Abstract
Caulobacter crescentus has a dimorphic life cycle composed of a motile stage and a sessile stage. In the sessile stage, C. crescentus is often found tightly attached to a surface through its adhesive holdfast. In this study, we examined the contribution of growth and external structures to the attachment of C. crescentus to abiotic surfaces. We show that the holdfast is essential but not sufficient for optimal attachment. Rather, adhesion in C. crescentus is a complex developmental process. We found that the attachment of C. crescentus to surfaces is cell cycle regulated and that growth or energy or both are essential for this process. The initial stage of attachment occurs in swarmer cells and is facilitated by flagellar motility and pili. Our results suggest that strong attachment is mediated by the synthesis of a holdfast as the swarmer cell differentiates into a stalked cell.
Aquatic bacteria can live in both planktonic and sessile states. In the planktonic state, a bacterium is free to move throughout the environment to find nutrients. However, in nature, bacteria are predominantly found in the sessile state attached to surfaces where they form communities called biofilms (14, 25). Biofilms are defined as matrix-enclosed bacterial populations adherent to each other and/or to surfaces or interfaces (7). Bacteria in a biofilm are more resistant to antibiotics and are able to form symbiotic relationships with other members of the biofilm community (4, 6, 22, 46).
The transition from a planktonic state to a sessile state is a developmental process involving different environmental cues and the coordination of various molecular pathways and extra-cellular structures (9, 25, 33). There are three stages of biofilm formation, early attachment, maturation, and detachment. Initially, bacteria are found swimming close to a surface until they are able to overcome surface tension and bind, forming a monolayer biofilm. This monolayer biofilm eventually becomes tightly packed with additional cells, and microcolonies begin to form. During the maturation stage, a three-dimensional structure emerges that is made up of a matrix of exopolysaccharide and cells. The last stage is that of detachment, where planktonic cells are released from the biofilm (25, 43).
Limited studies on a small number of γ-proteobacteria have been conducted to determine the genetic pathways and structures that play a role in biofilm development. Three structures show interspecies importance: pili, flagella, and exopolysaccharides. Flagella and type IV pili facilitate early attachment events in Pseudomonas aeruginosa (26) and Vibrio cholerae El Tor (44). Exopolysaccharides play a role in the initial stages of biofilm development in V. cholerae El Tor (44) as well as V. cholerae O139 (45), and contribute to biofilm maturation in Escherichia coli (8). Although there are overlaps in the requirements for adhesion to surfaces, some requirements are not common to all bacteria. For example, unlike the bacteria discussed above, V. cholerae O139 does not require type IV pili for initial attachment (45).
Caulobacter crescentus is a gram-negative α-purple bacterium commonly found in aquatic environments; it participates in forming biofilms that have a biofouling effect on a variety of surfaces (47). C. crescentus has a dimorphic life cycle, spending part of its life as a nonreplicating motile swarmer cell and part as a replicating sessile stalked cell (2). Each cell type has specific polar structures that predispose the cell to its distinct lifestyle. The swarmer cell is characterized by the presence of a flagellum and multiple pili at the swarmer pole. Eventually, the swarmer cell differentiates into a stalked cell, and the pili and flagellum are replaced with a stalk that is tipped by an adhesive organelle called a holdfast.
Little is known about the physical mechanism that results in stable attachment of C. crescentus to surfaces. The holdfast, composed in part of a polysaccharide containing N-acetylglucosamine, is essential for adhesion (20). All of the mutants identified so far that are completely deficient in surface adhesion lack detectable N-acetylglucosamine at the tip of the stalk (5, 21, 40, 41); this indicates that the holdfast N-acetylglucosamine plays a critical role in adhesion. Even though the holdfast N-acetylglucosamine cannot be detected in swarmer cells (15), swarmer cells can attach to surfaces (15, 23, 30). Therefore, other structures are likely to facilitate the attachment of swarmer cells. It has been suggested that the pilus may be one such structure (15).
In order to determine whether other polar structures besides the holdfast are necessary for attachment, we examined the attachment of mutant strains lacking pili or flagella. In this paper, we show that the presence of a holdfast is essential but not sufficient for the attachment of C. crescentus cells to surfaces. Flagella and pili facilitate attachment of cells to surfaces, attachment is cell cycle regulated, and it requires growth or energy or both. These results suggest that optimal attachment to surfaces involves an ordered series of developmental events during the C. crescentus cell cycle.
MATERIALS AND METHODS
Medium and strains.
Strains of C. crescentus were grown in peptone yeast extract (PYE) or 0.2 mM phosphate Hutner imidazole glucose glutamate medium (HIGG) at 30°C, unless otherwise specified (31). When required, antibiotics were used at the following concentrations: kanamycin, 5 or 20 μg/ml; and nalidixic acid, 20 μg/ml. E. coli was grown in Luria-Bertani (LB) medium at 37°C and supplemented with kanamycin (50 μg/ml) when necessary.
The mutant YB3756 (motB) was generated by amplifying a region of motB with the primers MotBXbaI and MotBPstI (oligonucleotide sequences are available from the authors upon request), and ligating the product into the nonreplicating plasmid pBGST18 (M. R. K. Alley, unpublished) at the XbaI and PstI sites. The plasmid was transformed into E. coli S17-1 (38) and introduced into the C. crescentus strain CB15 by conjugation. Homologous recombination occurred between the plasmid and genomic DNA, generating an insertional mutation in motB. The phenotype of the mutation was confirmed by flagellum staining and a swarm assay. motB does not appear to be part of an operon, and therefore the motB mutation should not be polar.
The mutant YB3373 (cpaA) was identified among a collection of φCBK-resistant (pilus minus) Mariner (37) transposon mutants (D. Klein and Y. V. Brun, unpublished). Mutations in cpaA may be polar on the cpaBCDE genes also involved in pilus synthesis (39). Transposon locations in the cpa region were shown by Southern hybridization and sequencing to be in the cpa region. Genomic DNA from the mutants was digested with BamHI, releasing a 5.5-kb fragment in the pilA-cpa region. The probe was a 614-bp fragment including pilA and was generated with the primers pilAup and pilAdn. The probe was labeled with digoxigenin according to the DIG HIGH prime kit (Boehringer Mannheim, Indianapolis, Ind.). Hybridization was done with QuikHyb (Stratagene, La Jolla, Calif.). An antidigoxigen-alkaline phosphatase conjugate (Boehringer Mannheim) diluted 1:5,000, along with a solution of nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP), was used to develop the blot.
PCR products of the regions adjacent to the estimated transposon locations were generated for sequencing purposes with the following primers: pilAHindIII and MarRseq, pilAXbaone and MarRseq, or pilAXbaone and MarLseq. PCR products were gel purified with the Qiagen kit QIAquick. Sequencing reactions of the PCR products were done in an MJ Research PTC-150 minicycler with MarRseq or MarLseq and the ABI Big Dye kit version 2. Sequencing was performed at the Institute for Molecular and Cellular Biology located at Indiana University in Bloomington, Ind., with the ABI 3700 capillary DNA sequencer. Sequence analysis was done with the NCBI Blast program (http://www.ncbi.nlm.nih.gov/). A transducing phage lysate was made of the cpaA mutant and was used to transduce the cpaA mutation into CB15 and SC305 (flaR) (11, 16).
Coverslip assay.
A coverslip cell binding assay was adapted from reference 24. Overnight cultures were diluted with fresh PYE to an optical density at 600 nm (OD600) of 0.15, and 1.5 ml was added to each well of a 6-well polystyrene cell culture dish (Corning Costar 3506) containing ethanol-flamed glass coverslips or ethanol-washed plastic coverslips (Corning Incorporated, catalog no. 3512). Samples were placed on an orbiter rotating between 90 and 100 rpm and incubated at 30°C for 1.5 to 4.5 h or overnight. After incubation, coverslips were removed from the wells and rinsed with a steady stream of deionized distilled water for 10 to 20 s. Coverslips were then placed on a slide containing 2 μl of SlowFade antifade reagent in 1× phosphate-buffered saline and glycerol (catalog S-2828, Molecular Probes, Eugene, Oreg.). At this time, the culture density of each sample was measured at 600 nm.
If the experiment required holdfast labeling, instead of being placed on a slide, coverslips were put in a darkened humid chamber cell side up and incubated with 50 μl of fluorescein isothiocyanate-labeled wheat germ agglutinin (FITC-wheat germ agglutinin) (Molecular Probes, Eugene, Oreg.) at a concentration of 0.05 μg/μl for 30 min. Coverslips were again rinsed with a steady stream of distilled H2O, and placed onto a slide.
Growth requirements for surface adhesion.
Overnight cell cultures were diluted to an OD600 of 0.40 in PYE with a final concentration of 2 μg of chloramphenicol/ml, 0.05% (wt/vol) sodium azide, 1.7% (wt/vol) formaldehyde, or no additives. The diluted cultures were incubated for 30 min at 30°C or placed in an ice slurry. The tissue culture dish was chilled at 4°C for 30 min for the change in temperature experiment. Coverslips and cell culture were added to each well and incubated for 4 h as described above.
To determine the effect of chemicals on cells already attached to glass coverslips, cells were grown in PYE with a coverslip for 5.5 h with a starting OD600 of 0.15. Chloramphenicol, sodium azide, or formaldehyde was added at the same concentrations as above and incubation at 30°C was continued overnight.
Microscopy.
Slides were examined with a Nikon Eclipse E800 microscope with a fluorescein isothiocyanate-HYQ filter (Ex 480/40 DM 505 BA 535/50) with a 40× objective or a 100× oil immersion objective. Images were captured with a Princeton Instruments cooled charge-coupled device camera and the software Metamorph Imaging versions 4.0 and 4.5.
Plastic binding assay.
Plastic binding assays were done as described (28) with modifications (C. Fuqua, personal communication). Briefly, overnight cultures were diluted with fresh PYE to an OD600 of 0.15 and incubated at 30°C until cultures reached an OD600 between 0.35 and 0.60. Cultures were diluted to an OD600 of 0.30 and added to the wells of a 12-well tissue culture dish (750 μl per well, four wells per strain). Dishes were incubated with shaking at room temperature for 15 min. The cell culture was removed, and the wells were washed twice with fresh PYE to remove any unattached cells. Each well was stained with 400 μl of 1% (wt/vol) crystal violet in H2O for 15 min. Wells were washed four times with H2O to remove excess crystal violet. After washing, the crystal violet was sequentially eluted with 200 μl and 400 μl of methanol. The samples were diluted with the addition of 400 μl of H2O. Color intensity was measured at 589 nm.
Synchrony.
Cells were synchronized by taking advantage of the fact that swarmer cells pellet more tightly than stalked cells, so that relatively pure cultures of swarmer cells can be obtained by centrifugation followed by the removal of loosely pelleted cells. A culture of C. crescentus ATCC 19089 (American Type Culture Collection, Manassas, Va.) (lab strain YB1360) was grown in HIGG at 30°C. The saturated culture was aliquoted into Corex tubes (20-ml volume) and centrifuged in a Beckman JA-20 rotor for 10 min at 9,000 rpm. The supernatant of each tube was then poured off until a 5- to 10-ml volume remained. The tubes were swirled gently by hand to resuspend the loose stalk pellet but not the tight swarmer pellet. The supernatant was poured off, and 5 ml of 1× M2 salts (29) was added to each tube. Any remaining stalk pellet was removed by gentle swirling.
The supernatant was poured off, and the swarmer cell pellet from each tube was resuspended in 1 ml of 1× M2 salts. The swarmer cells from five tubes were combined into one tube and centrifuged as described above. The loose stalk pellet was removed as described above. Resuspended swarmer cells were then examined for the presence of stalked cells. If the population of stalked cells exceeded 5%, the washing and centrifugation steps were repeated until the population of stalked cells diminished to below 5%. Once the population of swarmer cells was 95% pure, swarmer cells were resuspended in fresh medium to an OD600 of 0.30. Swarmer cells were allowed to recover at 30°C for 1 min with shaking and samples were taken for the assays described below.
The plastic binding assay was carried out as described above with two modifications. Cells were incubated for 20 min in a 12-well tissue culture dish instead of 15 min. Following the washes with fresh medium, cold 1× M2 salts were added to each well, and the tissue culture dish was stored at 4°C until the synchrony was completed.
Samples were taken at 0, 34, 60, 146, 180, 210, and 240 min for phage absorption assays, which were carried out as described (13). For immunoblots, 1-ml samples were taken at each time point, and centrifuged at 6,000 rpm in a microcentrifuge for 5 min. Cell pellets were then resuspended in 100 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading dye, boiled for 5 min, and stored at −20°C. An equivalent volume of cell lysate from each synchrony time point was loaded onto a 10% acrylamide gel and transferred onto nitrocellulose. The nitrocellulose was incubated for 2 h with an antiflagellin antibody at 1:40,000 or overnight with an anti-FtsZ antibody at 1:20,000. Blots were incubated for 1 h with the horseradish peroxidase-labeled secondary antibody, goat anti-rabbit immunoglobulin (InVitrogen Life Technologies, Carlsbad, Calif.) diluted 1:20,000 and preabsorbed with CB15 acetone powder (18). Blots were developed with the chemiluminescent substrate SuperSignal West Pico (Pierce Biotechnology, Inc., Rockford, Ill.) for 5 min.
Swarm and lectin binding assays.
We stabbed 0.5 μl of log-phase cells into 0.3% semisolid PYE medium and grew them at room temperature in a humid chamber for 3 to 5 days. Fluorescent lectin binding assays were performed as described (15) with fluorescein isothiocyanate-wheat germ agglutinin, except that cells were washed twice.
RESULTS
Presence of a holdfast is not sufficient for efficient attachment of C. crescentus to surfaces.
Although it has been shown that the holdfast is essential for attachment of C. crescentus to surfaces (21), it may not be the only requirement. In other bacteria, attachment has been shown to be a multistep process, requiring multiple structures such as pili, flagella, and exopolysaccharides (25). To determine if the presence of a holdfast is sufficient for C. crescentus attachment, we tested the ability of nongrowing C. crescentus cells to attach to glass. Cells were incubated at 30°C in PYE with chloramphenicol, sodium azide, or formaldehyde, or in PYE at 4°C to inhibit growth or to kill cells. We compared the ability of treated cells to attach to glass coverslips to that of untreated cells.
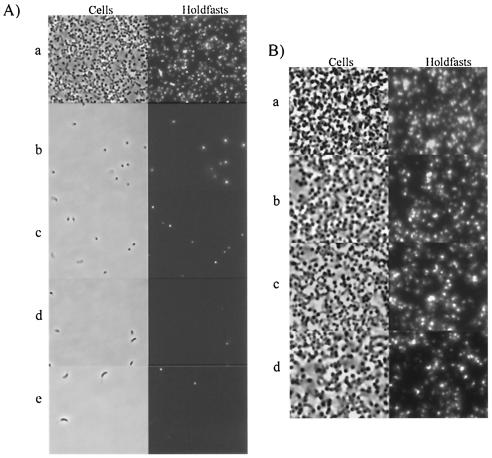
As shown in Fig. 1A, attachment to glass was severely affected by all four conditions. Under these conditions, approximately 75% of cells are stalked cells (42) and have a holdfast (15). These holdfast-bearing cells would be expected to randomly come into contact with the glass during the 5 h of incubation and should bind to glass if the holdfast is sufficient for attachment. In order to ensure that the lack of attachment was not due to a treatment-induced detachment of cells, cells already attached to glass were incubated with formaldehyde and sodium azide. The results in Fig. 1B show that the majority of cells remained bound after incubation with formaldehyde and sodium azide, suggesting that growth inhibition or cell death does not cause cells to detach. These results indicate that the presence of a holdfast is not sufficient for the efficient attachment of C. crescentus to a surface. One possibility to explain these results is that energy and/or growth is required for attachment.
FIG. 1.
Binding assays comparing cell attachment to and detachment from glass without growth and energy. (A) A binding assay was used to compare the effect of chemicals and temperature on the attachment of C. crescentus to glass coverslips. Cells were incubated under the following conditions: (a) no chemicals added; (b) chloramphenicol, 2 μg/ml; (c) sodium azide, 0.05% (wt/vol); (d) formaldehyde, 1.7% (wt/vol); (e) no chemicals added, 4°C. (B) A binding assay was used to determine the effect of 0.05% sodium azide and 1.7% formaldehyde on cells already attached to glass coverslips. Glass coverslips were exposed to exponentially growing cell cultures for 5.5 h starting at an OD600 of 0.15. At this time sodium azide and formaldehyde were added, and incubation was continued for the indicated time. (a) No chemicals; overnight incubation. (b) No chemicals and incubation for 5.5 h. (c) Sodium azide at 0.05% (wt/vol) and overnight incubation. (d) Formaldehyde at 1.7% (wt/vol) and overnight incubation.
Pili and flagellar motility are required for optimal attachment.
Since the holdfast is necessary but not sufficient for binding to a surface, it is likely that events preceding holdfast synthesis are required for the primary binding event. The fact that swarmer cells can bind to glass (23, 30) suggested the possibility that the flagellum and/or the pili could contribute to the primary binding stages. To analyze the contribution of various structures to attachment, we used a plastic binding assay (28) as a semiquantitative measure of the attachment of various mutants.
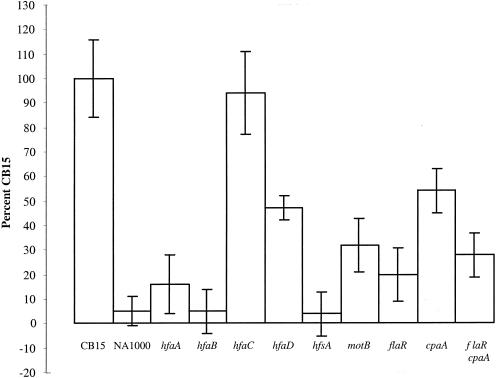
To validate this assay, we compared mutants whose relative binding efficiencies had already been evaluated with cellulose acetate and glass binding assays (5). We used six holdfast mutants with different degrees of attachment defects. NA1000 (12) and the YB3372 holdfast synthesis mutant (hfsA) (41) cannot synthesize a holdfast and are incapable of attachment according to cellulose acetate and glass binding assays (41). The results of the plastic binding assay confirmed that both strains were essentially unable to attach to surfaces; binding efficiencies were 4% for YB3372 (hfsA) and 5% for NA1000 compared to wild-type strain CB15 (100%) (Fig. 2).
FIG. 2.
Quantification of holdfast flagella and pilus mutants attached to polystyrene. Exponentially growing cells were allowed to attach to polystyrene wells, and the wells were washed and stained with crystal violet. The bound crystal violet was solubilized with methanol, and color intensity was measured at 589 nm.
Four of the mutants have insertional mutations located in the holdfast attachment hfa operon, YB2533 (hfaA), YB2536 (hfaB), YB2539 (hfaC), and YB2542 (hfaD) (5). The hfa genes play a role in securing the holdfast to the tip of the stalk (5, 17), and with the exception of YB2539 (hfaC), these mutants display a holdfast shedding phenotype (5). Of the three mutants, YB2536 (hfaB) has the most severe attachment defect, showing negligible binding to glass and cellulose acetate (5). The plastic binding assay indicated that YB2536 (hfaB) had a 5% binding efficiency compared to CB15, which is similar to YB3372 (hfsA) and NA1000 (Fig. 2). The mutant YB2533 (hfaA) bound more efficiently than YB2536 (hfaB) to glass and cellulose acetate (5). The plastic binding assay indicated that YB2533 (hfaA) had a 16% binding efficiency compared to wild-type CB15 (Fig. 2). Both the cellulose acetate and glass binding assays showed that although YB2542 (hfaD) was not capable of binding as well as CB15, it clearly bound more efficiently than YB2533 (hfaA) and YB2536 (hfaB) (5). Cultures of the hfaD mutant have the highest percentage of cells with holdfasts of the hfa mutants (5). The plastic binding assay showed that the YB2542 (hfaD) mutant had a binding efficiency of 47% compared to CB15 (Fig. 2). The mutant YB2539 (hfaC) consistently bound as well as CB15 in all three assays (5). Thus, the binding patterns of the holdfast mutants determined by the plastic binding assay mirror the results of the glass and cellulose acetate binding assays (5). This indicates that the plastic binding assay is at least an equivalent means of measuring attachment in C. crescentus compared to binding to glass or cellulose acetate.
As previously indicated, pili may play a role in attachment of C. crescentus to surfaces (15, 23, 30). We obtained nonpiliated mutants by selecting transposon mutants for resistance to the phage φCbK, which requires pili for infection, and used Southern hybridization to identify mutations in the pil gene cluster. We examined the binding efficiency of a mutant with a transposon insertion in the cpaA gene. CpaA is thought to be a prepilin peptidase (39). The plastic binding assay showed that YB3373 (cpaA) has a binding efficiency of 54% compared to the wild type (Fig. 2), indicating that the presence of pili contributes to the ability of C. crescentus to attach to surfaces but that the pili are not crucial binding elements.
Since the pilus mutant could still attach substantially to the surface in the plastic binding assay, it seemed likely that flagella might contribute to attachment. Flagella are key components in the initial stage of biofilm development in E. coli (32) and Pseudomonas aeruginosa (26) and facilitate attachment in V. cholerae El Tor (44). We used SC305, which has a point mutation located in the flaR gene required for flagellar filament assembly. According to the plastic binding assay, SC305 (flaR) had a binding efficiency of 20% compared to wild-type CB15 (Fig. 2). This indicates that the flagellum, while not as important as the holdfast for binding to surfaces, is more important than pili.
The flagellum could be playing a role through direct adherence to surfaces by acting as an adhesin, by breaking the electrostatic repulsion barrier at the surface, or perhaps by positioning the cell so that it is in the correct orientation for binding. Another possibility to explain the strong binding defect of the mutant SC305 (flaR) is that the flagellar mutation could reduce the efficiency of pilus biosynthesis. Indeed, flagellar mutants have a decreased rate of φCbK absorption, suggesting that normal flagellum synthesis is required for optimal pilus synthesis or function (1). SC305 (flaR) appears to have fewer pili than wild-type cells because the plaque-forming efficiency of φCbK was 2.3-fold less in SC305 than in wild-type CB15. Therefore, we examined the attachment ability of a double mutant, YB3757 (flaR cpaA). This double mutant had a binding efficiency of 28%, which is comparable to the binding efficiency of the SC305 (flaR) mutant (Fig. 2). The lack of an additive effect between the flagellar and pilus mutations suggests that flagella and pili act in the same attachment pathway and that part of the attachment defect of SC305 (flaR) is due to a deficiency in pilus synthesis.
The effect of a flagellar mutation on pilus synthesis is not sufficient to explain the strong adhesion defect of a flagellar mutant. The motility imparted by the flagellum may also be important for attachment. In order to test this possibility, a motB mutant, YB3756, was generated by insertional inactivation. As detected by flagellar staining and a swarm assay, YB3756 (motB) has a flagellum but lacks the ability to swim (data not shown). The results of the plastic binding assay showed that YB3756 (motB) has a binding efficiency of 32% compared to wild-type CB15 (Fig. 2). This result is comparable to the flaR mutant SC305, which suggests that it is not the presence of a flagellum per se that is necessary for attachment but rather it is the motility imparted by the flagellum. It is also possible that attachment relies on chemotactic ability.
A fluorescent lectin binding assay was done to ensure that the decrease in surface attachment observed in the flagellar and pilus mutants was not due to the absence of a holdfast. Fluorescent lectin binding observed for the mutant was equivalent to that seen in wild-type cells; this eliminates the possibility that any of these mutations have an indirect effect on holdfast synthesis (data not shown).
Time course attachment assays.
The plastic binding assay involves an attachment period of 15 min and thus provides a measure of primary adhesion. To observe the effect of mutations on the later stages of biofilm formation, we analyzed the attachment of cells to plastic coverslips during extended incubations. Coverslips were removed at each time point and washed extensively with water to remove loosely bound cells. Assays were repeated three times, and the different strains were consistently found to bind as described below.
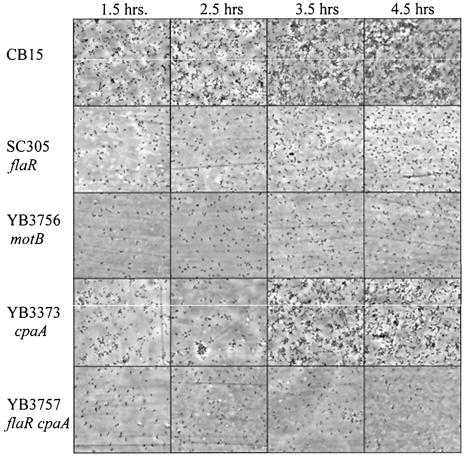
Figure 3 shows the binding patterns observed during 4.5 h of incubation. Wild-type cells initially bound to the surface as a dispersed monolayer of cells, as seen at 1.5-h. A few small aggregates of cells were also seen at that time. These small aggregates are probably not due to rosettes (groups of cells attached to each other holdfast-to-holdfast that form in liquid culture) since flagellar mutants, which make rosettes in liquid culture, do not form these aggregates (see below). The aggregates are most likely due to the binding of new cells from the culture, since the growth rate of attached cells (greater than or equal to 1.5-h doubling time) is not sufficient to explain their formation. By 2.5 to 3.5 -h, wild-type cells began to form larger aggregates, which continued to increase in size until the coverslip was covered by a high density of cells, as previously described (5, 40, 41).
FIG. 3.
Microscopic analysis comparing attachment patterns of wild-type cells and flagellar and pilus mutants over the course of 4.5 h. Overnight cell cultures were diluted to an OD600 of 0.15 and placed in the wells of a tissue culture dish containing plastic coverslips. Incubation proceeded at 30°C for 4.5 h with shaking. Beginning at 1.5 h, plastic coverslips were examined at 1-h intervals with a Nikon Eclipse E800 microscope (40×).
Although we established that flagellar motility contributes significantly to surface attachment in C. crescentus, the temporal contribution of flagella remained unclear. Microscopic analysis of SC305 (flaR), YB3757 (flaR cpaA), and YB3756 (motB) throughout the attachment time course showed that very few cells were able to bind compared to wild-type CB15 and that they never formed aggregates. YB3756 (motB) and SC305 (flaR) had very similar binding patterns and after 4.5 h exhibited less binding than wild-type CB15 after 1.5 h. YB3757 (flaR cpaA) exhibited the lowest binding of all the mutants tested. The pilus mutant YB3373 (cpaA) had a binding pattern similar to wild-type cells, starting with a dispersed monolayer of cells followed by some aggregation, but lagged slightly behind for the first 4.5 h. Note that the sudden increase in attached cells between 2.5 and 3.5 h for the cpaA mutant was not reproducibly observed in other experiments. When cells were allowed to bind to coverslips overnight, the cpaA mutant was unable to form the dense monolayer biofilms formed by wild-type cells (data not shown). These results suggest that flagellar motility and pili play a role in the primary attachment of C. crescentus to surfaces and that motility is important for the formation of aggregates in the subsequent stage of adhesion. In addition, pili seem to contribute to the binding process at a late stage.
Attachment occurs at a specific stage of the cell cycle.
The developmental cycle of C. crescentus involves the ordered appearance and disappearance of various polar structures. The flagellum is synthesized in predivisional cells, and this is followed by the production of pili at the same pole in swarmer cells. The differentiation of a swarmer cell into a stalked cell is marked by the ejection of the flagellum and the retraction of the pili; this is coordinated with the synthesis of a stalk and holdfast (2). Since primary adhesion is facilitated by the flagellum and pili and stable attachment is cemented by the holdfast, perhaps optimal attachment is dependent upon the completion of multiple ordered steps.
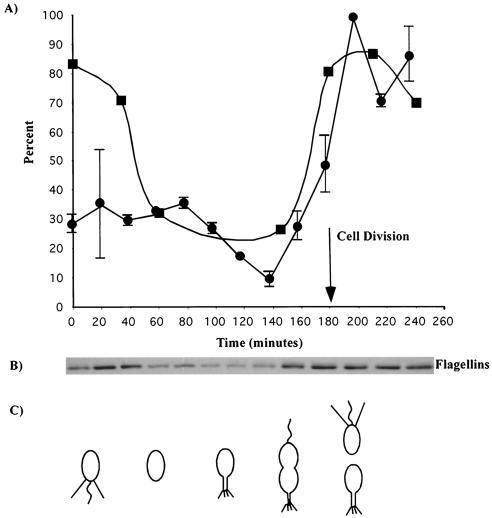
To determine the ability of various cell types to attach to surfaces, attachment was measured throughout the cell cycle of a synchronized population of C. crescentus. At each time point indicated, cells were allowed to bind to a cell culture dish for 20 min and attachment was measured with the plastic binding assay. Results are shown as a percentage of the maximum attachment and represent the average of two experiments (Fig. 4A). Attachment was relatively constant during the first 80 min of the cell cycle, with approximately 30% of the maximal attachment. This time period included the initial swarmer phase and the differentiation of swarmer cells into stalked cells. The time period between 80 and 140 min showed a marked decrease in attachment, with the lowest relative attachment of approximately 10% at 140 min. During this portion of the cell cycle, cells elongated and began to divide. The lowest point of attachment at 140 min correlated with the presence of late predivisional cells. Swarmer cells began to reappear at 160 min and steadily increased in number between 180 and 200 min. The reappearance of swarmer cells paralleled an increase in attachment, with the highest attachment occurring at 200 min.
FIG. 4.
Surface attachment throughout the cell cycle of a synchronized population of C. crescentus. (A) The crystal violet assay of bound cells is represented by circles (averages of two assays presented as a percentage of the maximum cell attachment). A phage absorbance assay measuring the binding of phage φCbK was used to measure the presence of pili throughout the cell cycle (squares). (B) Immunoblot of flagellins throughout the cell cycle. (C) Diagram of the cell cycle as observed microscopically throughout the synchrony.
An FtsZ immunoblot was used to establish the time of swarmer to stalked cell differentiation and of cell division (data not shown). FtsZ controls the initiation of cell division, is absent from swarmer cells, appears during swarmer cell differentiation, is expressed at its highest level as cell division begins, and is degraded when cells are at the end of the division process (34, 35). FtsZ appeared at 60 min, just prior to the decrease in the surface binding of cells, confirming that surface binding decreased after swarmer cell differentiation. The FtsZ level was highest between 140 and 160 min, which corresponded to the appearance of deeply constricted predivisional cells. An obvious decrease in the FtsZ level occurred at 180 min, which was marked microscopically by the appearance of numerous swarmer cells (data not shown). These results confirm that cell division took place between 160 and 180 min, coincident with the increase in surface adhesion.
Since the number of swarmer cells at time zero should be equivalent to the number of swarmer cells after cell division, we expected to see equivalent attachment at both stages. However, swarmer cells at the beginning of the cell cycle exhibited only one-third of the attachment observed after cell division. A possible explanation for this discrepancy is that the process of synchronization may have caused damage to flagella and/or pili, both of which play a role in attachment. In order to determine if the synchronization process affected the presence of pili, we performed a phage binding assay. The amount of φCbK absorbed by cells at different stages of the cell cycle is indicative of the level of piliation. According to the phage assay, pili were present in large amounts during the initial swarmer phase and during the reappearance of swarmer cells and the increase in surface adhesion between 160 and 200 min (Fig. 4A). Therefore, it appears that the lack of attachment seen at the beginning of the cell cycle is not due to a lack of pili.
Another possible explanation for the lack of binding by synchronized swarmer cells is that flagella might have been damaged and motility reduced during the synchrony process. Indeed, it is common to observe a loss of motility after the synchronization process. An antiflagellin antibody was used to assay for the presence of the flagellum throughout the cell cycle (Fig. 4B). The immunoblot revealed that the level of flagellin was lower at time zero compared to the end of the cell cycle. This correlated with microscopic observations that indicated that the cells were less motile at 0 to 40 min compared to 160 to 200 min (data not shown). This decrease in flagellin level and motility is probably due to a side effect of the synchronization process, which may cause flagella to shear off of the swarmer cells. We hypothesize that the reduced ability of swarmer cells to bind to surfaces at the beginning of the synchrony compared to after cell division is due to flagellar defects. Therefore, it appears that attachment is cell cycle regulated and is coordinated with appearance of swarmer cells and the differentiation of swarmer cells into stalked cells.
The possibility remained that cells attached at the predivisional stage. In order to test this possibility, exponentially growing cells from a mixed culture were allowed to attach to a coverslip for 10 min, and the attached cell types were quantitated. Swarmer cells accounted for 77% of the attached cells, 20% were stalked cells, and only 3% were predivisional cells (Table 1). The attached stalked cells had short stalks, suggesting that they were newborn stalked cells (data not shown). Attached cells were also quantitated after 4 h of attachment. In this case, predivisional cells predominated with 36%, and the percentage of swarmer cells dropped to 34% (Table 1). Furthermore, the stalks of attached cells were slightly longer than after 10 min (data not shown). This is expected because swarmer cells that attached early in the experiment would have differentiated into stalked and predivisional cells. These results clearly indicate that the strong increase in attachment around the time of cell division in the synchrony experiment was not due to the binding of predivisional cells or stalked cells and thus support the model that attachment is initiated by swarmer cells.
TABLE 1.
Quantitation of attachment by cell type from a mixed culture
| Attachment time (min) | No. of cells counted | % of cells
|
||
|---|---|---|---|---|
| Swarmer | Stalked | Predivisional | ||
| 10 | 223 | 77 | 20 | 3 |
| 240 | 109 | 34 | 30 | 36 |
DISCUSSION
The life cycle of C. crescentus involves the ordered synthesis of various polar structures, including the flagellum, the pili, and the holdfast. The holdfast is critical for attachment of C. crescentus cells to surfaces (3). Pili and flagella have been shown to be involved in the attachment of other types of bacteria (26, 27, 32, 44, 45). In this paper, we determined the relative contribution of these polar structures to surface adhesion in C. crescentus. We show that the presence of a holdfast is essential but not sufficient for attachment of C. crescentus to surfaces; optimal attachment also requires both pili and a motile flagellum. We show that growth and/or energy is essential for attachment and that attachment is cell cycle regulated. Specifically, we show that stalked cells are unable to attach efficiently and that attachment is highest following the production of swarmer cells. These results indicate that adhesion in C. crescentus is a developmental process whose initial stage occurs in swarmer cells, where it is mediated by motility, pili, and perhaps other properties, followed by cementing of the attachment by the holdfast during swarmer to stalked cell differentiation.
Attachment of C. crescentus to surfaces was severely affected by sodium azide, formaldehyde, chloramphenicol, and growth at 4°C, conditions that hinder the cell's ability to produce and use energy and to grow. Cells that were treated after their attachment to a surface still remained bound, indicating that the holdfast was able to maintain its adhesive properties. Since holdfast-bearing cells were still present during the various growth-inhibiting treatments and should undergo random collisions with surfaces, these results suggest that random collisions of holdfast-bearing cells with surfaces are not sufficient for attachment. These results correlate with the finding that showed that the initial cell attachment of Pseudomonas fluorescens was decreased by the inhibition of protein synthesis (27).
Since the presence of a holdfast is not sufficient for the efficient attachment of C. crescentus to surfaces, other structures may facilitate the initial binding events that lead to stable holdfast-to-surface interactions. The observation that swarmer cells, which lack a holdfast (15), are able to attach to glass (23, 30) suggested that flagella and pili may play a role in primary binding events. Plastic binding assays showed that a mutant lacking pili had a 54% binding efficiency compared to wild-type cells when exposed to a surface for 20 min, indicating that pili are contributing to the primary attachment process but are not essential. Microscopic examination over the course of hours revealed that the pilus mutant was deficient in early and late stages of attachment. Both the pilus mutant and wild-type cells first attached as individual cells seemingly randomly, but the accumulation of bound cells was slower in the pilus mutant. This was followed by the formation of tightly packed aggregates of cells.
The pilus mutant deviated from wild-type cells late in the attachment process; it appeared to lack the ability to go beyond the cell aggregation stage and to form the dense monolayers formed by wild-type cells. This attachment defect is similar to the type IV pilus mutants of P. aeruginosa (26). A time course attachment assay revealed that a mutant of P. aeruginosa defective in type IV pilus biosynthesis was able to form an initial monolayer similar to wild-type cells but was unable to form microcolonies or the dense cell populations seen with wild-type cells. The type IV pilus of P. aeruginosa may play a role in stabilizing initial interactions with a surface or between cells. The twitching motility imparted by type IV pili may be necessary for cells to migrate along the surface to form the microcolony cell aggregates (26). To our knowledge, twitching motility has not been observed in C. crescentus.
Bacteria usually have to overcome electrostatic repulsion with a surface in order to attach, since most bacteria and inert surfaces are negatively charged (10). During the initial stages of attachment, the C. crescentus pili may help attachment by reducing the radius of interaction between the cell and the surface. As the swarmer cell differentiates into a stalked cell, the pili retract, and a stalk and holdfast are synthesized at the pole that bore the pili (2). An initial attachment via pili may help position the cell so that a newly synthesized holdfast would be in close enough proximity to contact the surface, establishing a stronger, more permanent attachment than the one mediated by the pili (15). Pili have also been implicated in promoting the primary adhesion event in Hyphomonas spp., another prosthecate bacterium (36). Therefore, in cells lacking pili, holdfast contact with a surface would still be possible, but its efficiency would be reduced. In addition, it is possible that the pili of C. crescentus are capable of mediating twitching motility and act like the pili of P. aeruginosa to establish cell-to-cell interactions.
We found that the flagellum makes a substantial contribution to attachment, since a flagellar mutant had a binding efficiency of 20% compared to wild-type cells. This was four times higher than a holdfast mutant and 2.5 times less than a pilus mutant. While the flagellum is similar to a pilus in that it is a thin structure that can help break the electrostatic repulsion at a surface, it clearly has additional roles. Indeed, a motB mutant, which synthesizes a paralyzed flagellum, had a 32% binding efficiency compared to wild-type cells, slightly more than the 20% of the flagellar mutant. Therefore, we conclude that the force of motility, the ability to chemotax, or both is more important for efficient attachment than the role of the flagellum in breaking the electrostatic repulsion with the surface.
Motility plays an important role in binding of V. cholerae O139 (45), E. coli (32), and P. aeruginosa (26). Motility may also play a role in biofilm expansion (32). The direction of flagellum rotation can affect attachment and detachment of E. coli cells (19). It has been hypothesized that counterclockwise rotation may play an important role in cell detachment and that a clockwise rotation may play a role in anchoring cells to a surface. Since flagellar rotation is required for efficient binding of C. crescentus to surfaces and the proton motive force is needed for flagellar rotation, the lack of binding that we observed with nongrowing cells may in part be due to the lack of flagellar rotation.
We hypothesize that the lack of adhesion in nongrowing cells reflects the requirement for an ordered succession of structures at the adhesive pole of the cell. The lack of an additive effect between flagellum and pilus mutations suggests that flagella and pili are acting in the same attachment pathway. Furthermore, analysis of a synchronized population indicated that attachment correlated with the production of pilus-bearing swarmer cells. The elongating stalked cell and predivisional cell phases of the cell cycle exhibited the lowest attachment, confirming that holdfasts are not sufficient for attachment to a surface. The importance of flagellar motility for the attachment of swarmer cells was also indicated by the results of the cell synchrony experiment. Immunoblot analysis indicated that the synchronization procedure resulted in a reduction of flagellins, suggesting that flagella were broken off. When cells were examined by microscopy, they appeared to be less motile at the beginning of the synchrony than after cell division, correlating with the respective levels of adhesion at each step (data not shown).
Our results suggest a model for the succession of events that allow C. crescentus cells to bind efficiently to surfaces. Since binding correlates with the appearance of swarmer cells and since the pili and flagellar motility are required for efficient attachment, we hypothesize that the primary binding event occurs in swarmer cells. Indeed, when cells from a mixed culture were exposed to a surface for only 10 min, the vast majority of attached cells (77%) were swarmer cells. Motility is likely to increase the occurrence of contacts between the cell and a surface. In addition, motility probably provides the force that is necessary to overcome the repulsive barrier between the negatively charged surface and the negatively charged cell surface. The presence of thin polar structures such as the pili and flagellum may also help break this repulsive barrier.
After the initial association with the surface, the retraction of bound pili during swarmer to stalked cell differentiation would bring the pole at which the holdfast is about to be synthesized in close proximity to the surface. This would help ensure that cell orientation is optimal for the subsequent cementing of the adhesion by the holdfast. Export of the holdfast directly following flagellum ejection and pili retraction would then cement the permanent attachment to the surface. This succession of events explains why adhesion depends on growth; growth is required for the transition from the primary adhesion swarmer stage to the permanent adhesion stalked stage. Dissection of the mechanisms that coordinate motility, pilus retraction, flagellum ejection, and holdfast synthesis should provide major insight into the developmental process of biofilm formation.
Acknowledgments
We thank members of our laboratory and Clay Fuqua for critical reading of the manuscript and for helpful discussions.
This work was supported by National Institutes of Health grant GM51986 to Y.V.B.
REFERENCES
- 1.Bender, R. A., C. M. Refson, and E. A. O’Neill. 1989. Role of the flagellum in cell-cycle-dependent expression of bacteriophage receptor activity in Caulobacter crescentus. J. Bacteriol. 171:1035-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brun, Y., G. Marczynski, and L. Shapiro. 1994. The expression of asymmetry during cell differentiation. Annu. Rev. Biochem. 63:419-450. [DOI] [PubMed] [Google Scholar]
- 3.Brun, Y. V., and L. J. Shimkets. 2000. Prokaryotic development. American Society for Microbiology, Washington, D.C.
- 4.Ceri, H., M. E. Olson, C. Stremick, R. R. Read, D. Morck, and A. Buret. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, J., G. G. Hardy, D. Bodenmiller, E. Toh, A. Hinz, and Y. V. Brun. 2003. The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk. Mol. Microbiol. 49:1671-1683. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 8.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 10.Dunne, M. W. J. 2002. Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ely, B., R. H. Croft, and C. J. Gerardot. 1984. Genetic mapping of genes required for motility in Caulobacter crescentus. Genetics 108:523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda, A., K. Miyakawa, H. Iba, and Y. Okada. 1976. A flagellotropic bacteriophage and flagella formation in Caulobacter. Virology 71:583-592. [DOI] [PubMed] [Google Scholar]
- 14.Geesey, G. G., W. T. Richardson, H. G. Yeomans, R. T. Irvin, and J. W. Costerton. 1977. Microscopic examination of natural sessile bacterial populations from an alpine stream. Can. J. Microbiol. 23:1733-1736. [DOI] [PubMed] [Google Scholar]
- 15.Janakiraman, R. S., and Y. V. Brun. 1999. Cell cycle control of a holdfast attachment gene in Caulobacter. J. Bacteriol. 181:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, R. C., and B. Ely. 1979. Analysis of nonmotile mutants of the dimorphic bacterium Caulobacter crescentus. J. Bacteriol. 137:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurtz, H. D., Jr., and J. Smit. 1992. Analysis of a Caulobacter crescentus gene cluster involved in attachment of the holdfast to the cell. J. Bacteriol. 174:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 19.McClaine, J. W., and R. M. Ford. 2002. Reversal of flagellar rotation is important in initial attachment of Escherichia coli to glass in a dynamic system with high- and low-ionic-strength buffers. Appl. Environ. Microbiol. 68:1280-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merker, R. I., and J. Smit. 1988. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl. Environ. Microbiol. 54:2078-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, D., and J. Smit. 1990. Identification of genes affecting production of the adhesion organelle of Caulobacter crescentus CB2. J. Bacteriol. 172:5425-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton, A. 1972. Role of transcription in the temporal control of development in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 69:447-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ong, C. J., M. L. Y. Wong, and J. Smit. 1990. Attachment of the adhesive holdfast organelle to the cellular stalk of Caulobacter crescentus. J. Bacteriol. 172:1448-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 26.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 27.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 28.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 29.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poindexter, J. S. 1981. The caulobacters: ubiquitous unusual bacteria. Microbiol. Rev. 45:123-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poindexter, J. S. 1978. Selection for nonbuoyant morphological mutants of Caulobacter crescentus. J. Bacteriol. 135:1141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 33.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quardokus, E., N. Din, and Y. V. Brun. 1996. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc. Natl. Acad. Sci. USA 93:6314-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quardokus, E. M., N. Din, and Y. V. Brun. 2001. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol. Microbiol. 39:949-959. [DOI] [PubMed] [Google Scholar]
- 36.Quintero, E. J., K. Busch, and R. M. Weiner. 1998. Spatial and temporal deposition of adhesive extracellular polysaccharide capsule and fimbriae by Hyphomonas strain MHS-3. Appl. Environ. Microbiol. 64:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon, R., U. Prieffer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 39.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19:3223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smit, J., C. S. Sherwood, and R. F. Turner. 2000. Characterization of high density monolayers of the biofilm bacterium Caulobacter crescentus: evaluating prospects for developing immobilized cell bioreactors. Can. J. Microbiol. 46:339-349. [PubMed] [Google Scholar]
- 41.Smith, C. S., A. Hinz, D. Bodenmiller, D. E. Larson, and Y. V. Brun. 2003. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J. Bacteriol. 185:1432-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tax, R. 1978. Age distribution of Caulobacter cells in an exponential population. J. Bacteriol. 135:16-17. [Google Scholar]
- 43.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfaardt, G. M., J. R. Lawrence, R. D. Robarts, S. J. Caldwell, and D. E. Caldwell. 1994. Multicellular organization in a degradative biofilm community. Appl. Environ. Microbiol. 60:434-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yun, C., B. Ely, and J. Smit. 1994. Identification of genes affecting production of the adhesive holdfast of a marine caulobacter. J. Bacteriol. 176:796-803. [DOI] [PMC free article] [PubMed] [Google Scholar]