Abstract
Trop-2, a cell surface glycoprotein, contains both extracellular epidermal growth factor-like and thyroglobulin type-1 repeat domains. Low TROP2 expression was observed in lung adenocarcinoma tissues as compared with their normal counterparts. The lack of expression could be due to either the loss of heterozygosity (LOH) or hypermethylation of the CpG island DNA of TROP2 upstream promoter region as confirmed by bisulphite sequencing and methylation-specific (MS) polymerase chain reaction (PCR). 5-Aza-2′-deoxycytidine treatment on lung cancer cell (CL) lines, CL1-5 and A549, reversed the hypermethylation status and elevated both TROP2 mRNA and protein expression levels. Enforced expression of TROP2 in the lung CL line H1299 reduced AKT as well as ERK activation and suppressed cell proliferation and colony formation. Conversely, silencing TROP2 with shRNA transfection in the less efficiently tumour-forming cell line H322M enhanced AKT activation and increased tumour growth. Trop-2 could attenuate IGF-1R signalling-mediated AKT/β-catenin and ERK activation through a direct binding of IGF1. In conclusion, inactivation of TROP2 due to LOH or by DNA methylation may play an important role in lung cancer tumourigenicity through losing its suppressive effect on IGF-1R signalling and tumour growth.
Keywords: epigenetic, IGF-1R, lung cancer, slug, TROP2
INTRODUCTION
Aberrant expression and/or mutation of membrane proteins may lead to persistent activation of cell-survival signalling, cellular transformation and invasion as seen in various lung tumours. The EGFR (Grandis & Sok, 2004) and IGF-1R (Ludovini et al, 2009) signalling pathways are typical examples of pathways whose dysregulation may drive lung cancer growth and malignancy. Recently, researchers successfully developed an effective strategy for treating non-small cell lung cancer (NSCLC) by targeting the protein tyrosine kinases responsible for activating EGFR and IGF-1R signalling (Lee et al, 2007; Shepherd et al, 2005). This development further illustrates the importance of membrane proteins in lung cancer carcinogenesis.
In this study, we focused on the functional role of Trop-2, a membrane glycoprotein, in lung cancer malignancy. The TROP2 gene encodes a 323-amino acid cell-surface glycoprotein with a conserved phosphatidylinositol 4,5-bisphosphate (PIP2)-binding domain containing one phosphorylation site (S303) (Cubas et al, 2009) as well as extracellular EGF-like and thyroglobulin type-1 repeat domains. These domains may potentially bind IGF1 and, as a result, may suppress IGF1 binding to its receptor, IGF-1R, and consequently downstream signalling. IGF-1R mediates IGF-1 signalling and plays a critical role in cell growth, differentiation, transformation and metastasis (Larsson et al, 2005; Rosenzweig & Atreya, 2010). Recent studies have shown that IGF-1R is upregulated in lung cancer tissues (Ouban et al, 2003), and activation of IGF-1R signalling may enhance lung tumour development in vivo through β-catenin/slug expression (Frankel et al, 2005; Moorehead et al, 2003; Playford et al, 2000). Silencing of IGF-1R signalling by antagonist or shRNA-mediated downregulation of the receptor was found to diminish the anchorage-independent colony formation ability of lung cancer cells (CL; Lee et al, 1996), to induce apoptosis among these cells (Ma et al, 2007), and also to restore the ability of TKI (gefitinib) to induce apoptosis in mucinous adenocarcinoma (Hurbin et al, 2011).
Recent studies have shown that Trop-2 is overexpressed in colorectal, gastric, oral and pancreatic cancers (Fong et al, 2008a, 2008b; Muhlmann et al, 2009; Ohmachi et al, 2006), and TROP2 overexpression has been associated with cancer progression and decreased survival in colon cancers (Wang et al, 2008). Other studies have demonstrated that TROP2 may be downregulated by DNA methylation in CL (Kim et al, 2006; Shames et al, 2006) and that forced expression of TROP2 can suppress cell proliferation and colony formation (Kim et al, 2006). Cumulating evidence has suggested that TROP2 expression is mediated by microenvironmental cues (e.g. Fgf10 expression) and that it acts during lung development to maintain epithelial cells in a progenitor-like state; as such, it has been proposed to be a marker for undifferentiated epithelial cells (Lu et al, 2005). TROP2 is also upregulated by increased foetal lung expansion (Sozo et al, 2006). These previous findings indicate that TROP2 may be genetically and epigenetically regulated. However, the precise biological function of Trop-2 in lung cancer is not yet known.
Here, we investigated the role of Trop-2 in lung CL development, focusing on its epigenetic regulation and the potential mechanisms mediating lung adenocarcinoma growth and malignancy. We also investigated the potential interaction between Trop-2 and IGF-1R signalling in lung adenocarcinoma cells. Our data, for the first time, show that TROP2 is epigenetically downregulated and affected by loss of heterozygosity (LOH) in most of NSCLC tissues and cells. These associations are relevant to the function of Trop-2 to downregulate IGF-1R/Akt signalling pathway-mediated β-catenin/slug expression.
RESULTS
Genetics of TROP2 and its expression in lung adenocarcinoma
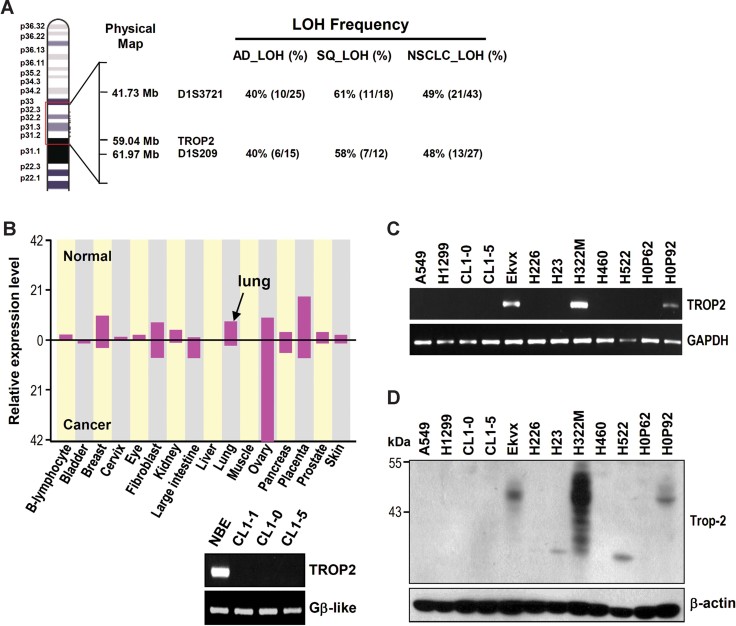
TROP2 is located at human chromosome 1p32-p31, which showed a high frequency of LOH and microsatellite instability (MI) in the region (Fig 1A). We investigated the presence of the genomic instability frequency in the TROP2 region in lung tumours and CL lines. Two different sets of lung cancer tissues and paired normal specimens were studied (Tsai et al, 2006; Tseng et al, 2005). Using a pair of microsatellite markers that flank TROP2, namely D1S3721 and D1S209, we showed that the first set of tissues from 37 adeno- and 34 squamous carcinomas contains 40 and 58–61% of LOH frequency in the region, respectively (Fig 1A inserted Table). Overall, we detected a 48 and 49% LOH frequency at microsatellite markers D1S228 and D1S1728, respectively, for all 71 lung tumours in the study. In the second set that composed 48 lung adenocarcinoma tissues, LOH frequency was 60–67%, while MI frequency was near 100% at microsatellite markers D1S228 and D1S1728 (Fig S1 of Supporting Information). Thus, TROP2 appeared to be frequently lost from the genomes of lung adenocarcinoma and squamous carcinoma.
Figure 1. LOH and downregulation of TROP2 expression in lung adenocarcinomas.
- TROP2 is located on chromosome 1p32-p31, between markers D1S3721 and D1S209, which show frequent LOH in lung cancer samples. AD, adenocarcinoma; SQ, squamous cell carcinoma; NSCLC, non-small cell lung cancer.
- The expression patterns of TROP2 in normal and cancer tissues, as modified from the ECgene database (http://genome.ewha.ac.kr/ECgene). The black arrow indicates the expression level of TROP2 in normal lung tissue. The expression of TROP2 in human NBE cells and lung CL were confirmed by RT-PCR.
- The expression levels of TROP2 in various lung CL lines were measured by RT-PCR.
- The protein levels of Trop-2 were determined by Western blot analysis.
The expression levels of TROP2 in normal and cancerous cells were analysed using the expressed sequence tag (EST) clustering from the ECgene database (http://genome.ewha.kr/ECgene/). The results showed that TROP2 mRNA expression was upregulated in large intestine and ovary cancer, but downregulated in most other cancers such as breast, kidney, lung and placenta when compared to their normal tissues (Fig 1B). RT-PCR analysis of various cell lines demonstrated that TROP2 was highly expressed in normal bronchial epithelial cells (NBE) but was downregulated in most of the lung CL lines, except H322M (adenocarcinoma), EKVX (adenocarcinoma) and HOP92 (large cell) (Fig 1C). Western blot analysis further showed that the levels of Trop-2 protein were low or not detectable in most lung CL lines, except very high expression in H322M cells and modest expression in EKVX and HOP92 cell lines (Fig 1D).
The multiple protein bands of Trop-2 detected in the Western blots may be related to different glycosylation patterns. As shown in Fig S2 of Supporting Information, Trop-2-Flag transfection of H1299 cells also resulted in detection of multiple protein bands in the Western blots, while tunicamycin treatment reduced the ectopic Trop-2-flag protein band to 35 kD, the molecular weight predicted from the coding sequence for an unglycosylated native Trop-2 protein.
Downregulation of TROP2 expression by promoter hypermethylation
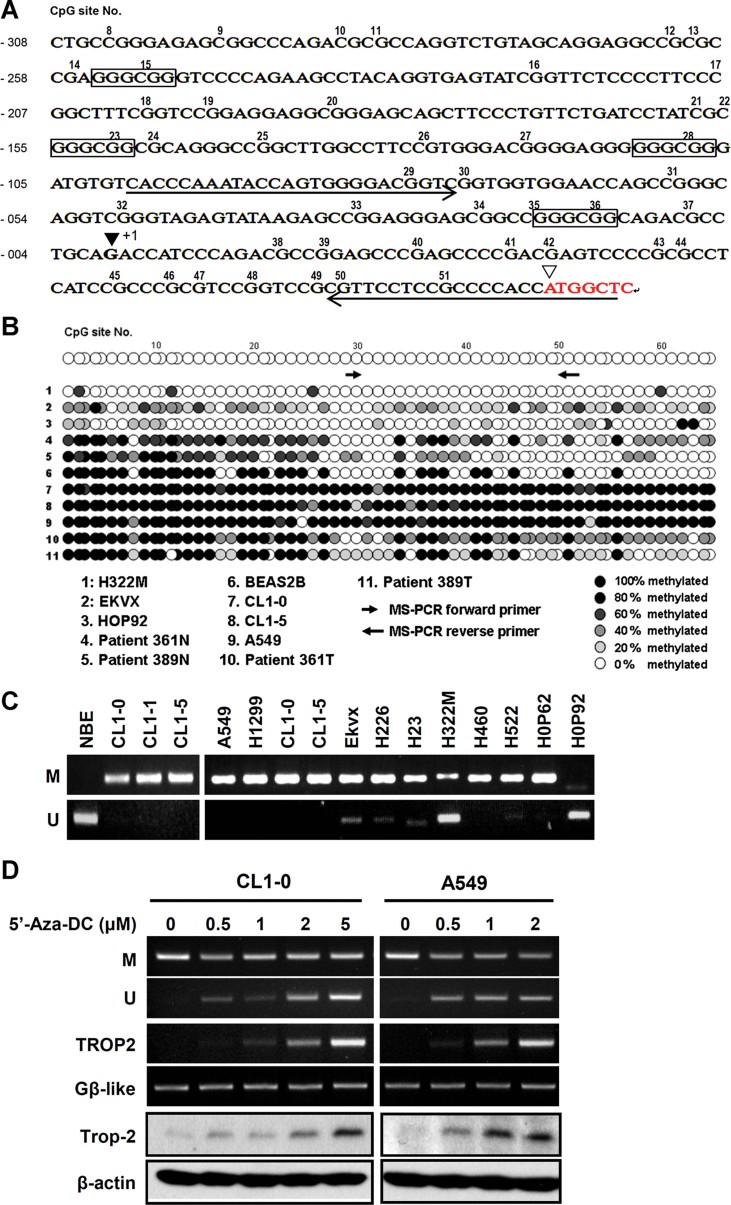
We carried out CpG island DNA analysis on TROP2 based on the following definition: CpG island is defined as DNA sequence of at least 200 bp with a GC content above 50% and an observed/expected ratio of CpG dinucleotide above 0.6 (Gardiner-Garden & Frommer, 1987). Based on this analysis, we found an CpG island in TROP2, which covers a DNA fragment larger than 4 kb located upstream of the transcriptional start site. Since it was too long for the current lab technique, we decided to look at a portion of this long CpG island by designing primers flanking only a 769 bp promoter fragment for bisulphite sequencing analysis. As shown in Fig 2A, only a partial DNA sequence of the 769 bp promoter fragment was obtained. The sequencing information confirmed the presence of 78 CpGs, 4 SP1 sites (GGGCGG) and the transcriptional start site (+1) as well as the translation start site. Figure 2B illustrates the bisulphite sequencing results of two paired normal and tumour tissues, lung CL lines and the immortalized NBE cell line BEAS-2B. Methylation occurs in most of cancer tissues (361T and 389T) as well as in several lung CL lines such as CL1-0, CL1-5 and A549. Non-methylated Cs were largely detected in normal tissues and the BEAS-2B cells line with three exceptions. H322M cells were largely unmethylated in the investigated region, while there was reduced methylation in EKVX and HOP92. Since methylation occurred frequently between the 29/30th and 50/51st CpG, MS and un-methylation-specific (UMS) primers flanking this region were designed for PCR quantification of the methylation status. As shown in Fig 2C, both MS- and UMS-PCR results were consistent with the above bisulphite sequencing. Taken together, we conclude that TROP2 undergoes epigenetic inactivation in most lung CL lines and tissues, and this phenomenon may result in lower or absent protein and mRNA expression in these cells (Fig 1C and D).
Figure 2. Downregulation of TROP2 is associated with DNA hypermethylation of the TROP2 promoter region in lung CL lines.
- The putative promoter region of TROP2 was predicted using the promoter scan website (http://www-bimas.cit.nih.gov/molbio/proscan). The transcription and translation start sites are marked as ▾ and ▿, respectively. The numbers above the sequence indicate the CpGs that are predicted to be methylation sites. The boxed sequence (GGGCGG) is a putative Sp1 binding site. The underline ‘→’ and ‘←’ indicate the designed forward and reverse primers site on promoter region for MS-PCR analysis.
- The methylation status of the putative promoter region in various cell lines was examined by bisulphite sequencing analysis. Each spot indicates one methylation site (CpG). The forward primer contains 29th and 30th CpG sites. The reverse primer contains 50th, 51st and 52nd CpG sites.
- MS-PCR analysis of methylation in the various lung cancer lines by using specific methyl- and unmethyl- primer sets.
- Treatment of cells with 5-aza-2′-DC dose-dependently restored TROP2 expression in A549 and CL1-0 cells. M and U indicated amplified by using methyl- and unmethyl- detecting primers. Panels 3 and 4 were detected by RT-PCR and panels 5 and 6 were measured by Western blot.
To further confirm the significance of epigenetic regulation, 5-aza-2′-deoxycytidine (5 Az-DC) was used in A549 and CL1-0 cells. As shown in Fig 2D, low doses of 5 Az-DC at less than 5 µM were able to change the methylation status of the TROP2 promoter CpG island based on MS- and UMS-PCR analyses. This change was accompanied by an increase of Trop-2 mRNA and protein synthesis in these cells.
Correlation between hypermethylation and downregulation of TROP2 expression in lung adenocarcinoma tissues
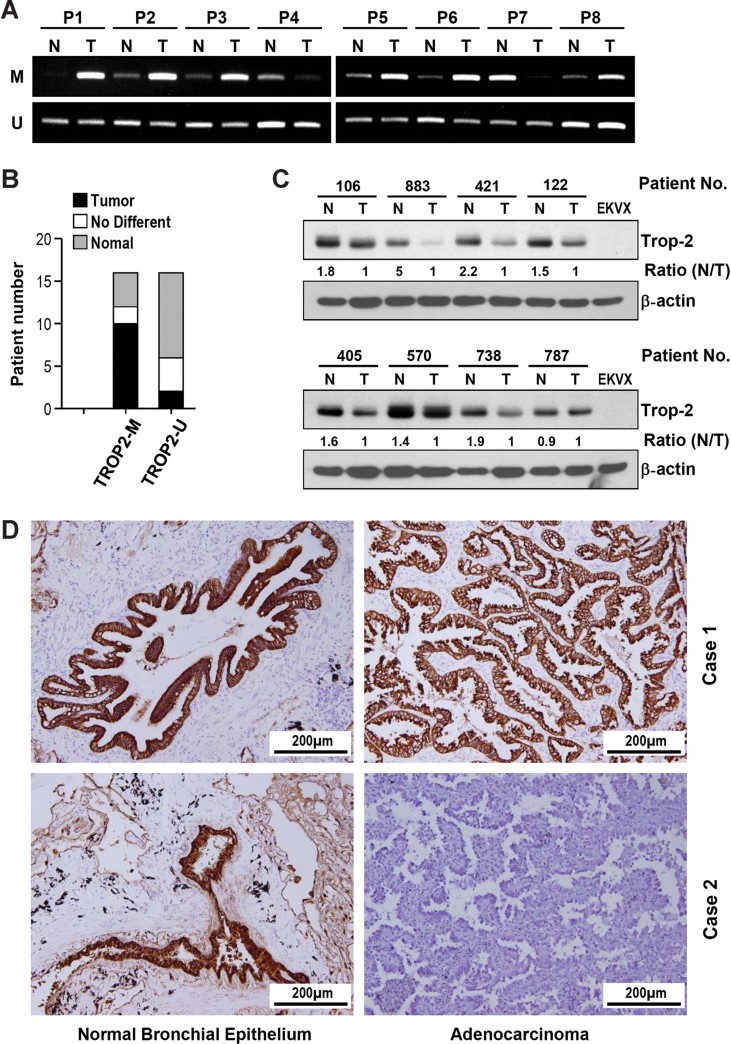
Using the MS- and UMS-PCR approach, we examined the TROP2 methylation status and Trop-2 expression in 16 paired normal and adenocarcinoma tissues. We found that about 60% (10/16) of the examined lung adenocarcinoma patients showed hypermethylation at the TROP2 promoter region in tumour tissues versus paired normal lung tissues as assessed by MS-PCR (Fig 3B). Examples of such a PCR analysis are shown in Fig 3A. Western blots were carried out using protein extracts of eight additional paired normal and tumour tissues (Fig 3C). These data confirmed the differential expression levels with high Trop-2 expression in normal tissues as compared to the low expression in tumours. Immunohistochemical characterization of 55 lung adenocarcinoma tissues further supported this conclusion. 33 out of 55 tumour tissues (60%) had either very low or no expression of Trop-2, while normal bronchial epithelia were rich in expression (Table 1). There was no significant difference among these patients in terms of age, sex or disease stage. Representative images of Trop-2-positive and -negative lung cancer specimen are shown in Fig 3D. In general, TROP2 was highly expressed in NBE cells and the protein accumulated at the cell membranes, but decreased expression was noted in the tumour cells. Taken together, our results suggest that Trop-2 protein expression is downregulated in lung adenocarcinoma tissues versus paired normal specimen and that this is commonly associated with DNA hypermethylation of the TROP2 promoter region.
Figure 3. Epigenetic downregulation of TROP2 in lung cancer tissues.
- The methylation patterns in a number of lung cancer and normal tissue pairs were determined by MS-PCR.
- In nearly 60% (10/16) of the pairs, the cancer tissues showed promoter region hypermethylation compared to the normal tissues.
- Trop-2 protein expression in a number of paired samples was assessed by Western blot analysis. The densitometry was analysed by TL100 Gel Analysis Software (Nonlinear Dynamics Ltd, UK).
- Immunohistochemical analysis of paired tissues (normal bronchial epithelium vs. adenocarcinoma) from two lung adenocarcinoma patients. In patient No. 1, the cancer tissue and normal bronchial epithelium showed nearly equal amounts of Trop-2. In patient No. 2, the NBE cells showed TROP2 expression, but the cancer tissue did not.
Table 1.
Characteristics of 55 patients with lung adenocarcinoma analysed by immunohistochemistry
| Characteristics | Patient no. (%) with higher Trop-2 expression | Patient no. (%) with lower Trop-2 expression | p-Value* |
|---|---|---|---|
| Age (mean ± SD) (years) | 64.4 ± 10.3 | 59.6 ± 10 | 0.093 |
| Sex | |||
| Male | 11 (50) | 12 (36) | 0.406 |
| Female | 11 (50) | 21 (64) | |
| Stage | |||
| I and II | 18 (82) | 27 (82) | 1 |
| III | 4 (18) | 6 (18) | |
Higher and lower Trop-2 expression refers to the expression of Trop-2 in the tumor compared to corresponding normal tissue.
Between-group data comparisons were made using the t-test. A p-value of <0.05 was considered statistically significant.
Inhibition of cell proliferation and colony formation by ectopic Trop-2 expression
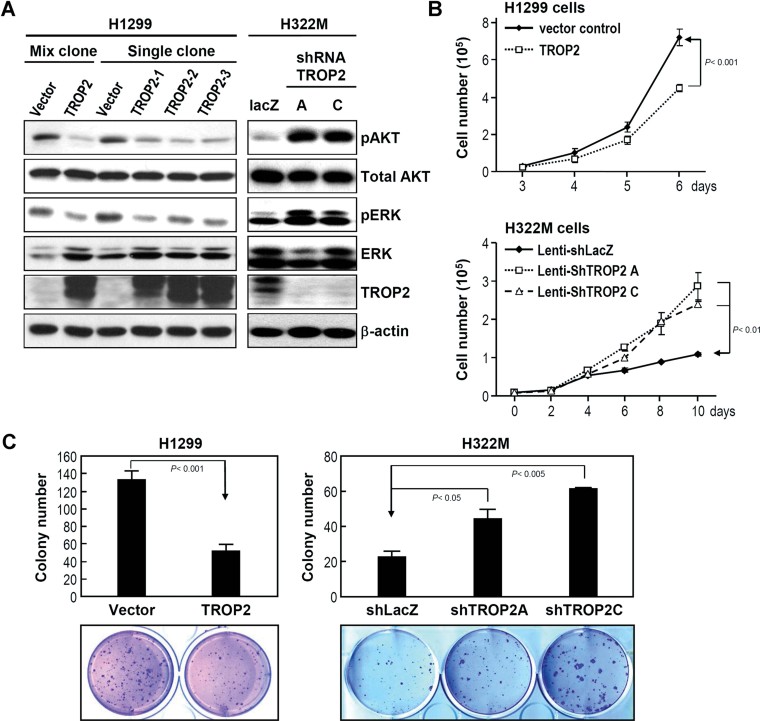
To further examine the role of Trop-2 in lung adenocarcinoma malignancy, Trop-2-encoding plasmid DNA was constructed and transfected into H1299 cells, which lack endogenous TROP2 expression. Forced expression of Trop-2 in H1299 cells inhibited the phosphorylation of AKT and ERK (Fig 4A), attenuated cell proliferation (Fig 4B), and suppressed colony formation (Fig 4C). Similar results were obtained in CL1-0 and H23 lung CL lines (Fig S3 of Supporting Information). To examine the reverse situation, we used lenti-shRNAs to specifically knock down TROP2 expression in H322M cells, which have high levels of endogenous Trop-2. Our results revealed that silencing TROP2 expression with either lenti-shTROP2A or lenti-shTROP2C upregulated the activities of AKT and ERK (Fig 4A), promoted cell proliferation (Fig 4B) and increased colony formation (Fig 4C).
Figure 4. TROP2 expression modulates cell proliferation and colony formation.
- Trop-2 was overexpressed in H1299 cells and knocked down in H322M cells, and the activities of AKT and ERK were determined by Western blotting with anti-pAKT and anti-p-ERK antibodies, respectively.
- Forced expression of Trop-2 in H1299 cells inhibited cell proliferation compared to the vector control, as determined by cell counting. Conversely, Trop-2 knockdown by either lenti-shTROP2A or lenti-shTROP2C enhanced cell proliferation.
- The tumour growth ability of lung CL was measured by the formation of colonies in soft agar. The results are presented as means ± SD, data were compared between groups using the t-test, and p < 0.05 was considered statistically significant when compared to the control.
TROP2 knockdown promotes xenograft tumour growth in mice
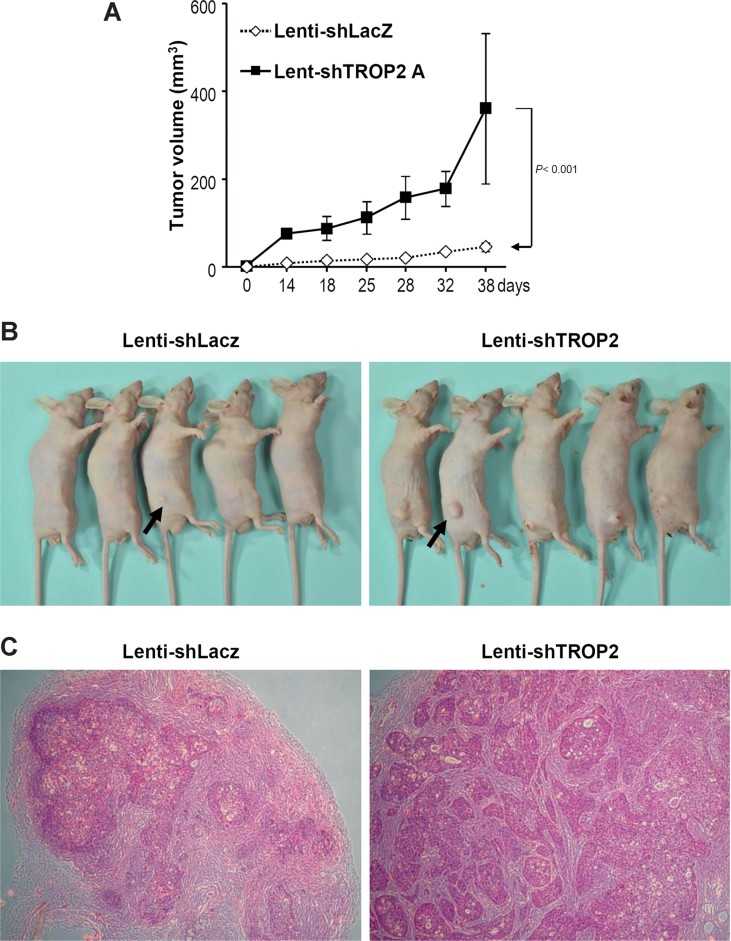
To explore whether TROP2 knockdown could promote lung adenocarcinoma progression, we used lenti-shTROP2A to decrease TROP2 expression in H322M cells, and then subcutaneously inoculated nude mice with these cells. We found that tumours arising from TROP2-silenced H322M cells time-dependently increased in volume at a greater rate compared to those arising from lenti-shLacZ-infected control cells (Fig 5A). There were no body weight differences between the two groups (n = 5) of mice (Fig 5B and C).
Figure 5. TROP-2 knockdown promotes xenograft tumour growth in mice.
- H322M cells were infected with lenti-shTROP2A or lenti-shLacZ (control) and injected into nude mice, and tumour volumes were monitored for 6 weeks. The results are presented as means±SEM, data were compared between groups using the t-test, and p < 0.05 was considered statistically significant when compared to the control.
- Photos of tumour-bearing mice, with tumours indicated by black arrows.
- H&E staining of tumour tissues obtained from mice inoculated with lenti-shTROP2A or lenti-shLacZ.
Trop-2 binds to IGF-1 and interferes with IGF-1R signalling mediated β-catenin/slug expression
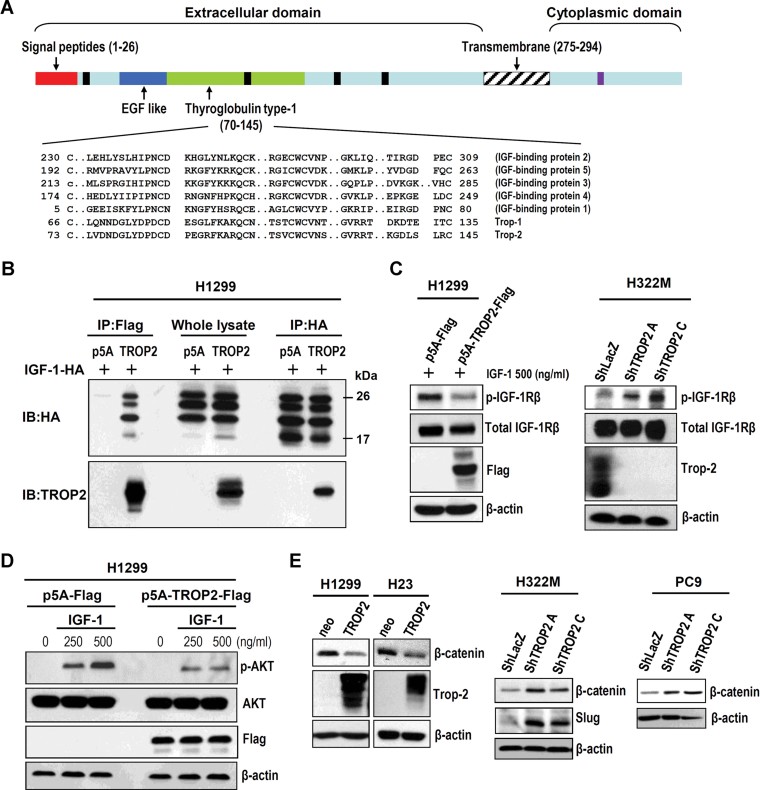
We carried out a conserved domain analysis using the conserved domain database on the NCBI website. Interestingly, we found that members of the IGF-binding proteins (IGFBP1 to −5) and TROP families (TROP1 and −2) all have a thyroglobulin type-1 domain (Fig 6A). We therefore hypothesized that Trop-2 may potentially attenuate IGF-1/IGF-1R signal transduction by competing with the receptor for IGF-1 binding via the thyroglobulin type-1 domain. To examine this possibility, we performed a co-immunoprecipitation (co-IP) assay to investigate whether Trop-2 can interact with IGF-1. As shown in Fig 6B, mIGF-1HA and Trop-2-Flag interacted in cells co-transfected with these two plasmids (Fig 6B). Deletion analysis showed that the interaction depends on the presence of the region between EGF-like and thyroglobulin type-1 domains of Trop-2 protein (Fig S4 of Supporting Information). Furthermore, the activation of IGF-1R signalling (pIGF-1Rβ) was attenuated when Trop-2-Flag was overexpressed in H1299 cells, which have low or no endogenous Trop-2 expression (Fig 6C). A similar suppression of β-catenin expression was seen in H23 and H1299 after ectopic Trop-2 expression (Fig 6E). This attenuation was also seen in IGF-1/IGF-1R downstream AKT phosphorylation (Fig 6D). Conversely, TROP2 knockdown in H322M cells upregulated IGF-1R signalling (Fig 6C) and expression of both β-catenin and slug (Fig 6E). A similar observation regarding β-catenin expression was made in PC9 cells transfected with shTROP2A and shTROP2C (Fig 6E). These results indicate that the cell-surface protein Trop-2 may trap IGF-1 in the surrounding microenvironment, thereby, inhibiting the activation of IGF-1R signalling-mediated gene expression.
Figure 6. Trop-2 can bind to IGF-1 and interfere with IGF-1R signalling.
- BLAST analysis of Trop-2 against the NCBI database (http://www.ncbi.nlm.nih.gov/) shows that the members of the TROP and insulin growth factor binding protein (IGFBP) families share a conserved thyroglobulin type-1 domain. Black and purple boxes indicated the glycosylation and phosphorylation sites, respectively.
- Vectors encoding TROP2 and mIGF-1 were co-transfected into H1299 lung CL, and the interaction of TROP2 and mIGF-1 was examined by co-IP. WB, western blot.
- Overexpression of TROP2 in H1299 cells reduced the activation of IGF-1R signalling, while TROP2 knockdown activated IGF-1R signalling in H322M cells.
- Forced expression of TROP2 in H1299 cells suppressed the IGF-1-induced phosphorylation of AKT.
- The expression of downstream mediators of IGF-1R signalling (e.g. β-catenin) decreased when TROP2 was overexpressed in H1299 and H23 cells, and increased following TROP2 knockdown in H322M and PC-9 cells. Slug expression was induced by TROP2 knockdown.
Together, our findings suggest a model in which epigenetic silencing of TROP2 by DNA methylation or LOH in lung CL suppresses the expression of Trop-2 in the membrane, thereby, attenuating its suppressive effect on IGF-1R signalling and promoting tumour progression via activation of the signalling molecules downstream of IGF-1R, including β-catenin and slug (Fig 7).
Figure 7. The proposed role of TROP2 in lung tumourigenesis.
When TROP2 is silenced by DNA methylation, Trop-2-mediated suppression of IGF-1R signalling decreases. This may lead to cancer progression (e.g. invasion, metastasis and/or angiogenesis) via activation of IGF-1R signalling and its downstream mediators β-catenin and slug.
DISCUSSION
In this study, we demonstrated that TROP2 is either epigenetically inactivated by DNA methylation or due to LOH in most lung adenocarcinoma tissues and cells. This suppression may promote CL proliferation and tumourigenicity. On the contrary, elevated expression of Trop-2 can inhibit cell proliferation and colony forming efficiency. We further demonstrated that Trop-2 can interact with IGF-1 to suppress IGF-1/IGF-1R signalling and its downstream target genes, AKT and ERK activation, and the suppression of β-catenin and slug expression. This finding suggests that high level expression of Trop-2 in lung CL can suppress cell proliferation by binding IGF-1 and reducing the activation of IGF-1R signalling. Conversely, the loss of Trop-2 expression in lung CL may increase the binding of IGF-1 to IGF-1R, thereby, enhancing the activation of IGF-1R signal transduction, promoting cell proliferation and increasing tumour growth. These results suggest a very important regulatory role of Trop-2 in negatively interfering with IGF-1R signalling that is needed for lung cancer development and malignancy (Fig 7).
Gene expression may be regulated epigenetically or genetically. Epigenetic changes, which are defined as modifications of the chromatin structure without alteration of the primary DNA sequence (Jaenisch & Bird, 2003), may be induced by external factors and are potentially reversible. LOH at human chromosome 1p32 is frequent in various cancers, including lung cancer (Chizhikov et al, 2001; Fong et al, 1996), breast cancer (el-Rifai et al, 1999), meningioma (Kim et al, 2009; Sulman et al, 1998), adenoid cystic carcinoma (Rao et al, 2008) and oligodendroglia (Husemann et al, 1999). Thus, this region of chromosome 1 is expected to contain one or more tumour suppressor genes. In this study, we demonstrated that TROP2 is located in this region of chromosome 1p32, and the expression of this gene is downregulated in most lung CL lines due to increased DNA methylation of the TROP2 promoter region. These two-hit findings, DNA hypermethylation and high frequency of LOH, strongly support the notion that TROP2 may potentially serve as a tumour suppressor gene in lung cancer development (Yang et al, 2002). Although the main focus in this study was on lung adenocarcinoma, we also find high LOH frequency associated with some lung squamous carcinomas (Fig 1 and Fig S1 of Supporting Information). Thus, Trop-2 may play a generalized role in suppressing most of lung cancer development and malignancy. Further studies on the functional role of Trop-2 in lung squamous carcinoma are needed.
Recently, low level expression of Trop-2 in NSCLC cells was found to be associated with EGFR-TKI resistance (Frederick et al, 2007). However, the mechanism underlying this effect remains unknown. In addition, some studies have indicated that gefitinib, an EGFR-TKI inhibitor, resistance can be acquired by lung CL when they undergo an alternative signalling switch from EGFR to IGF-1R (Guix et al, 2008). Blockade of IGF-1R signalling by specific inhibitors can decrease tumour growth in a xenograft mouse model by disrupting the association of IRS-1 with PI3K and downregulating PI3K/AKT signalling (Guix et al, 2008). These inhibitors also inhibit heterodimerization and signalling through IGFR/EGFR driven by amphiregulin in mucinous lung adenocarcinoma (Hurbin et al, 2011).
Overexpression of IGF binding protein-3 (IGFBP3), a natural inhibitor of IGF signalling, was shown to inhibit the growth of xenograft tumours derived from lung CL by downregulating PI3K/AKT signalling (Lee et al, 2002). Here, we found that both IGFBP3 and Trop-2 have conserved thyroglobulin type-1 domains. A deletion approach demonstrated the requirement of this domain for IGF-1 binding. Thus, this domain may allow Trop-2 to interfere with IGF-1R signalling by interacting with IGF-1. In addition, we showed that downregulation of Trop-2 induced the expression of slug, which was recently identified as an anti-apoptotic factor in EGFR-TKI-resistant cells (Chang et al, 2011). These findings indicate that there may be an important connection between Trop-2 expression and EGFR-TKI/gefitinib sensitivity. From this study and previous ones, we conclude that Trop-2 is a potential therapeutic target for lung cancer treatment and this potential is related to the dual effects, IGF-1R signalling and EGFR-TKI/gefitinib resistance, associated with a lowering of Trop-2 expression in most of lung cancers.
Several studies have shown that Trop-2 is a malignant factor and is overexpressed in colorectal, gastric, oral and pancreatic cancer (Fong et al, 2008a, 2008b; Muhlmann et al, 2009; Ohmachi et al, 2006). Interestingly, Tsujikawa et al identified that TROP2 mRNA is expressed in many normal human tissues such as cornea, kidney, lung, placenta, pancreas and prostate by Northern analysis (Tsujikawa et al, 1999). In addition, we found that TROP2 mRNA expression is downregulated in some cancer tissues such as breast, kidney, lung, prostate and placenta in EST database analysis (Fig 2). Differences in expression in different tumours may suggest diverse roles of Trop-2 in cancer development and malignancy. Kobayashi et al demonstrated that Trop-2 was expressed in the cytoplasm when cells became malignant in some cases of cancer metastasis and recurrence, but the precise role of this protein in the cytoplasm is not yet understood (Kobayashi et al, 2010). Terrinoni et al (2001) identified a novel chimeric cyclin D1-TROP2 protein in some human cancers and showed that its expression could induce cell transformation. Silencing of this chimeric protein was shown to inhibit tumour growth (Guerra et al, 2008). Thus, incomplete or aberrant production of Trop-2 may cause the protein to lose its function and be internalized from the membrane to the cytoplasm resulting in cancer progression. Tsujikawa et al demonstrated that four genetic mutations (Q118X, 632delA, Q207X and S170X) of TROP2 may lead to the synthesis of a truncated, non-functional protein that causes gelatinous droplike corneal dystrophy (Tsujikawa et al, 1999). Although there is not yet any evidence linking this Trop-2 mutation with cancer metastasis, we suspect that the differential subcellular localization and genetic mutation of TROP2 could also be involved in cancer progression.
Trop-2 is generally believed to be a receptor, but its intracellular signalling pathways and the involved ligands have not yet been fully elucidated. Cubas et al (2010) demonstrated that mouse Trop-2 could activate ERK and promote cell proliferation in pancreatic and colon CL in low serum condition and in a mouse animal model (Cubas et al, 2010). It is reasonable because Trop-2 contains an EGF-like domain that may likely activate EGFR signalling and ERK activation in serum-free condition. Our current data are in contrast to this notion. As shown in Fig 4, we observed that pERK activation was suppressed by Trop-2 overexpression and enhanced by shRNA knockdown in lung CL lines. In addition, Wang et al (2011) recently showed that loss of Trop-2 (in knockout mice) could enhance tumour formation and epithelial to mesenchymal transition (EMT) of keratinocytes through activation of mitogen-activated protein kinase (MAPK) and downregulation of E-cadherin expression (Wang et al, 2011). These results may support our current findings in this study. It may be difficult to explain the difference, however, different cancer histotypes and culture conditions among others may be involved. To further investigate this, we silenced TROP2 in highly Trop-2 expressing cell lines including T74-D (mammary), HT29 (colon), and OVCAR3 (ovary) human cancer cells using lentiviral transduction (Supporting Information Fig. S5) and found that knockdown of Trop-2 in T47-D cells can promote the activation of AKT and ERK slightly but has no or an inhibitory effect on human colon (HT29) and ovary (OVCAR3) cancer cells, respectively (Supporting Information Fig. S5). This finding suggested that Trop-2 may regulate the activities of AKT and ERK dependend on the histotypes of cancer cells.
Here, we provide the first report suggesting that Trop-2 can interact with IGF-1 to interfere with its binding to IGF-1R, thereby, decreasing the activation of IGF-1R signalling. However, we cannot rule out the possibility that this effect could be due to the ability of downstream mediators of Trop-2 to attenuate the activation of AKT and ERK. Under this paradigm, we see two plausible mechanisms: (1) IGF-1 might be a ligand of Trop-2, allowing it to activate its putative downstream mediators (e.g. PIP2 and Ca2+) and modulate IGF-1R signalling (El Sewedy et al, 1998; Ripani et al, 1998) or (2) Trop-2 may form a complex with IGF-1 and thereby mediate the activation of IGF-1R signalling. Further studies are needed to test this hypothesis.
In conclusion, we herein show that inactivation of TROP2 may play an important role in lung carcinogenesis. The endogenous expression of TROP2 in lung epithelial cells can suppress IGF-1-mediated activation of IGF-1R, β-catenin and slug signalling. Conversely, TROP2 downregulation by DNA hypermethylation may decrease its suppressive effect on IGF-1R signalling, potentially leading to lung carcinogenesis.
MATERIALS AND METHODS
Cell lines, chemicals and antibodies
Human lung adenocarcinoma cell lines with increasing invasive ability were previously established in our laboratory and designated CL1-0, CL-1-1 and CL1-5 (Chu et al, 1997). Lung CL lines (i.e. H322M, EKVX, HOP62, HOP92, A549, H522, H460, H226 and H23) were purchased from the National Cancer Institute (Bethesda, MD). The cells were grown in RPMI 1640 culture medium (GIBCO-Life Technologies, Inc., Gaithersburg, MD) or Dulbecco's modified Eagle's medium (GIBCO-Life Technologies), supplemented with 1.5 g/L of NaHCO3, and 10% foetal bovine serum (FBS; GIBCO-Life Technologies). rIGF-1 and 5-aza-2′-DC were obtained from R&D Systems, Inc. (Minneapolis, MN) and Sigma, Inc. (Saint Louis, MO), respectively. The antibodies used in this study were obtained as follows: anti-Trop-2 from R&D Systems, Inc.; anti-p-AKT from Cell Signalling Inc. (Danvers, MA); and anti-AKT, -ERK, -p-ERK, -p-IGF-1R, -IGF-1R and -β-actin from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
The paper explained
PROBLEM:
NSCLC remains a major cause of cancer mortality worldwide. The etiology of NSCLC, especially in non-smokers, is still unknown and effective therapy is not available. Trop-2 is a cell-surface glycoprotein that contains extracellular epidermal growth factor-like and thyroglobulin type-1 repeat domains. Its cytoplasmic tail contains a conserved PIP2-binding domain with one phosphorylation site (S303). The TROP2 expression has been found to associate with tumour formation and progression of various cancers, but positive and negative effects were both reported. However, the role of TROP2 in lung cancer remains unclear. Improving our understanding of the molecular role played by Trop-2 in lung carcinogenesis may help us to develop new strategies for more effective treatment and prevention of lung cancer.
RESULTS:
We explored the potential role of TROP2 gene in lung cancer with several different aspects including cancer genomics, epigenetics, in vitro, in vivo and clinical studies. These results suggested that TROP2 gene was downregulated in lung cancer patients via DNA hypermethylation on its promoter region. Re-expression of Trop-2 in lung CL attenuates cell proliferation and colony formation while, conversely, TROP2 silencing promotes tumour growth. We also showed that Trop-2 may act on IGF-1R signalling pathway to suppress the activation of AKT and ERK through disrupting the interaction between IGF-1 and IGF-1R. The crosstalk between Trop-2 and IGF-1R signalling may play an important role on the tumour progression in lung adenocarcinoma.
IMPACT:
We showed that TROP2 expression is downregulated in lung adenocarcinoma through DNA hypermethylation on its promoter region. Both IGFBP3 and Trop-2 have conserved thyroglobulin type-1 domains, which may allow Trop-2 to interfere with IGF-1R signalling by interacting with IGF-1. In addition, downregulation of Trop-2 induces the expression of slug, which is one of the key players in lung cancer progression, invasion/metastasis and EGFR-TKI resistance. The findings indicate the existence of important crosstalk between Trop-2 signalling and EGFR-TKI sensitivity, suggesting that downregulation of Trop-2 in lung adenocarcinoma may associate with the activation of IGF-1R signalling and EGFR-TKI resistance. Trop-2 may be a potential biomarker and therapeutic target in NSCLC.
Patients, loss of heterozygosity analysis and immunohistochemistry
Paired lung adenocarcinoma and normal lung tissues were obtained from 55 patients with a histologically confirmed diagnosis who underwent surgical resection at the National Taiwan University (NTU) Hospital (Taipei, Taiwan) between 2000 and 2003. None of the patients had received neoadjuvant chemotherapy or radiation therapy prior to surgery. This study was approved by the Institutional Review Board of the NTU Hospital, and written informed consent was obtained from all patients. Genomic DNA isolation and LOH analysis using microsatellite markers close to TROP2 were performed as previously described (Tsai et al, 2006; Tseng et al, 2005). For immunohistochemical analysis of Trop-2 protein expression, tissue sections were first autoclaved in Trilogy (Cell Marque Corp., Rocklin, CA) or Antigen Retrieval Citra Solution (Biogenex, San Ramon, CA) at 121°C for 10 min. The samples were then treated with 3% H2O2–methanol, and sequentially incubated with the following: DakoCytomation Dual Endogenous Enzyme Block (DakoCytomation, Inc., Carpinteria, CA) for 10 min; Ultra V Block (LAB VISION Corp., Fremont, CA) for 10 min; and antibody dilution buffer (Ventana Medical Systems, Inc., Tucson, AZ) for 10 min. The sections were incubated overnight at 4°C with anti-Trop-2 (1:100 dilution), and immunoreactive staining was detected using the Super Sensitive Non-Biotin Polymer HRP Detection System (Biogenex).
Reverse transcriptase-polymerase chain reaction
RNA was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA), and 1 µg of total RNA was subjected to reverse transcription by using Superscript III (Invitrogen). The resulting cDNA was then taken as the template for PCR amplification using primers for TROP2 (sense, 5′-GACAACTGCACGTGTCCCAC-3′; and antisense 5′-AGAGGCCATCGTTGTCCACG-3′) or the Gβ-like protein (internal control; sense, 5′-GTATGGAACCTGGCTAACTG-3′; and antisense 5′-TACTGATAACTTCTTGCTTC-3′). All results were visualized on an ethidium bromide-stained agarose gel.
Bisulphite sequencing
Genomic DNA (1 µg) was treated with EZ DNA Methylation Kit (Zymo Research, Irvine, CA) as described in the manufacturer's instructions, and then eluted in 40 µl of elution buffer. The eluted DNA (2 µl) was PCR amplified with TROP2-specific bisulphate sequencing primers (TROP2-BSF 5′-TGTGTTATTGTGAGAATTGGATAAAG-3′; and TROP2-BSR 5′-AAACAACAAACACTTAAAAATCAAC-3′). The resulting PCR product (∼1 kb) was obtained by 1% agarose gel electrophoresis, cloned using an RBC TA Cloning Vector kit (RBC Bioscience, New Taipei, Taiwan), and then analysed by DNA sequencing.
Methylation specific-polymerase chain reaction
Genomic DNA was isolated using a QIAamp DNA mini kit (Qiagen, Hilden, Germany), 1 µg of genomic DNA was treated with sodium bisulphate and eluted as described above and 2 µl of the treated DNA was amplified with two set of MS primers (TROP2-M-F 5′-TATTTAAATATTAGTGGGGACGGTC-3′ and TROP2-M-R 5′-GAACCATAATAAAACGAAAAAACG-3′; and TROP2-U-F 5′-ATTTAAATATTAGTGGGGATGGTTG-3′ and TROP2-U-R 5′-CCAAACCATAATAAAACAAAAAAACA-3′). The forward primer contains 29th and 30th CpG sites. The reverse primer contains 50th, 51st and 52nd CpG sites. The PCR conditions were as follows: 45 cycles of 94°C for 30 s, 62°C for 30 s and 72°C for 45 s. The resulting PCR products with 191 bp were resolved using 2% agarose gel electrophoresis.
Cell proliferation assay
Cells were transferred to triplicate wells of a 24-well plates (1 × 104 cells/well in 500 µl of complete medium) and incubated at 37°C in 5% CO2 for 6 or 10 days. Cell proliferation was measured every 24 or 48 h using a haemocytometer and the trypan blue method.
Colony formation assays
Cells (500 or 1000) were plated in triplicate wells of 6-well plates and grown for 10–14 days, and then fixed and stained with 0.05% methylene blue for 1 h. The plates were washed and dried, and the colonies were counted.
Western blot analysis
Equal amounts of cell lysate were separated by 10% SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to a polyvinylidene (PVD) membrane (Millipore, Billerica, MA). The membranes were probed with the appropriate antibody diluted in TBS (pH 7.5) containing 0.05% v/v Tween 20 (Sigma) and 5% w/v dried milk, followed by incubation with the appropriate horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences, Uppsala, Sweden). The bound antibodies were visualized by electrochemical luminescence (ECL) staining followed by autoradiography using Kodak X-Omat Blue film (PerkinElmer Life Science, Boston, MA).
Gene knockdown and viral transdcution
TROP2-shRNA-containing lentiviral vectors were obtained from the National RNAi Core Facility (Academia Sinica, Taiwan); lenti-shTROP2-A was designed to target the sequence GAGAAAGGAACCGAGCTTGTA, while lenti-shTROP2-C targeted CGTGGACAACGATGGCCTCT. Cells were infected with lentivirus (multiplicity of infection = 3) in serum-free medium containing polybrene (8 µg/ ml). Beginning 24 h post-infection, the cells were selected for 14 days with 2 µg/ml puromycin.
Animal models
Female nude mice were purchased from BioLasco Taiwan Co., Ltd (Taipei, Taiwan), housed in laminar flow cabinets under specific pathogen-free conditions, and provided with sterilized food and water. H322M cells were infected with either lenti-shLacZ or lenti-shTROP2A (9 × 106 cells per injection site) and injected subcutaneously into the right hind legs of 7-week-old nu/nu mice (n = 7 mice per group). Tumour volumes were determined by measuring the longest (length) and shortest (width) diameters and using the formula: tumour volume = (length) × (width)2/2. After 35 days, the mice were sacrificed and the tumour xenografts were removed and photographed.
Immunoprecipitation
IP was performed as previously described (Wang et al, 2009). In brief, cells were treated with IP lysis buffer containing 1× complete protease inhibitor cocktail (EDTA-free) and passed several times through a 21-gauge needle. The samples were centrifuged, and the obtained protein lysate (∼800 µg) was incubated overnight at 4°C with either 2 µg anti-HA antibody (HA-11; Covance) or 2 µg of anti-Flag antibody (Santa Cruz Biotechnology). The samples were incubated for 1 h at 4°C with protein A Sepharose (Sigma), and the bound beads were washed three times with TBS buffer. The protein samples were separated by SDS–PAGE, transferred to a PVDF membrane, immunoblotted with the indicated antibodies and visualized by chemiluminescence.
Statistical analyses
Results are presented as means ± standard deviations (SD). All statistical analyses were performed using SPSS for Windows, version 10.0 (SPSS Inc., Chicago, IL). Between-group data comparisons were made using the t-test. A p-value of <0.05 was considered statistically significant.
Acknowledgments
This study was supported from grants DOH 98-TD-G-111-007 and DOH99-TD-G-111-005 (Department of Health, Executive Yuan, Taiwan), and grant, NSC-97-2314-B-002-146-MY3, NSC-98-2628-B-002-086-MY3, NSC-99-2314-B-030-002, NSC-100-3112-B-006-005 and NSC-100-2321-B-002-071 (National Science Council, Taiwan). We thank Dr. Yu-Ting Yan (Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan) for providing mIGF-1-expressing plasmid, and Professor Reen Wu (University of California at Davis) for comments and editing of the manuscript.
Supporting Information is available at EMBO Molecular Medicine online.
The authors declare that they have no conflict of interest.
Author contributions
TMH and PCY conceptualized and designed the project and contributed equally to this work; JCL, JYW, YYW and TCL performed and analysed most of the experiments; CTW, YLC and YSJ performed IHC and LOH data analysis; JCL, TMH and PCY prepared the manuscript.
For more information
ECgene database (http://genome.ewha.kr/ECgene/)
Human DNA sequence clone RP4-592A1 http://www.ncbi.nlm.nih.gov/nuccore/AL035411.28
Conserved domain database in the NCBI website http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
TROP2 (Human) http://www.ncbi.nlm.nih.gov/nuccore/BC009409
Web Promoter Scan Service http://www-bimas.cit.nih.gov/molbio/proscan/
MethPrimer http://www.urogene.org/methprimer/index1.html
PhosphoSitePlus http://www.phosphosite.org/
Supplementaary material
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
References
- Chang TH, Tsai MF, Su KY, Wu SG, Huang CP, Yu SL, Yu YL, Lan CC, Yang CH, Lin SB, et al. Slug confers resistance to the epidermal growth factor receptor tyrosine kinase inhibitor. Am J Respir Crit Care Med. 2011;183:1071–1079. doi: 10.1164/rccm.201009-1440OC. [DOI] [PubMed] [Google Scholar]
- Chizhikov V, Zborovskaya I, Laktionov K, Delektorskaya V, Polotskii B, Tatosyan A, Gasparian A. Two consistently deleted regions within chromosome 1p32-pter in human non-small cell lung cancer. Mol Carcinog. 2001;30:151–158. doi: 10.1002/mc.1023. [DOI] [PubMed] [Google Scholar]
- Chu YW, Yang PC, Yang SC, Shyu YC, Hendrix MJ, Wu R, Wu CW. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am J Respir Cell Mol Biol. 1997;17:353–360. doi: 10.1165/ajrcmb.17.3.2837. [DOI] [PubMed] [Google Scholar]
- Cubas R, Li M, Chen C, Yao Q. Trop2: a possible therapeutic target for late stage epithelial carcinomas. Biochim Biophys Acta. 2009;1796:309–314. doi: 10.1016/j.bbcan.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Cubas R, Zhang S, Li M, Chen C, Yao Q. Trop2 expression contributes to tumor pathogenesis by activating the ERK MAPK pathway. Mol Cancer. 2010;9:253. doi: 10.1186/1476-4598-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sewedy T, Fornaro M, Alberti S. Cloning of the murine TROP2 gene: conservation of a PIP2-binding sequence in the cytoplasmic domain of TROP-2. Int J Cancer. 1998;75:324–330. doi: 10.1002/(sici)1097-0215(19980119)75:2<324::aid-ijc24>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- el-Rifai W, Tarmo L, Hemmer S, Forsti A, Pedersen N, Lichtenstein P, Ahlbom A, Soderberg M, Knuutila S, Hemminki K. DNA copy number losses at 1p32-pter in monozygotic twins concordant for breast cancer. Cancer Genet Cytogenet. 1999;112:169–172. doi: 10.1016/s0165-4608(98)00274-x. [DOI] [PubMed] [Google Scholar]
- Fong KM, Kida Y, Zimmerman PV, Smith PJ. MYCL genotypes and loss of heterozygosity in non-small-cell lung cancer. Br J Cancer. 1996;74:1975–1978. doi: 10.1038/bjc.1996.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong D, Moser P, Krammel C, Gostner JM, Margreiter R, Mitterer M, Gastl G, Spizzo G. High expression of TROP2 correlates with poor prognosis in pancreatic cancer. Br J Cancer. 2008a;99:1290–1295. doi: 10.1038/sj.bjc.6604677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong D, Spizzo G, Gostner JM, Gastl G, Moser P, Krammel C, Gerhard S, Rasse M, Laimer K. TROP2: a novel prognostic marker in squamous cell carcinoma of the oral cavity. Mod Pathol. 2008b;21:186–191. doi: 10.1038/modpathol.3801001. [DOI] [PubMed] [Google Scholar]
- Frankel SK, Moats-Staats BM, Cool CD, Wynes MW, Stiles AD, Riches DW. Human insulin-like growth factor-IA expression in transgenic mice promotes adenomatous hyperplasia but not pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2005;288:L805–812. doi: 10.1152/ajplung.00420.2004. [DOI] [PubMed] [Google Scholar]
- Frederick BA, Helfrich BA, Coldren CD, Zheng D, Chan D, Bunn PA, Jr, Raben D. Epithelial to mesenchymal transition predicts gefitinib resistance in cell lines of head and neck squamous cell carcinoma and non-small cell lung carcinoma. Mol Cancer Ther. 2007;6:1683–1691. doi: 10.1158/1535-7163.MCT-07-0138. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Grandis JR, Sok JC. Signaling through the epidermal growth factor receptor during the development of malignancy. Pharmacol Ther. 2004;102:37–46. doi: 10.1016/j.pharmthera.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Guerra E, Trerotola M, Dell' Arciprete R, Bonasera V, Palombo B, El-Sewedy T, Ciccimarra T, Crescenzi C, Lorenzini F, Rossi C, et al. A bicistronic CYCLIN D1-TROP2 mRNA chimera demonstrates a novel oncogenic mechanism in human cancer. Cancer Res. 2008;68:8113–8121. doi: 10.1158/0008-5472.CAN-07-6135. [DOI] [PubMed] [Google Scholar]
- Guix M, Faber AC, Wang SE, Olivares MG, Song Y, Qu S, Rinehart C, Seidel B, Yee D, Arteaga CL, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurbin A, Wislez M, Busser B, Antoine M, Tenaud C, Rabbe N, Dufort S, de Fraipont F, Moro-Sibilot D, Cadranel J, et al. Insulin-like growth factor-1 receptor inhibition overcomes gefitinib resistance in mucinous lung adenocarcinoma. J Pathol. 2011;225:83–95. doi: 10.1002/path.2897. [DOI] [PubMed] [Google Scholar]
- Husemann K, Wolter M, Buschges R, Bostrom J, Sabel M, Reifenberger G. Identification of two distinct deleted regions on the short arm of chromosome 1 and rare mutation of the CDKN2C gene from 1p32 in oligodendroglial tumors. J Neuropathol Exp Neurol. 1999;58:1041–1050. doi: 10.1097/00005072-199910000-00002. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Kim TY, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–7501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- Kim NR, Cho SJ, Suh YL. Allelic loss on chromosomes 1p32, 9p21, 13q14, 16q22, 17p, and 22q12 in meningiomas associated with meningioangiomatosis and pure meningioangiomatosis. J Neurooncol. 2009;94:425–430. doi: 10.1007/s11060-009-9879-3. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Minami Y, Anami Y, Kondou Y, Iijima T, Kano J, Morishita Y, Tsuta K, Hayashi S, Noguchi M. Expression of the GA733 gene family and its relationship to prognosis in pulmonary adenocarcinoma. Virchows Arch. 2010;457:69–76. doi: 10.1007/s00428-010-0930-8. [DOI] [PubMed] [Google Scholar]
- Larsson O, Girnita A, Girnita L. Role of insulin-like growth factor 1 receptor signalling in cancer. Br J Cancer. 2005;92:2097–2101. doi: 10.1038/sj.bjc.6602627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CT, Wu S, Gabrilovich D, Chen H, Nadaf-Rahrov S, Ciernik IF, Carbone DP. Antitumor effects of an adenovirus expressing antisense insulin-like growth factor I receptor on human lung cancer cell lines. Cancer Res. 1996;56:3038–3041. [PubMed] [Google Scholar]
- Lee HY, Chun KH, Liu B, Wiehle SA, Cristiano RJ, Hong WK, Cohen P, Kurie JM. Insulin-like growth factor binding protein-3 inhibits the growth of non-small cell lung cancer. Cancer Res. 2002;62:3530–3537. [PubMed] [Google Scholar]
- Lee YJ, Imsumran A, Park MY, Kwon SY, Yoon HI, Lee JH, Yoo CG, Kim YW, Han SK, Shim YS, et al. Adenovirus expressing shRNA to IGF-1R enhances the chemosensitivity of lung cancer cell lines by blocking IGF-1 pathway. Lung Cancer. 2007;55:279–286. doi: 10.1016/j.lungcan.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Lu J, Izvolsky KI, Qian J, Cardoso WV. Identification of FGF10 targets in the embryonic lung epithelium during bud morphogenesis. J Biol Chem. 2005;280:4834–4841. doi: 10.1074/jbc.M410714200. [DOI] [PubMed] [Google Scholar]
- Ludovini V, Bellezza G, Pistola L, Bianconi F, Di Carlo L, Sidoni A, Semeraro A, Del Sordo R, Tofanetti FR, Mameli MG, et al. High coexpression of both insulin-like growth factor receptor-1 (IGFR-1) and epidermal growth factor receptor (EGFR) is associated with shorter disease-free survival in resected non-small-cell lung cancer patients. Ann Oncol. 2009;20:842–849. doi: 10.1093/annonc/mdn727. [DOI] [PubMed] [Google Scholar]
- Ma Z, Dong A, Kong M, Qian J. Silencing of the type 1 insulin-like growth factor receptor increases the sensitivity to apoptosis and inhibits invasion in human lung adenocarcinoma A549 cells. Cell Mol Biol Lett. 2007;12:556–572. doi: 10.2478/s11658-007-0022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorehead RA, Sanchez OH, Baldwin RM, Khokha R. Transgenic overexpression of IGF-II induces spontaneous lung tumors: a model for human lung adenocarcinoma. Oncogene. 2003;22:853–857. doi: 10.1038/sj.onc.1206188. [DOI] [PubMed] [Google Scholar]
- Muhlmann G, Spizzo G, Gostner J, Zitt M, Maier H, Moser P, Gastl G, Muller HM, Margreiter R, Ofner D, et al. TROP2 expression as prognostic marker for gastric carcinoma. J Clin Pathol. 2009;62:152–158. doi: 10.1136/jcp.2008.060590. [DOI] [PubMed] [Google Scholar]
- Ohmachi T, Tanaka F, Mimori K, Inoue H, Yanaga K, Mori M. Clinical significance of TROP2 expression in colorectal cancer. Clin Cancer Res. 2006;12:3057–3063. doi: 10.1158/1078-0432.CCR-05-1961. [DOI] [PubMed] [Google Scholar]
- Ouban A, Muraca P, Yeatman T, Coppola D. Expression and distribution of insulin-like growth factor-1 receptor in human carcinomas. Hum Pathol. 2003;34:803–808. doi: 10.1016/s0046-8177(03)00291-0. [DOI] [PubMed] [Google Scholar]
- Playford MP, Bicknell D, Bodmer WF, Macaulay VM. Insulin-like growth factor 1 regulates the location, stability, and transcriptional activity of beta-catenin. Proc Natl Acad Sci USA. 2000;97:12103–12108. doi: 10.1073/pnas.210394297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PH, Roberts D, Zhao YJ, Bell D, Harris CP, Weber RS, El-Naggar AK. Deletion of 1p32-p36 is the most frequent genetic change and poor prognostic marker in adenoid cystic carcinoma of the salivary glands. Clin Cancer Res. 2008;14:5181–5187. doi: 10.1158/1078-0432.CCR-08-0158. [DOI] [PubMed] [Google Scholar]
- Ripani E, Sacchetti A, Corda D, Alberti S. Human Trop-2 is a tumor-associated calcium signal transducer. Int J Cancer. 1998;76:671–676. doi: 10.1002/(sici)1097-0215(19980529)76:5<671::aid-ijc10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Rosenzweig SA, Atreya HS. Defining the pathway to insulin-like growth factor system targeting in cancer. Biochem Pharmacol. 2010;80:1115–1124. doi: 10.1016/j.bcp.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shames DS, Girard L, Gao B, Sato M, Lewis CM, Shivapurkar N, Jiang A, Perou CM, Kim YH, Pollack JR, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006;3:e86. doi: 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- Sozo F, Wallace MJ, Zahra VA, Filby CE, Hooper SB. Gene expression profiling during increased fetal lung expansion identifies genes likely to regulate development of the distal airways. Physiol Genomics. 2006;24:105–113. doi: 10.1152/physiolgenomics.00148.2005. [DOI] [PubMed] [Google Scholar]
- Sulman EP, Dumanski JP, White PS, Zhao H, Maris JM, Mathiesen T, Bruder C, Cnaan A, Brodeur GM. Identification of a consistent region of allelic loss on 1p32 in meningiomas: correlation with increased morbidity. Cancer Res. 1998;58:3226–3230. [PubMed] [Google Scholar]
- Terrinoni A, Dell'Arciprete R, Fornaro M, Stella M, Alberti S. Cyclin D1 gene contains a cryptic promoter that is functional in human cancer cells. Genes Chromosomes Cancer. 2001;31:209–220. doi: 10.1002/gcc.1137. [DOI] [PubMed] [Google Scholar]
- Tsai MF, Wang CC, Chang GC, Chen CY, Chen HY, Cheng CL, Yang YP, Wu CY, Shih FY, Liu CC, et al. A new tumor suppressor DnaJ-like heat shock protein, HLJ1, and survival of patients with non-small-cell lung carcinoma. J Natl Cancer Inst. 2006;98:825–838. doi: 10.1093/jnci/djj229. [DOI] [PubMed] [Google Scholar]
- Tseng RC, Chang JW, Hsien FJ, Chang YH, Hsiao CF, Chen JT, Chen CY, Jou YS, Wang YC. Genomewide loss of heterozygosity and its clinical associations in non small cell lung cancer. Int J Cancer. 2005;117:241–247. doi: 10.1002/ijc.21178. [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Kurahashi H, Tanaka T, Nishida K, Shimomura Y, Tano Y, Nakamura Y. Identification of the gene responsible for gelatinous drop-like corneal dystrophy. Nat Genet. 1999;21:420–423. doi: 10.1038/7759. [DOI] [PubMed] [Google Scholar]
- Wang J, Day R, Dong Y, Weintraub SJ, Michel L. Identification of Trop-2 as an oncogene and an attractive therapeutic target in colon cancers. Mol Cancer Ther. 2008;7:280–285. doi: 10.1158/1535-7163.MCT-07-2003. [DOI] [PubMed] [Google Scholar]
- Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, Yuan A, Lin CW, Yang SC, Chan WK, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of slug. Nat Cell Biol. 2009;11:694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang K, Grabowska D, Li A, Dong Y, Day R, Humphrey P, Lewis J, Kladney RD, Arbeit JM, et al. Loss of trop2 promotes carcinogenesis and features of epithelial to mesenchymal transition in squamous cell carcinoma. Mol Cancer Res. 2011;9:1686–1695. doi: 10.1158/1541-7786.MCR-11-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Nakamura M, Nakamura Y, Yoshimura G, Suzuma T, Umemura T, Shimizu Y, Mori I, Sakurai T, Kakudo K. Two-hit inactivation of FHIT by loss of heterozygosity and hypermethylation in breast cancer. Clin Cancer Res. 2002;8:2890–2893. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.