of DNA cross-links in FA cells. Our results extend these findings and also suggest that inappropriate engagement of NHEJ is responsible for the toxicity of PARP inhibitors in ATM-deficient cells. Thus, in response to PARP inhibition in ATM-deficient cells, DNA-PK can act to both maintain cellular viability, presumably through p53 phosphorylation/stabilization, and also induce cell death, via NHEJ (Fig 6). This balance of pro- and anti-survival effects of DNA-PK might explain the decreased toxicity of PARP inhibitors in ATM-deficient cells that retain p53 activity.
In summary, our work extends the understanding of how the exploitation of synthetic lethality might be applied clinically. Given the large number of p53-deficient malignancies, our demonstration of a role for p53 in the synthetic lethality of PARP inhibitors might have widespread implications beyond MCL. The specific reasons for the failure of the addition of PARP inhibition to chemotherapy for triple negative breast cancer have not yet been identified, but our data strongly suggest that more detailed molecular typing may better define the patient population most likely to benefit from treatment with these agents.
MATERIALS AND METHODS
Cell culture
C35ABR (BT; ATM-proficient) cells were kindly provided by Dr. M. Lavin (Queensland Institute of Medical Research, Australia). UPN1 and UPN2 cell lines were a kind gift from Dr A. Turhan (University of Poitiers, France). Other cell lines were purchased from ATCC. Granta-519, HBL-2, JVM-2, Z138, C35ABR (BT) and L3 cells were cultured in suspension in RPMI 1640 medium (Invitrogen, Carlsbad, CA) containing 10% v/v Hyclone III serum (FBS, Hyclone, Logan, UT). UPN1 and UPN2 cells were cultured in suspension in MEM-α medium (Invitrogen) containing 10% FBS and antibiotics as above. HeLa cells were cultured in DMEM medium with 5% v/v foetal bovine serum. BJ cells were cultured in DMEM with 10% v/v foetal bovine serum. All cells were cultured in the presence of 50 units/ml penicillin and 50 µg/ml streptomycin at 37°C under 5% CO2.
The paper explained
PROBLEM:
MCL is an aggressive B-cell lymphoma that, despite recent advances, remains incurable with current strategies. This study explores the concept of improving therapeutic outcomes in MCL and other cancers by exploiting the molecular signature of the tumour. In recent years, a synthetic lethal approach using poly-ADP ribose polymerase (PARP-1) inhibitors has shown promise in the treatment of cancers with mutation in the breast and ovarian cancer susceptibility genes, BRCA1 and BRCA2. We recently showed that MCL cells lacking the ataxia telangiectasia mutated protein kinase (ATM) are more sensitive to PARP inhibitors than their ATM-proficient counterparts, suggesting that the synthetic lethal concept can be extended to MCL. However, how other genetic factors influence the sensitivity of cells to PARP inhibitors is not known.
RESULTS:
We show that the sensitivity of ATM-deficient MCL cell lines to the PARP inhibitor olaparib is enhanced when p53 is inactivated. Furthermore, the combination of small molecule inhibitors of ATM and PARP was significantly more cytotoxic in both MCL cells and normal human fibroblasts with inactivation of p53 than in cells with wild-type p53. In contrast, inhibition of the ATM-related protein kinase, DNA-PKcs, provided protection against olaparib cytotoxicity.
IMPACT:
Our results highlight the importance of p53 and DNA-PK in the synthetic lethal interaction between ATM and PARP and confirm that functional non-homologous end-joining is required for PARP inhibitor-induced cell death. Moreover, our findings suggest that p53-deficiency, a molecular aberration characteristic of a large proportion of malignancies, can be exploited using a combination of ATM and PARP inhibitors. Together, our results suggest that p53 status may be an important indictor of response to PARP inhibitors and that detailed molecular typing may better define the patient population most likely to benefit from treatment with these agents.
Cell extracts and immunoblotting
Irradiation, preparation of whole cell extracts and immunoblotting was as described previously (Williamson et al, 2010). Antibodies specific for ATM were a kind gift from Dr. M. Lavin, while antibodies to DNA-PKcs and P-S824 KAP-1 (Douglas et al, 2002; Goodarzi et al, 2008) were generated in house. Antibodies to GADD45α (Abcam), p21 (Santa Cruz), p53 (Santa Cruz), SMC-1 (Novus), KAP-1 (Abcam), Actin (Sigma), NOXA (Calbiochem), PUMA (Cell Signaling), PARP (Cell Signaling) and Caspase-3 (Cell Signaling) and phosphospecific antibodies to P-S15 p53 (Cell Signaling) and P-S2056 DNA-PKcs (Abcam) were purchased commercially.
Quantitative reverse transcriptase polymerase chain reaction
qRT-PCR was performed as previously described (Melanson et al, 2011). Briefly, total RNA was isolated using the RNeasy RNA isolation kit (Qiagen, Valencia, CA) and 5 µg of RNA was reverse-transcribed using the first-strand cDNA synthesis kit (MBI Fermentas, Burlington, ON). Primers were used to amplify 1/200th of the cDNA product per reaction in the presence of Sybr green fluorescent DNA stain (Invitrogen) using the LightCycler 2 quantitative PCR machine with Light-Cycler software version 3 (Roche Diagnostics, Switzerland; Melanson et al, 2011). The expression of each transcript was normalized to both ACTB and GAPDH. See Supporting Information for primer sequences and additional information.
In vivo studies
All animal procedures were carried out by a trained animal technician in accordance with established procedures at the Animal Resource Center at the University of Calgary. Female RAG2−/− mice (Taconic, Hudson, NY) were injected subcutaneously in the right flank with 5 × 106 cells in a 1:1 emulsion of Matrigel (BD Biosciences Cat. # 354234) as described previously (Williamson et al, 2010). Five days following xenograft implantation mice were injected intraperitoneally daily with either vehicle alone (10% v/v DMSO, 10% w/v 2-hydroxy-propyl-β-cyclodextrin (HPBCD) in PBS) or 50 mg/kg olaparib, as described previously (Rottenberg et al, 2008; Williamson et al, 2010). Injections were given daily until sacrifice. Tumour volume (0.5 × (width) × (length)2) was measured manually using digital callipers thrice weekly. Mice were sacrificed when tumours reached >1500 mm3, weight loss exceeded 20% of initial weight, or at the first obvious signs of distress. Statistical significances of differences in tumour volume were determined using the Student t-test. Kaplan–Meier survival was analysed by the Log-Rank (Mantel–Cox) test to determine statistical significance.
Quantitative fluorescent immunohistochemistry
The HistoRx AQUA® platform and fluorescent IHC (Camp et al, 2002) were used to quantify the expression of p21 in formalin fixed paraffin embedded (FFPE) xenografts generated using Granta-519 and UPN2 cells. Tissue microarrays (TMAs) were created from triplicate 0.6 mm cores from 77 FFPE mouse xenograft tumour samples. TMA sections (4 µm) were deparaffinized in xylene, rinsed in ethanol and rehydrated. Heat-induced epitope retrieval was performed by heating slides to 121°C in a citrate-based buffered (pH 6.0) Target Retrieval Solution (DAKO) for 3 min in a decloaking chamber (Biocare Medical). TMA slides were then stained for p21 (rabbit monoclonal anti-p21, 1:100 dilution, Epitomics) using a DAKO autostainer. Rabbit Envision+ kit (DAKO) was used in conjunction with tyramide-Cy5 (Perkin-Elmer) to visualize p21 expression. The tumour compartment was identified by staining with a mouse anti-CD20 antibody (mouse monoclonal, 1:100 dilution, DAKO) and an Alexa555 conjugated anti-mouse secondary antibody. Cross-reactivity with endogenous mouse IgG was minimized by a 30 min incubation with Rodent Block M (Biocare Medical). Slides were scanned on an Aperio Scanscope FL and analysed using AQUAnalysis software (version 2.3.4.1): the tumour compartment was defined as the CD20-positive area for each TMA core. AQUAnalysis software calculated the p21 expression from the average p21 pixel intensity density within the CD21 positive tumour compartment (AQUA score). Values for p21 expression were normalized to vehicle treated tumour samples, n = 25–29 tumour samples per group.
Transfection of Granta-519 cells
Granta-519 cells were transfected with vectors expressing GFP or DN p53 (Clontech) both under the control of a CMV promoter. Cells were transfected using Nucleofector Kit T (AMAXA) with electroporation program O-17. Cells were incubated 24 h following electroporation prior to further experimental treatment. UPN2 cells were transfected with vectors expressing GFP, DN p53 or WT p53 (Clontech). Cells were transfected using Nucleofector Kit T (AMAXA) with electroporation program O-17.
Acknowledgments
We thank Dr Y. Shiloh, Dr. M. Lavin and Dr. A. Turhan for cell lines; Dr. P. Forsyth, Dr. S. Xueqing, Ms. M. Chisholm and the University of Calgary Animal Resource Centre for assistance with in vivo experiments; Gita Pokherl, Mie Konno and Stephanie Petrillo at the Functional Tissue Imaging Unit for their assistance with the IHC staining and tumour microarray; and Dr. Mark O'Connor (AstraZeneca) for olaparib, PIKK inhibitors and helpful discussions and comments on the manuscript. We also thank members of the Lees-Miller laboratory, Dr. G. Moorhead and Dr. L. Williamson for helpful discussions. CTW was supported by graduate studentships from the Alberta Cancer Foundation and the Translational Research Training in Cancer program. SPLM is a scientist of Alberta Innovates-Health Solutions and holds the Engineered Air Chair in Cancer Research at the University of Calgary. This work was supported by grant 0011053 from the National Cancer Institute of Canada/Canadian Cancer Research Institute, with funds from the Canadian Cancer Society (to SPLM), and grants from the Leukemia and Lymphoma Society of Canada (to DGB and SPLM) and the Canadian Institutes of Health Research (to BCM).
Supporting Information is available at EMBO Molecular Medicine online.
The authors declare that they have no conflict of interest.
Author contributions
AK, MD and AMM were responsible for the IHC analysis; JDH and BCM performed qPCR experiments; EK, LRT and DGB performed experiments using BJ cells; HM grew cells for animal experiments; RY carried out FACS experiments; CTW and EK performed all other experiments; CTW, DGB and SPLM planned experiments and wrote the paper; All authors contributed to editing the manuscript.
Supplementaary material
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
References
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-Mcguinn KM, Scott C, Weitzel JN, Oaknin A, Loman N, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou L-VF, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Boultwood J. Ataxia telangiectasia gene mutations in leukaemia and lymphoma. J Clin Pathol. 2001;54:512–516. doi: 10.1136/jcp.54.7.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34:1685–1691. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Callén E, Jankovic M, Wong N, Zha S, Chen H-T, Difilippantonio S, Di Virgilio M, Heidkamp G, Alt FW, Nussenzweig A, et al. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell. 2009;34:285–297. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, Meek K, Alessi DR, Lees-Miller SP. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem J. 2002;368:243–251. doi: 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers B, Helleday T, Jonkers J. Targeting homologous recombination repair defects in cancer. Trends Pharmacol Sci. 2010;31:372–380. doi: 10.1016/j.tips.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Fang NY, Greiner TC, Weisenburger DD, Chan WC, Vose JM, Smith LM, Armitage JO, Mayer RA, Pike BL, Collins FS, et al. Oligonucleotide microarrays demonstrate the highest frequency of ATM mutations in the mantle cell subtype of lymphoma. Proc Natl Acad Sci USA. 2003;100:5372–5377. doi: 10.1073/pnas.0831102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Kozlov S, Mckay MJ, Woods R, Birrell G, Sprung CN, Murrell DF, Wangoo K, Teng L, Kearsley JH, et al. Low levels of ATM in breast cancer patients with clinical radiosensitivity. Genome Integr. 2010;1:9. doi: 10.1186/2041-9414-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt AN, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Greiner TC, Dasgupta C, Ho VV, Weisenburger DD, Smith LM, Lynch JC, Vose JM, Fu K, Armitage JO, Braziel RM, et al. Mutation and genomic deletion status of ataxia telangiectasia mutated (ATM) and p53 confer specific gene expression profiles in mantle cell lymphoma. Proc Natl Acad Sci USA. 2006;103:2352–2357. doi: 10.1073/pnas.0510441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M. PARP inhibitors stumble in breast cancer. Nat Biotechnol. 2011;29:373–374. doi: 10.1038/nbt0511-373. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, Bartek J, Yaffe MB, Hemann MT. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–1909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Kang B, Guo R-F, Tan X-H, Zhao M, Tang Z-B, Lu Y-Y. Expression status of ataxia-telangiectasia-mutated gene correlated with prognosis in advanced gastric cancer. Mutat Res. 2008;638:17–25. doi: 10.1016/j.mrfmmm.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- Lord CJ, McDonald S, Swift S, Turner NC, Ashworth A. A high-throughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair (Amst) 2008;7:2010–2019. doi: 10.1016/j.dnarep.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Maclaine NJ, Hupp TR. How phosphorylation controls p53. Cell cycle (Georgetown, Tex) 2011;10:916–921. doi: 10.4161/cc.10.6.15076. [DOI] [PubMed] [Google Scholar]
- Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka MZ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- Melanson BD, Bose R, Hamill JD, Marcellus KA, Pan EF, McKay BC. The role of mRNA decay in p53-induced gene expression. RNA. 2011;17:2222–2234. doi: 10.1261/rna.030122.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M'kacher R, Bennaceur A, Farace F, Laugé A, Plassa LF, Wittmer E, Dossou J, Violot D, Deutsch E, Bourhis J, et al. Multiple molecular mechanisms contribute to radiation sensitivity in mantle cell lymphoma. Oncogene. 2003;22:7905–7912. doi: 10.1038/sj.onc.1206826. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, Koo IC, Sherman BM, Bradley C. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011;364:205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci USA. 2011;108:3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, Jiang H, Hemann MT, Yaffe MB. Exploiting synthetic lethal interactions for targeted cancer therapy. Cell cycle (Georgetown, Tex) 2009;8:3112–3119. doi: 10.4161/cc.8.19.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AOH, Zander SAL, Derksen PWB, de Bruin M, Zevenhoven J, Lau A, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci USA. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Schaffner C, Stilgenbauer S, Rappold GA, Döhner H, Lichter P. Somatic ATM mutations indicate a pathogenic role of ATM in B-cell chronic lymphocytic leukemia. Blood. 1999;94:748–753. [PubMed] [Google Scholar]
- Schaffner C, Idler I, Stilgenbauer S, Döhner H, Lichter P. Mantle cell lymphoma is characterized by inactivation of the ATM gene. Proc Natl Acad Sci USA. 2000;97:2773–2778. doi: 10.1073/pnas.050400997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Löbrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- Tommiska J, Bartkova J, Heinonen M, Hautala L, Kilpivaara O, Eerola H, Aittomäki K, Hofstetter B, Lukas J, von Smitten K, et al. The DNA damage signalling kinase ATM is aberrantly reduced or lost in BRCA1/BRCA2-deficient and ER/PR/ERBB2-triple-negative breast cancer. Oncogene. 2008;27:2501–2506. doi: 10.1038/sj.onc.1210885. [DOI] [PubMed] [Google Scholar]
- Turner NC, Lord CJ, Iorns E, Brough R, Swift S, Elliott R, Rayter S, Tutt AN, Ashworth A. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 2008;27:1368–1377. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt AN, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- Weston VJ, Oldreive CE, Skowronska A, Oscier DG, Pratt G, Dyer MJ, Smith G, Powell JE, Rudzki Z, Kearns P, et al. The PARP inhibitor olaparib induces significant killing of ATM deficient lymphoid tumour cells in vitro and in vivo. Blood. 2010;116:4578–4587. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- Williams ME, Connors JM, Dreyling MH, Gascoyne RD, Kahl BS, Leonard JP, Press OW, Wilson WH. Mantle cell lymphoma: report of the 2010 Mantle Cell Lymphoma Consortium Workshop. Leukemia Lymphoma. 2011;52:24–33. doi: 10.3109/10428194.2010.532893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson CT, Muzik H, Turhan AG, Zamò A, O'Connor MJ, Bebb DG, Lees-Miller SP. ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors. Mol Cancer Ther. 2010;9:347–357. doi: 10.1158/1535-7163.MCT-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Misri S, Shay JW, Pandita TK, Wright WE. Altered states of telomere deprotection and the two-stage mechanism of replicative aging. Mol Cell Biol. 2009;29:2390–2397. doi: 10.1128/MCB.01569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.