Abstract
The variable predisposition to cachexia may, in part, be due to the interaction of host genotype. We analyzed 129 single nucleotide polymorphisms (SNPs) in 80 genes for association with cachexia based on degree of weight loss (>5, >10, >15%) as well as weight loss in the presence of systemic inflammation (C-reactive protein, >10 mg/l). 775 cancer patients were studied with a validation association study performed on an independently recruited cohort (n = 101) of cancer patients. The C allele (minor allele frequency 10.7%) of the rs6136 (SELP) SNP was found to be associated with weight loss >10% both in the discovery study (odds ratio (OR) 0.52; 95% confidence intervals (CI), 0.29–0.93; p = 0.026) and the validation study (OR 0.09, 95% CI 0.01–0.98, p = 0.035). In separate studies, induction of muscle atrophy gene expression was investigated using qPCR following either tumour-induced cachexia in rats or intra-peritoneal injection of lipopolysaccharide in mice. P-selectin was found to be significantly upregulated in muscle in both models. Identification of P-selectin as relevant in both animal models and in cachectic cancer patients supports this as a risk factor/potential mediator in cachexia.
Keywords: animal models, cachexia, inflammation, polymorphisms, P-selectin
→See accompanying article 10.1002/emmm.201200232
INTRODUCTION
Cachexia is a wasting condition that manifests itself in several life-threatening diseases, including cancer, AIDS, congestive heart failure and sepsis (Argiles et al, 2003; Tisdale, 2004). Patients exhibit a loss of both adipose tissue and lean body mass (Fearon & Preston, 1990), which is resistant to conventional nutritional support (Tisdale et al, 1987). Cachexia is typically characterized by severe weight loss, anorexia, early satiety, weakness, anaemia and oedema (Fearon & Preston, 1990). The cachectic state is particularly problematic in cancer, typified by poor prognosis and often associated with a lower response to chemotherapy and radiotherapy than might be expected (Tisdale, 2002). Patients are also more likely to report decreased quality of life (QoL) scores (Fearon et al, 2006). More than half of cancer patients suffer from cachexia, and it is responsible for death in up to 20% of cases (Tisdale, 2002). Cachexia is therefore a significant cause of morbidity and mortality in cancer patients.
Based on our current knowledge of demographic and clinical factors, we are unable to predict, for any given cohort of patients, who will develop cancer cachexia and who will not. Such variation may, in part, be due to the patient's genotype. Knowledge of genotypic variation associated with cachexia would contribute to early identification of patients at risk and allow institution of prophylactic measures. The wealth of known genetic polymorphisms in genes controlling pro/anti-inflammatory pathways, neuronal melanocortin signalling pathways and muscle and adipose tissue catabolic pathways suggest their exploitable potential as biomarkers of inter-individual predictability of developing cachexia.
We utilized a candidate gene approach to evaluate the association between genetic polymorphisms and the risks of developing cachexia in patients recruited across three centres. Patients recruited from a fourth centre were used as a validation cohort. To further corroborate the most significantly related single nucleotide polymorphism (SNP) to cancer cachexia in the gene association study, we tested the same gene for participation in the induction of the skeletal muscle atrophy gene program, either by intra-peritoneal administration of lipopolysaccharide (LPS) in mice or in a rat model of cancer cachexia (methylcholanthrene (MCA)-induced sarcoma). LPS is known to induce acutely a number of catabolic factors in sepsis, suppress anabolic factors and result in muscle atrophy (Dehoux et al, 2003; Vary et al, 1998). The MCA model is a preclinical cancer cachexia model, and is known to reliably induce loss of lean body mass (Sato et al, 2001).
RESULTS
Following the relevant quality control checks, 129 SNPs in 80 genes (Supporting Information Table S1) were available for analysis in 775 patients. The overall completion rate of genotyping was 95.6%.
The general characteristics of the study population are presented in Table 1. Average age of the patient cohort at diagnosis was 65.5 ± 11.8 years (mean ± SD). The majority of patients were diagnosed with stage III or IV cancers. Average weight loss was 6.9 ± 9.8% with a mean body mass index (BMI) of 24.9 ± 4.9 at diagnosis. Of the patients in whom C-reactive protein (CRP) levels were assessed (n = 569), 58.7% had a CRP concentration of >10 mg/l. There were no significant differences in age, stage of disease, pre-diagnosis BMI and percentage weight loss between patients with CRP measured and the entire cohort.
Table 1.
Patient demographics (main cohort). Patients were recruited from (2004 to 2008) from the NHS Lothian, UK, Cross Cancer Institute, Edmonton, Canada, and McGill University Health Centre, Montreal, Canada
| No. of patients (n = 775) | |
|---|---|
| Age (years) † | 65.5 ± 11.8 |
| Range | 27–97 |
| Sex | |
| M | 476 (61.4) |
| F | 299 (38.6) |
| Tumour type | |
| Oesophageal or gastric | 389 (50.2) |
| Pancreatic | 114 (14.7) |
| Non-small cell lung cancer | 232 (29.9) |
| Other | 40 (5.2) |
| Stage | |
| I | 38 (4.9) |
| II | 95 (12.3) |
| III | 216 (27.9) |
| IV | 392 (50.5) |
| Unknown | 34 (4.4) |
| Body mass index (kg/m2) † | 24.9 ± 4.9 |
| Range | 12.9–46.7 |
| Percentage weight loss † | 7.95 ± 8.16 |
| Range | 0–43.8 |
| C-reactive protein (mg/l)† (n = 569) | 23.0 ± 35.9 |
| CRP > 10 mg/l | 235 (41.3) |
| CRP ≤ 10 mg/l | 334 (58.7) |
Values are number of patients with percentages in parentheses unless indicated otherwise.
values are mean ± SD. Characteristics were measured at first presentation to a surgical or oncology clinic.
Table 2a lists the detailed results for SNPs significantly associated with cancer cachexia in patients classified according to weight loss alone. Table 2b lists the detailed results for SNPs significantly associated with cancer cachexia in patients classified according to weight loss with systemic inflammation (CRP >10 mg/l). In total, eight SNPs have associations of p < 0.02 with various cachexia phenotypes. Three of these SNPs are found within chromosome 1 in the genes selectin P (SELP), leptin receptor (LEPR) and deiodinase, iodothyronine, type I (DIO1); three within chromosome 3 in the genes N-acylaminoacyl-peptide hydrolase (APEH) and ghrelin (GHRL), one within chromosome 12 in the TNFRSF1A gene and one within chromosome 19 in the ICAM1 gene. SNPs found on the same chromosomal region (within 10 000 kb) were grouped together to form haplotypes. The haplotypes formed by the rs4855881 and rs2960548 SNPs in the APEH gene failed to show any significant association with weight loss.
Table 2a.
Genes with variants significantly associated with cancer cachexia in patients classified according to weight loss alone
| Weight loss >15%. Number affected: 145/775 (18.7%) | |||||
|---|---|---|---|---|---|
| Gene | SNP | Risk allele | OR (95% CI) | p-Value | Permutated p |
| SELP | rs6136 | C | 0.31 (0.14–0.72) | 0.006615 | 0.008062 |
| ICAM1 | rs281432 | G | 1.53 (1.06–2.20) | 0.02163 | 0.01652 |
| DIO1 | rs11206244 | T | 1.54 (1.06–2.24) | 0.0226 | 0.02164 |
| ADIPOR2 | rs16928751 | A | 0.53 (0.29–0.96) | 0.03521 | 0.03053 |
| APEH | rs2960548 | G | 1.48 (1.03–2.11) | 0.03384 | 0.03768 |
| Weight loss >10%. Number affected: 266/775 (34.3%) | |||||
|---|---|---|---|---|---|
| Gene | SNP | Risk allele | OR (95% CI) | p-Value | Permutated p |
| LEPR | rs1137100 | G | 0.66 (0.47–0.92) | 0.01494 | 0.013 |
| DIO1 | rs11206244 | T | 1.52 (1.09–2.11) | 0.0129 | 0.01512 |
| SELP | rs6136 | C | 0.52 (0.29–0.93) | 0.02746 | 0.02581 |
| HYLS1 | rs3088241 | C | 0.72 (0.53–0.97) | 0.02829 | 0.02709 |
| CAMK2B | rs10441113 | A | 0.73 (0.54–0.99) | 0.04096 | 0.03419 |
| Weight loss >5%. Number affected: 415/775 (53.5%) | |||||
|---|---|---|---|---|---|
| Gene | SNP | Risk allele | OR (95% CI) | p-Value | Permutated p |
| TNFRSF1A | rs4149570 | T | 1.42 (1.08–1.87) | 0.01134 | 0.01759 |
| TNFRSF1A | rs767455 | C | 0.71 (0.53–0.95) | 0.02034 | 0.02275 |
| TNFRSF1B | rs976881 | A | 0.76 (0.57–1.00) | 0.04804 | 0.04324 |
| IL18 | rs1946519 | A | 1.35 (1.02–1.79) | 0.03895 | 0.04969 |
Table 2b.
Genes with variants significantly associated with cancer cachexia in patients classified according to weight loss with systemic inflammation (CRP >10 mg/l)
| Weight loss >15% & CRP >10 mg/l. Number affected: 76/569 (13.4%) | |||||
|---|---|---|---|---|---|
| Gene | SNP | Risk allele | OR (95% CI) | p-Value | Permutated p |
| APEH | rs2960548 | G | 2.17 (1.36–3.47) | 0.001125 | 0.000997 |
| GHRL | rs42451 | T | 2.04 (1.25–3.31) | 0.004031 | 0.004058 |
| TNFRSF1A | rs4149570 | T | 1.84 (1.16–2.92) | 0.009322 | 0.01031 |
| SELP | rs6136 | C | 0.26 (0.08–0.79) | 0.01765 | 0.01103 |
| CNR1 | rs1049353 | A | 1.82 (1.08–3.06) | 0.02366 | 0.02254 |
| IRS1 | rs1025333 | A | 2.24 (1.07–4.69) | 0.03257 | 0.03183 |
| APEH | rs4855881 | C | 1.64 (1.04–2.59) | 0.03431 | 0.03191 |
| FOXO1 | rs17446593 | G | 0.49 (0.26–0.92) | 0.02704 | 0.03239 |
| ICAM1 | rs281432 | G | 1.63 (1.04–2.54) | 0.03276 | 0.03941 |
| Weight loss >10% & CRP >10 mg/l. Number affected: 123/569 (21.6%) | |||||
|---|---|---|---|---|---|
| Gene | SNP | Risk allele | OR (95% CI) | p-Value | Permutated p |
| APEH | rs2960548 | G | 1.80 (1.21–2.68) | 0.003528 | 0.003499 |
| GHRL | rs42451 | T | 1.79 (1.18–2.72) | 0.006219 | 0.00467 |
| TNFRSF1A | rs4149570 | T | 1.51 (1.04–2.18) | 0.02958 | 0.01998 |
| HYLS1 | rs3088241 | C | 0.66 (0.46–0.95) | 0.02374 | 0.02074 |
| APEH | rs4855881 | C | 1.57 (1.06–2.32) | 0.02334 | 0.02847 |
| TSC2 | rs7187438 | C | 0.64 (0.43–0.95) | 0.0265 | 0.03438 |
| TNFRSF1B | rs3397 | C | 0.67 (0.46–0.97) | 0.03527 | 0.04286 |
| Weight loss >5% & CRP >10 mg/l. Number affected: 166/569 (29.2%) | |||||
|---|---|---|---|---|---|
| Gene | SNP | Risk allele | OR (95% CI) | p-Value | Permutated p |
| APEH | rs2960548 | G | 1.67 (1.17–2.38) | 0.004924 | 0.004533 |
| APEH | rs4855881 | C | 1.56 (1.10–2.21) | 0.01321 | 0.01212 |
| TNFRSF1A | rs4149570 | T | 1.51 (1.08–2.10) | 0.01559 | 0.02074 |
| ADIPOR2 | rs16928751 | A | 0.56 (0.33–0.95) | 0.03308 | 0.02096 |
| ADIPOR2 | rs35854772 | T | 0.57 (0.33–0.97) | 0.03733 | 0.02667 |
| TNFRSF1B | rs3397 | C | 0.70 (0.50–0.98) | 0.03944 | 0.02923 |
| LTBP1 | rs817529 | G | 0.70 (0.49–0.98) | 0.03719 | 0.03791 |
| TNFRSF1A | rs767455 | C | 0.68 (0.48–0.96) | 0.02682 | 0.03846 |
Analyses of candidate gene groups based on functional similarity revealed three groups that were associated with at least one cachexia phenotype at the p < 0.05 level (Table 3).
Table 3.
Candidate gene groups associated with cancer cachexia phenotypes
| Phenotype | Candidate gene group function | Number of genes† | Number of SNPs | p-Values |
|---|---|---|---|---|
| Weight loss >10% & CRP >10 mg/l | Appetite regulation | 2 | 3 | 0.0155 |
| Glucocorticoid signalling | 4 | 9 | 0.0351 | |
| MAPK activity regulation | 7 | 14 | 0.0481 | |
| Weight loss >15% & CRP >10 mg/l | Appetite regulation | 2 | 3 | 0.008499 |
| Glucocorticoid signalling | 4 | 9 | 0.0181 | |
| MAPK activity regulation | 7 | 14 | 0.0264 |
The genes in each candidate gene group are listed in Supporting Information Table S2.
Validation study
Patient demographics of the validation cohort (n = 101) are presented in Table 4. Although, patients in the validation cohort did not have an identical distribution of cancer types as the main cohort, the distribution of BMI and weight loss remain quite similar between the two cohorts. Approximately 60% of the patients in the validation cohort had other cancer types which also had tendency to develop cachexia like prostate cancer and colorectal cancer (it is estimated that 30% of patients suffering from these cancers have a weight loss of 5% or more (Dewys et al, 1980)).
Table 4.
Patient demographics (validation cohort). Patients recruited from (2007 to 2008) from the Oncology & Palliative Medicine, Cantonal Hospital, St. Gallen, Switzerland
| No. of patients (n = 101) | |
|---|---|
| Age (years) † | 62.0 ± 11.5 |
| Range | 35–88 |
| Sex | |
| M | 60 (59.4) |
| F | 41 (40.6) |
| Tumour type | |
| Oesophageal or gastric | 18 (17.8) |
| Pancreatic | 6 (5.9) |
| Non-small cell lung cancer | 19 (18.8) |
| Other | 58 (57.4) |
| Stage | |
| I | 0 |
| II | 3 (3.0) |
| III | 2 (2.0) |
| IV | 96 (95.0) |
| Body mass index (kg/m2) † | 23.7 ± 4.3 |
| Range | 15.4–37.8 |
| Percentage weight loss † | 5.54 ± 7.91 |
| Range | 0–43.1 |
| C-reactive protein (mg/l)† (n = 95) | 75.5 ± 76.4 |
| CRP > 10 mg/l | 78 (82.1) |
| CRP ≤ 10 mg/l | 17 (17.9) |
Values are number of patients with percentages in parentheses unless indicated otherwise.
Values are mean ± SD. Characteristics were measured at first presentation to an oncology clinic.
Study subjects were genotyped for SNPs with p < 0.05 in the main study. One replication of the main study was found. The C allele of the rs6136 SNP was inversely associated with weight loss >10% in the main study (odds ratio, OR 0.52; 95% confidence intervals, 95% CI 0.29–0.93; p = 0.026) as well as in the validation study (OR 0.09, 95% CI 0.01–0.98, p = 0.035).
Changes in skeletal muscle gene expression following either intra-peritoneal injection of LPS in mice or in rats bearing the MCA sarcoma
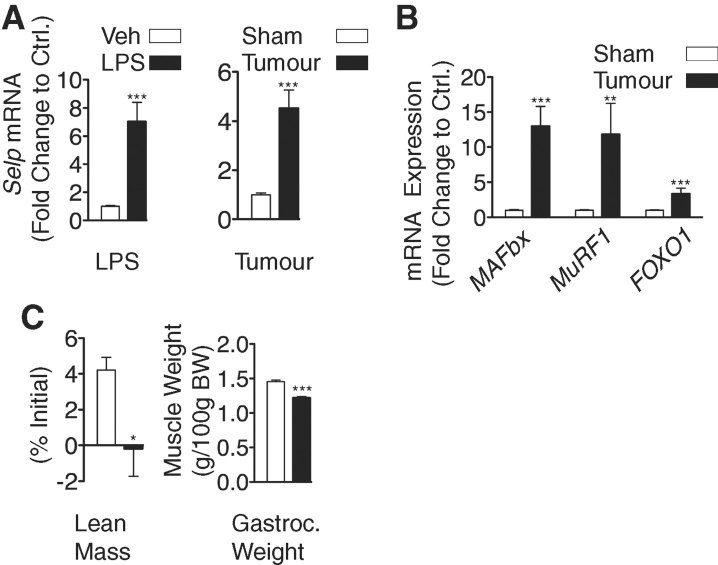
qPCR analysis of mouse skeletal muscle RNA performed after intra-peritoneal LPS injection revealed that the SELP (P-selectin) transcript was significantly differentially expressed compared with control (Fig 1a). In a separate study, rats with net loss of lean body mass and gastrocnemius mass due to growth of the MCA tumour (Fig 1c), showed similar upregulation of the SELP transcript. The latter was associated with significant upregulation of the ‘atrogen’ E3 ligases muscle atrophy F-box (MAFBx) and muscle ring finger 1 (MuRF1) along with forkhead box O1 (FOXO1), a transcription factor associated with muscle atrophy (Fig 1b).
Figure 1. Changes in skeletal muscle gene expression following either intra-peritoneal injection of LPS in mice or in rats bearing the MCA sarcoma.
Wild type mice received either intra-peritoneal injections of LPS or vehicle alone. Food was removed from the cages at the time of injection, and animals were sacrificed at 8 h after the injection (n = 6–7/group). Veh = vehicule. *Student's t-test p < 0.05, **p < 0.01, ***p < 0.001.
- Intra-peritoneal LPS treatment in mice or solid tumour growth in rats induces dynamic changes in P-selectin mRNA levels.
- In tumour-bearing rats the changes in P-selectin expression are accompanied by concomitant upregulation of the E3-ligases (MAFBx and MuRF1) and transcription factor FOXO1.
- The growth of the MCA sarcoma in rats is associated with net loss of lean body mass and muscle mass.
DISCUSSION
This study has identified that individuals who carry the C-allele of the rs6136 polymorphism in SELP gene which encodes P-selectin, are at reduced risk of developing cachexia as defined by weight loss >10%. The C allele of the non-synonymous intronic variant, rs6136 has been previously associated with decreased serum P-selectin levels (Miller et al, 2004; Volcik et al, 2006). Information on P-selectin genotypes may eventually prove useful in the risk stratification of pre-cachectic cancer patients. Further evidence for the role of P-selectin in the development of cachexia is highlighted in the studies involving the induction of muscle atrophy in mice/rats. Strikingly, P-selectin was highly upregulated following either intra-peritoneal injection of LPS or in tumour-bearing (TB) animals. Furthermore, preliminary studies also indicate that P-selectin show a similar striking upregulation (10-fold) 2 h after intra-cerebroventricular (ICV) injection of interleukin-1 beta (IL-1β) in mice (Braun et al, 2011). Acute and chronic infusion of IL-1β into the brain leads to muscle breakdown, anorexia, weight loss and negative nitrogen balance (Hill et al, 1996) and is a potential central mediator of LPS effects. Therefore, we have confirmed this gene target in three separate murine models of cachexia representing both acute and chronic inflammatory insults.
It could be argued that P-selectin expression in skeletal muscle is simply an endothelial event reflecting the presence of systemic inflammation. However, both identification of P-selectin as a top early induced gene of the mouse/rat muscle atrophy program and the significant association of the rs6136 SNP in the P-selectin gene with wasting in cancer patients provide supportive evidence for the likely involvement of P-selectin in muscle wasting. The role of P-selectin in the genesis of cachexia remains to be determined. The human P-selectin gene spans over 50 kbp on chromosome 1, containing 17 exons, almost all of which encode distinctive domain structures (Johnston et al, 1990; Watson et al, 1990). It has both membrane and soluble forms in platelets and endothelial cells (Johnston et al, 1990). Both the membrane and soluble forms of P-selectin bind to leukocytes. In certain inflammatory conditions, the plasma concentrations of soluble P-selectin is highly elevated (Dunlop et al, 1992). It is suggested, that the membrane and soluble forms of P-selectin may work co-ordinately in vivo for the regulation of their cell adhesion and, perhaps, signalling functionality. P-selectin has been characterized previously by approaches such as gene knockout or the use of specific inhibitors to be involved in the recruitment of neutrophils and macrophages in inflammatory responses (Borges et al, 1997; Chen & Geng, 2006). P-selectin may also participate in intra-tumoural regulation of the genesis of systemic inflammation via the innate immune system and/or regulation of the complex interaction within muscle between the endothelium and signalling pathways in muscle fibres (Wagenmakers et al, 2006).
CRP is a marker of systemic inflammation that has been studied in a wide variety of tumour types and has been linked to poorer survival (Mahmoud & Rivera, 2002; McMillan et al, 2003). To reflect that cachexia represents a spectrum and that the presence of systemic inflammation with weight loss may represent a unique sub-phenotype of cachexia which confers an increased mortality risk, we have chosen to study cachexia across three different percentage weight loss categories alone and with the presence of an increased CRP concentration in comparison with a weight-stable phenotype (i.e. ≤5% weight loss). Clearly, much work is required before fully validated definitions of cachexia are available. Until then, it appears reasonable to investigate cachexia based on the present definitions.
One limitation of the study is that patients were recruited at various stages of the disease process therefore there may be significant variation in time frame for weight loss. We have attempted to address this issue by adjusting the analyses for tumour stage at the time of recruitment assuming that patients who are diagnosed with more advanced disease would present with greater amount of weight loss. The amount of weight lost during the cancer journey may be affected by patients' pre-diagnosis BMI. The initially overweight/obese cancer patient may be more likely to lose a greater amount weight compared with a patient with the same cancer type in the normal BMI range over the same period of time. To account for this variation we have also adjusted the analyses for pre-diagnosis BMI. Another limitation of the study is that patients with upper GI malignancy often report dysphagia which may contribute to secondary malnutrition and influence the degree of weight loss. However, a previous study suggest that dysphagia may not be the sole contributing factor to weight loss in gastro-oesophageal malignancy as patients without dysphagia still report a median 4.4% weight loss at diagnosis. Moreover, in a multivariate model of the same cohort, dietary intake accounted for only 38% of variation in weight loss (Deans et al, 2009b).
The present study represents the first large scale candidate gene association study of cancer cachexia spanning a wide variety of genes such as genes that regulate inflammation, muscle and adipose tissue metabolism and appetite. SNPs chosen for the study were based on a literature review of SNPs with known functional effects and/or clinical relevance with regard to the development of cachexia (Tan et al, 2011). We also chose to analyze SNPs based on 18 genes identified in a gene expression study on muscle wasting in patients with cancer cachexia (Stephens et al, 2010). Instead of utilizing a tag SNP approach, as it was not realistic to analyze all possible gene variants and combinations, we selected SNPs that were most likely to be functional (i.e. within exons, non-synonymous and with a minor allele frequency (MAF) of >0.1) and hence more likely to be associated with the development of cachexia.
To further add strength to the study, we also attempted to validate the results by replicating the association study in an independently recruited group of patients. In the initial exploratory cohort we identified 21 SNPs in 17 genes with significant associations with cachexia phenotypes. However, when both the exploratory and validation cohorts were considered, only cancer patients carrying the minor allele (C) of rs6136 were found to be at reduced risk of developing cachexia as defined by weight loss >10% (main study (OR 0.52, 95% CI 0.29–0.93, p = 0.026); validation study (OR 0.09, 95% CI 0.01–0.98, p = 0.035)). We were unable to confirm other significant associations from the main cohort in the validation study. This may be due to the small sample size of the validation study which is a key limitation.
This study included a variety of cancer types, with significant numbers of patients with cancers of the digestive tract, lung and pancreas. Validation in larger independent cohorts is required to fully establish the generalizability of our findings, however the significant association with the rs6136 polymorphism and cachexia across both the main group and an independent validation cohort suggest that our results may apply across numerous cancer types.
Due to the small sample size of the validation cohort, we chose only to perform gene group analysis on the main cohort. The gene group analysis performed provides one way of summarizing the evidence between cachexia traits and multiple genetic variants across groups of genes that share functional similarity. Appetite regulation was found to be most significantly associated with the cachexia trait weight loss >15% and CRP >10 mg/l (p = 0.008). There has been some evidence to date that negative regulators of appetite are elevated in cachexia (Doehner et al, 2001; le Roux et al, 2005). A number of animal studies have also shown prevention or reversal of cachexia by deletion or blockade of specific appetite pathways (Marks et al, 2001; Nicholson et al, 2006; Wisse et al, 2001).
In addition to the above link, the glucocorticoid signalling pathway was also found to be associated with cachexia (weight loss >15% and CRP >10 mg/l) (p = 0.0181). There has been evidence that glucocorticoids and its associated signalling pathway are involved in accelerating protein degradation in muscle, which results in loss of lean body mass in cachexia (Tisdale, 2009). Glucocorticoids work through a permissive effect on the upregulation of messenger RNA and the subsequent synthesis of components of the ubiquitin–proteasome system in muscle. Glucocorticoids inhibit protein synthesis and promote gluconeogenesis, and suppress glucose and amino acid muscle uptake by inhibiting cellular transporters (Lecker et al, 2006). Mitogen activated protein kinases (MAPK) activity regulation was also found to be associated with cachexia (weight loss >15% and CRP >10 mg/l) (p = 0.0264). MAPKs are known to mediate lipolysis in cancer cachexia (Ryden & Arner, 2007), and are also potential regulators of muscle catabolism in cachexia (Keren et al, 2006).
Previous genetic studies on cancer cachexia have identified associations with cachexia and polymorphisms in cytokine genes such as the IL1B 3954C/T polymorphism (rs1143634) in patients with gastric cancer (Zhang et al, 2007), and the IL10-1082A/G polymorphism (rs1800896) in patients with gastro-oesophageal cancer (Deans et al, 2009a). Cancer related anorexia has been associated with the TNF-308G/A polymorphism (rs1800629) in patients with non-small cell lung cancer (Jatoi et al, 2009). Despite some significant associations with other polymorphisms in pro-inflammatory cytokines genes (Table 2), we were unable to confirm the previous specific associations in the present study. However, all these studies have focused only on one particular type of cancer and on a small number of genetic variants. More widely applicable biomarkers may prove more useful. One of the strengths of the present study is the analysis of a wide variety of candidate genes that may influence the development of cachexia in patients with various cancer types.
The nature of cancer cachexia dictates that there are fewer individuals who develop the most severe aspects of the syndrome. At the severe end of the cachexia spectrum, the power in the present study to detect weak associations with uncommon variants was low. It may be that a larger sample size may be required to fully elucidate the effects of such variants in individuals with severe or refractory cachexia.
The diverse cachexia phenotypes we investigated represent various stages in the cachexia journey with potential genetic influences at each stage. The present study suggests that multiple pathways are likely to be involved in the pathogenesis of cancer cachexia and, in particular, P-selectin, appetite regulation, glucocorticoid signalling and MAPK activity regulation may have central roles in this process and should be further investigated. The animal data presented herein suggests that upregulation of P-selectin in skeletal muscle accompanies muscle atrophy in different circumstances. It remains to be determined if modulation of P-selectin might alter the development of cachexia in such models and therefore be a candidate therapeutic target in human cancer cachexia.
MATERIALS AND METHODS
Main study population
Study subjects were recruited from three centres from 2004 to 2008: NHS Lothian, UK; Cross Cancer Institute, Edmonton, Canada; and McGill University Health Centre, Montreal, Canada.
All subjects recruited had participated in clinical or research studies at the host institutions under ethically approved protocols. Recruitment was conducted at first presentation to surgical or oncology clinics at each institution. Recruitment was performed sequentially with the following exclusion criteria: (i) under 18 years of age; (ii) learning disability, and mental health problems; (iii) inability to give written, informed consent; (iv) presence of underlying infection; (v) on corticosteroids.
Patients recruited generally had cancer types with propensity to develop cachexia (e.g. gastric/oesophageal, pancreatic, lung). Overall, 855 patients were recruited. More than 98% of the study subjects were of European descent. Information collected on each patient included date of birth, date of diagnosis, type and stage of cancer. All patients underwent measurements of height and weight at the time of recruitment to the study. Pre-morbid weight was recalled by the patient and verified where possible from the medical notes. Although there may be recall bias, evidence to support the reliability of self-reported weight and weight history (Perry et al, 1995; Stunkard & Albaum, 1981) is well documented. Individual weight loss was calculated and expressed as percentage of pre-morbid body weight lost. Height and weight data were subsequently used to compute a common anthropometric descriptor, BMI (kg/m2).
Serum CRP concentration was measured with an automated immunoturbidimetric assay by each institution's clinical chemistry department using blood collected from patients at the time of recruitment and before any therapeutic intervention. CRP measurement was not available from patients recruited from the Cross Cancer Institute, Edmonton, Canada.
Stage of disease was based on the American Joint Committee on Cancer stage groupings I, II, III and IV.
All patients provided written informed consent to allow analysis of their DNA.
Phenotype definitions
There is currently no consensus diagnostic criteria for cancer cachexia, however two recent international consensus groups (Evans et al, 2008; Fearon et al, 2011) provide a conceptual framework for the classification of this condition. Cachexia is defined by the presence of involuntary weight loss. Varying thresholds of weight loss have been used, the most common being >5% (Fox et al, 2009; Knoll et al, 2008; Maltoni et al, 2001) and >10% (Gordon et al, 2005; Skipworth et al, 2010; Zhang et al, 2007). A weight loss of >15% has been linked to major complications in cancer patients undergoing surgery (Antoun et al, 2009). Evans et al (2008) suggested classifying cachexia as mild or greater, moderate or greater or severe depending on whether the observed weight loss is >5, >10 or >15%, respectively.
The presence of underlying disease and pro-inflammatory catabolic signals discriminate cachexia from malnutrition (Evans et al, 2008; Fearon et al, 2011). The presence of systemic inflammation (serum CRP >10 mg/l) has also been linked to decreased survival (Mahmoud & Rivera, 2002; McMillan et al, 2003), and has also been correlated positively with weight loss in human cancer patients (Deans et al, 2009b; O'Gorman et al, 1999). CRP was incorporated into a three-factor model of cachexia for patients with pancreatic cancer (Fearon et al, 2006). The latter multi-profile definition was found to have more prognostic value compared with weight loss alone.
To take into account the above, we classified cachexia as a spectrum, represented by cut-offs of >5, >10 and >15% weight loss and we also examined weight loss in the presence of systemic inflammation.
Candidate gene and SNP selection
Initial candidate gene and SNP selection was based on a systematic literature review of SNPs with either putative functional or clinical relevance in the development of cancer cachexia (Tan et al, 2011). A further 18 candidate genes were selected based on the results of a gene expression analysis array study on muscle samples of cancer patients with cachexia (Stephens et al, 2010). From these genes were selected non-synonymous coding SNPs with MAF of >0.05. Overall 191 SNPs in 99 genes were considered for the association study.
Genotyping
The Applied Biosystems SNPlex™ Genotyping System (Applied Biosystems, California, USA) was employed for SNP genotyping. All DNA samples were processed and assayed without regard to phenotype. DNA samples were separated electrophoretically on a 3730 DNA Genetic Analyzer (Applied Biosystems, California, USA), and automated allele calls and genotype clustering of each individual sample was performed by Applied Biosystems' GeneMapper® Software (version 4.0). All automatic calls by the software were evaluated by one researcher. Any SNPs with less than 90% of the sample auto-called by the software were either rescored manually or discarded if clustering confidence was low. Reproducibility was determined by rerunning entire plates of DNA samples and a reproducibility rate of 99.7% was achieved.
Individual samples were removed if more than 10% of SNPs failed genotyping, and individual SNPs were removed if more than 10% of samples failed. As an additional genotyping quality-control check, SNPs with significant deviation from Hardy–Weinberg equilibrium (HWE) (p < 0.01) were removed from the final analysis. SNPs with a MAF <0.03 were also removed from the final analysis.
Power calculations
Power calculations were performed using Quanto. For the most prevalent cachexia phenotype (i.e. >5% weight loss, 54% affected), the present study has between 43 and 97% power to detect an OR of 1.5 for SNPs with a MAF of 0.05–0.35.
For the least prevalent cachexia phenotype (i.e. >15% weight loss & CRP >10 mg/l, 14% affected), the present study has between 12 and 40% power to detect an OR of 1.5 for SNPs with a MAF of 0.05–0.35.
Statistical analysis
Statistical analyses were performed using PLINK (version 1.06) (Purcell et al, 2007). Patients who met the criteria for each of the proposed cachexia phenotypes were compared with patients who have lost ≤5% body weight as control. Unconditional logistic regression was employed to calculate ORs and their 95% CI for the minor allele of individual SNPs and its association with each proposed cachexia phenotype. All analyses were adjusted for covariates that may affect weight loss, i.e. age at diagnosis, sex, pre-diagnosis BMI, tumour type and stage.
To account for multiple testing, permutation testing was performed by running the adaptive permutation test in PLINK within each proposed phenotype. Permutation tests are often employed to adjust groups of correlated tests for multiple testing, since conventional methods such as Bonferroni correction are overly conservative when tests are correlated (Conneely & Boehnke, 2007). The adaptive permutation test in PLINK gives up permuting SNPs that are clearly going to be non-significant. This greatly speeds up the permutation procedure, as SNPs that are not significant will drop out quite quickly, making it possible to properly evaluate significance for the handful of SNPs that require millions of permutations.
SNPs with a permuted p-value of <0.02 within the same chromosomal region (within 10 000 kb) were then analyzed for any possible haplotype associations. Only haplotypes that had a frequency greater than 5% were considered for further analysis. Each identified haplotype and significant SNPs were then tested for association with percentage weight loss as a continuous variable.
Finally, candidate genes (and the SNPs in the corresponding gene regions) were grouped based on known functional similarity according to gene ontology using AmiGO (Supporting Information Table S2). The set-based test in PLINK was used to analyze association between grouped SNPs and cachexia phenotypes. The set-based test selects the best set of SNPs whose mean of these single SNP statistics is significant after permutation, which is particularly suited to large-scale candidate gene studies (Ott & Hoh, 2003). The empirical p-values of the set-based test were obtained by a permutation of 10 000 times of phenotype labels.
Validation study
Subjects from the validation study were recruited from an independent centre, Oncology & Palliative Medicine, Cantonal Hospital, St. Gallen, Switzerland from 2007 to 2008. All patients with proven cancer diagnosis were considered. Patients were recruited sequentially at first presentation to the oncology clinic. Exclusion criteria were identical to the main study.
In total, 101 cancer patients were recruited, all of whom were of European descent. Like the main study, all patients underwent measurements of height and weight at the time of recruitment. Pre-morbid weight was recalled by the patient and verified where possible from the medical notes. Individual weight loss was calculated and expressed as percentage of pre-illness body weight lost. Height and weight data were subsequently used to compute a common anthropometric descriptor, BMI (kg/m2). Serum CRP concentration was measured with an automated immunoturbidimetric assay at the institution's clinical chemistry department using blood collected from patients at the time of recruitment and before any therapeutic intervention.
The paper explained
PROBLEM:
More than half of cancer patients suffer from cachexia, and it is responsible for death in up to 20% of cases. Cachexia is also a significant cause of morbidity in cancer patients. Based on our current knowledge of demographic and clinical factors, we are unable to predict, for any given cohort of patients, who will develop cancer cachexia and who will not. Such variation may, in part, be due to the patient's genotype. Knowledge of genotypic variation associated with cachexia would contribute to early identification of patients at risk and allow institution of prophylactic measures.
RESULTS:
In a large scale genetic association study, the C allele of the rs6136 (P-selectin) SNP was found to be associated with weight loss >10% both in the discovery study and the validation study. To further corroborate the P-selectin SNP to cancer cachexia in the gene association study, we tested the same gene for participation in the induction of the skeletal muscle atrophy gene program in animal models of cachexia. P-selectin was found to be significantly upregulated in muscle following both tumour-induced cachexia in rats and intra-peritoneal injection of LPS in mice.
IMPACT:
The C-allele of the rs6136 polymorphism is associated with reduced risk of developing cachexia. Identification of P-selectin as relevant in both animal models and in cachectic cancer patients supports this as a risk factor/potential mediator in cachexia.
Patients were genotyped for SNPs found to have permuted p < 0.05 in the main study and quality control checks were carried out as described previously. As with the main study, patients in each of the proposed cachexia phenotypes were compared with patients with ≤5% weight loss as control, and association analyses were adjusted for age at diagnosis, sex, pre-diagnosis BMI, tumour type and stage.
Animal studies
Wild type C57BL/6J mice (20–25 g) (Jackson Laboratories) and male F344/NTacfBR rats were maintained on a normal 12:12 h light/dark cycle and provided ad libitum access to water and food. Animals were anaesthetized at the time of tumour implantation or sacrifice using a ketamine cocktail. Experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and approved by the Animal Care and Use Committees of Oregon Health & Science University.
Intra-peritoneal injection of LPS
Lipopolysaccharide was dissolved in 0.5% bovine serum albumin (BSA)/0.9% saline and injected intra-peritoneally at 250 µg/kg. Food was removed from cages at the time of injection, and animals were sacrificed 8 h after injection.
Cancer cachexia model
The MCA sarcoma does not metastasize and has a curvilinear growth pattern (Sato et al, 2001). On day 0, TB rats (n = 8) had 0.2–0.3 g tumour tissue implanted subcutaneously into the flank (Ramos et al, 2004) whilst controls (n = 7) underwent sham operation (SH). On day 13, tumour growth was within the pre-determined end-points of the study, according to OHSU IACUC Policy on tumour burden and the animals were sacrificed. Body composition was determined by magnetic resonance (EchoMRI, Echo Medical Systems, Houston, TX) at the time of tumour implantation and again at the time of sacrifice. The gastrocnemius muscles were immediately removed, weighed, preserved in RNAlater solution (Ambion, Inc.) and stored at −80°C until RNA extraction and qPCR analysis.
qPCR analysis
Total gastrocnemius muscle RNA was extracted using the RNeasy fibrous tissue mini kit (Qiagen, Valencia, CA). The total RNA was quantified and checked for integrity using standard protocols. Complementary DNA (cDNA) was transcribed using Taqman reverse transcription reagents according to the manufacturer's instructions. PCR reactions were run on an ABI 7300, using Taqman universal PCR master mix, using Taqman gene expression assays. Relative expression was calculated by the ΔΔCt method using GAPDH as an endogenous control.
Acknowledgments
This work was supported by the European Palliative Care Research Collaborative (EPCRC), an EU framework 6 funded consortium (LSHC-CT-2006-037777).
Supporting Information is available at EMBO Molecular Medicine online.
The authors declare that they have no conflict of interest.
Author contributions
KCHF, FSk, VEB, DLM, SK and JAR designed the study. BHLT, AV, FSt, DACD, RJES, SD and TSS recruited the patients and collected data. BHLT and TF performed the genotyping and overall genetic analysis. TPB performed the animal studies. All authors contributed towards the interpretation of the data, critically reviewed and commented on the report and approved the final version.
Supplementaary material
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
References
- Antoun S, Rey A, Beal J, Montange F, Pressoir M, Vasson MP, Dupoiron D, Gourdiat-Borye A, Guillaume A, Maget B, et al. Nutritional risk factors in planned oncologic surgery: what clinical and biological parameters should be routinely used. World J Surg. 2009;33:1633–1640. doi: 10.1007/s00268-009-0033-3. [DOI] [PubMed] [Google Scholar]
- Argiles JM, Moore-Carrasco R, Fuster G, Busquets S, Lopez-Soriano FJ. Cancer cachexia: the molecular mechanisms. Int J Biochem Cell Biol. 2003;35:405–409. doi: 10.1016/s1357-2725(02)00251-0. [DOI] [PubMed] [Google Scholar]
- Borges E, Eytner R, Moll T, Steegmaier M, Campbell MA, Ley K, Mossmann H, Vestweber D. The P-selectin glycoprotein ligand-1 is important for recruitment of neutrophils into inflamed mouse peritoneum. Blood. 1997;90:1934–1942. [PubMed] [Google Scholar]
- Braun T, Zhu XX, Szumowski M, Scott G, Grossberg A, Graham K, Khan A, Damaraju S, Colmers W, Baracos V, et al. Interleukin 1beta triggers muscle catabolism via a central nervous system-mediated pathway. Endocr Rev. 2011;32:P3–528. [Google Scholar]
- Chen M, Geng JG. P-selectin mediates adhesion of leukocytes, platelets, and cancer cells in inflammation, thrombosis, and cancer growth and metastasis. Arch Immunol Ther Exp (Warsz) 2006;54:75–84. doi: 10.1007/s00005-006-0010-6. [DOI] [PubMed] [Google Scholar]
- Conneely KN, Boehnke M. So many correlated tests, so little time!Rapid adjustment of P values for multiple correlated tests. Am J Hum Genet. 2007;81:1158–1168. doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans DA, Tan BH, Ross JA, Rose-Zerilli M, Wigmore SJ, Howell WM, Grimble RF, Fearon KC. Cancer cachexia is associated with the IL10-1082 gene promoter polymorphism in patients with gastroesophageal malignancy. Am J Clin Nutr. 2009a;89:1164–1172. doi: 10.3945/ajcn.2008.27025. [DOI] [PubMed] [Google Scholar]
- Deans DA, Tan BH, Wigmore SJ, Ross JA, de Beaux AC, Paterson-Brown S, Fearon KC. The influence of systemic inflammation, dietary intake and stage of disease on rate of weight loss in patients with gastro-oesophageal cancer. Br J Cancer. 2009b;100:63–69. doi: 10.1038/sj.bjc.6604828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehoux MJ, van Beneden RP, Fernandez-Celemin L, Lause PL, Thissen JP. Induction of MafBx and Murf ubiquitin ligase mRNAs in rat skeletal muscle after LPS injection. FEBS Lett. 2003;544:214–217. doi: 10.1016/s0014-5793(03)00505-2. [DOI] [PubMed] [Google Scholar]
- Dewys WD, Begg C, Lavin PT, Band PR, Bennett JM, Bertino JR, Cohen MH, Douglass HO, Jr, Engstrom PF, Ezdinli EZ, et al. Prognostic effect of weight loss prior to chemotherapy in cancer patients. Eastern Cooperative Oncology Group. Am J Med. 1980;69:491–497. doi: 10.1016/s0149-2918(05)80001-3. [DOI] [PubMed] [Google Scholar]
- Doehner W, Pflaum CD, Rauchhaus M, Godsland IF, Egerer K, Cicoira M, Florea VG, Sharma R, Bolger AP, Coats AJ, et al. Leptin, insulin sensitivity and growth hormone binding protein in chronic heart failure with and without cardiac cachexia. Eur J Endocrinol. 2001;145:727–735. doi: 10.1530/eje.0.1450727. [DOI] [PubMed] [Google Scholar]
- Dunlop LC, Skinner MP, Bendall LJ, Favaloro EJ, Castaldi PA, Gorman JJ, Gamble JR, Vadas MA, Berndt MC. Characterization of GMP-140 (P-selectin) as a circulating plasma protein. J Exp Med. 1992;175:1147–1150. doi: 10.1084/jem.175.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WJ, Morley JE, Argiles J, Bales C, Baracos V, Guttridge D, Jatoi A, Kalantar-Zadeh K, Lochs H, Mantovani G, et al. Cachexia: a new definition. Clin Nutr. 2008;27:793–799. doi: 10.1016/j.clnu.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Fearon KC, Preston T. Body composition in cancer cachexia. Infusionstherapie. 1990;17:63–66. doi: 10.1159/000222558. [DOI] [PubMed] [Google Scholar]
- Fearon KC, Voss AC, Hustead DS. Definition of cancer cachexia: effect of weight loss, reduced food intake, and systemic inflammation on functional status and prognosis. Am J Clin Nutr. 2006;83:1345–1350. doi: 10.1093/ajcn/83.6.1345. [DOI] [PubMed] [Google Scholar]
- Fearon K, Strasser F, Anker SD, Bosaeus I, Bruera E, Fainsinger RL, Jatoi A, Loprinzi C, Macdonald N, Mantovani G, et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol. 2011;12:489–495. doi: 10.1016/S1470-2045(10)70218-7. [DOI] [PubMed] [Google Scholar]
- Fox KM, Brooks JM, Gandra SR, Markus R, Chiou CF. Estimation of cachexia among cancer patients based on four definitions. J Oncol. 2009;2009:693458. doi: 10.1155/2009/693458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JN, Trebble TM, Ellis RD, Duncan HD, Johns T, Goggin PM. Thalidomide in the treatment of cancer cachexia: a randomised placebo controlled trial. Gut. 2005;54:540–545. doi: 10.1136/gut.2004.047563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AG, Jacobson L, Gonzalez J, Rounds J, Majzoub JA, Wilmore DW. Chronic central nervous system exposure to interleukin-1 beta causes catabolism in the rat. Am J Physiol. 1996;271:R1142–R1148. doi: 10.1152/ajpregu.1996.271.5.R1142. [DOI] [PubMed] [Google Scholar]
- Jatoi A, Qi Y, Kendall G, Jiang R, McNallan S, Cunningham J, Mandrekar S, Yang P. The cancer anorexia/weight loss syndrome: exploring associations with single nucleotide polymorphisms (SNPs) of inflammatory cytokines in patients with non-small cell lung cancer. Support Care Cancer. 2009;18:1299–1304. doi: 10.1007/s00520-009-0748-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GI, Bliss GA, Newman PJ, McEver RP. Structure of the human gene encoding granule membrane protein-140, a member of the selectin family of adhesion receptors for leukocytes. J Biol Chem. 1990;265:21381–21385. [PubMed] [Google Scholar]
- Keren A, Tamir Y, Bengal E. The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol Cell Endocrinol. 2006;252:224–230. doi: 10.1016/j.mce.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Knoll S, Zimmer S, Hinney A, Scherag A, Neubauer A, Hebebrand J. Val103Ile polymorphism of the melanocortin-4 receptor gene (MC4R) in cancer cachexia. BMC Cancer. 2008;8:85. doi: 10.1186/1471-2407-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Roux CW, Ghatei MA, Gibbs JS, Bloom SR. The putative satiety hormone PYY is raised in cardiac cachexia associated with primary pulmonary hypertension. Heart. 2005;91:241–242. doi: 10.1136/hrt.2003.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- Mahmoud FA, Rivera NI. The role of C-reactive protein as a prognostic indicator in advanced cancer. Curr Oncol Rep. 2002;4:250–255. doi: 10.1007/s11912-002-0023-1. [DOI] [PubMed] [Google Scholar]
- Maltoni M, Nanni O, Scarpi E, Rossi D, Serra P, Amadori D. High-dose progestins for the treatment of cancer anorexia–cachexia syndrome: a systematic review of randomised clinical trials. Ann Oncol. 2001;12:289–300. doi: 10.1023/a:1011156811739. [DOI] [PubMed] [Google Scholar]
- Marks DL, Ling N, Cone RD. Role of the central melanocortin system in cachexia. Cancer Res. 2001;61:1432–1438. [PubMed] [Google Scholar]
- McMillan DC, Canna K, McArdle CS. Systemic inflammatory response predicts survival following curative resection of colorectal cancer. Br J Surg. 2003;90:215–219. doi: 10.1002/bjs.4038. [DOI] [PubMed] [Google Scholar]
- Miller MA, Kerry SM, Dong Y, Strazzullo P, Cappuccio FP. Association between the Thr715Pro P-selectin gene polymorphism and soluble P-selectin levels in a multiethnic population in South London. Thromb Haemost. 2004;92:1060–1065. doi: 10.1160/TH04-04-0228. [DOI] [PubMed] [Google Scholar]
- Nicholson JR, Kohler G, Schaerer F, Senn C, Weyermann P, Hofbauer KG. Peripheral administration of a melanocortin 4-receptor inverse agonist prevents loss of lean body mass in tumor-bearing mice. J Pharmacol Exp Ther. 2006;317:771–777. doi: 10.1124/jpet.105.097725. [DOI] [PubMed] [Google Scholar]
- O'Gorman P, McMillan DC, McArdle CS. Longitudinal study of weight, appetite, performance status, and inflammation in advanced gastrointestinal cancer. Nutr Cancer. 1999;35:127–129. doi: 10.1207/S15327914NC352_5. [DOI] [PubMed] [Google Scholar]
- Ott J, Hoh J. Set association analysis of SNP case-control and microarray data. J Comput Biol. 2003;10:569–574. doi: 10.1089/10665270360688192. [DOI] [PubMed] [Google Scholar]
- Perry GS, Byers TE, Mokdad AH, Serdula MK, Williamson DF. The validity of self-reports of past body weights by U.S. adults. Epidemiology. 1995;6:61–66. doi: 10.1097/00001648-199501000-00012. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos EJ, Middleton FA, Laviano A, Sato T, Romanova I, Das UN, Chen C, Qi Y, Meguid MM. Effects of omega-3 fatty acid supplementation on tumor-bearing rats. J Am Coll Surg. 2004;199:716–723. doi: 10.1016/j.jamcollsurg.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Ryden M, Arner P. Fat loss in cachexia-is there a role for adipocyte lipolysis. Clin Nutr. 2007;26:1–6. doi: 10.1016/j.clnu.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Sato T, Meguid MM, Fetissov SO, Chen C, Zhang L. Hypothalamic dopaminergic receptor expressions in anorexia of tumor-bearing rats. Am J Physiol Regul Integr Comp Physiol. 2001;281:R1907–R1916. doi: 10.1152/ajpregu.2001.281.6.R1907. [DOI] [PubMed] [Google Scholar]
- Skipworth RJ, Stewart GD, Bhana M, Christie J, Sturgeon CM, Guttridge DC, Cronshaw AD, Fearon KC, Ross JA. Mass spectrometric detection of candidate protein biomarkers of cancer cachexia in human urine. Int J Oncol. 2010;36:973–982. doi: 10.3892/ijo_00000577. [DOI] [PubMed] [Google Scholar]
- Stephens NA, Gallagher IJ, Rooyackers O, Skipworth RJ, Tan BH, Marstrand T, Ross JA, Guttridge DC, Lundell L, Fearon KC, et al. Using transcriptomics to identify and validate novel biomarkers of human skeletal muscle cancer cachexia. Genome Med. 2010;2:122. doi: 10.1186/gm122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunkard AJ, Albaum JM. The accuracy of self-reported weights. Am J Clin Nutr. 1981;34:1593–1599. doi: 10.1093/ajcn/34.8.1593. [DOI] [PubMed] [Google Scholar]
- Tan BH, Ross JA, Kaasa S, Skorpen F, Fearon KC. Identification of possible genetic polymorphisms involved in cancer cachexia: a systematic review. J Genet. 2011;90:165–177. doi: 10.1007/s12041-011-0027-4. [DOI] [PubMed] [Google Scholar]
- Tisdale MJ. Cachexia in cancer patients. Nat Rev Cancer. 2002;2:862–871. doi: 10.1038/nrc927. [DOI] [PubMed] [Google Scholar]
- Tisdale MJ. Cancer cachexia. Langenbecks Arch Surg. 2004;389:299–305. doi: 10.1007/s00423-004-0486-7. [DOI] [PubMed] [Google Scholar]
- Tisdale MJ. Mechanisms of cancer cachexia. Physiol Rev. 2009;89:381–410. doi: 10.1152/physrev.00016.2008. [DOI] [PubMed] [Google Scholar]
- Tisdale MJ, Brennan RA, Fearon KC. Reduction of weight loss and tumour size in a cachexia model by a high fat diet. Br J Cancer. 1987;56:39–43. doi: 10.1038/bjc.1987.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary TC, Dardevet D, Grizard J, Voisin L, Buffiere C, Denis P, Breuille D, Obled C. Differential regulation of skeletal muscle protein turnover by insulin and IGF-I after bacteremia. Am J Physiol. 1998;275:E584–E593. doi: 10.1152/ajpendo.1998.275.4.E584. [DOI] [PubMed] [Google Scholar]
- Volcik KA, Ballantyne CM, Coresh J, Folsom AR, Wu KK, Boerwinkle E. P-selectin Thr715Pro polymorphism predicts P-selectin levels but not risk of incident coronary heart disease or ischemic stroke in a cohort of 14595 participants: the Atherosclerosis Risk in Communities Study. Atherosclerosis. 2006;186:74–79. doi: 10.1016/j.atherosclerosis.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Wagenmakers AJ, van Riel NA, Frenneaux MP, Stewart PM. Integration of the metabolic and cardiovascular effects of exercise. Essays Biochem. 2006;42:193–210. doi: 10.1042/bse0420193. [DOI] [PubMed] [Google Scholar]
- Watson ML, Kingsmore SF, Johnston GI, Siegelman MH, Le Beau MM, Lemons RS, Bora NS, Howard TA, Weissman IL, McEver RP, et al. Genomic organization of the selectin family of leukocyte adhesion molecules on human and mouse chromosome 1. J Exp Med. 1990;172:263–272. doi: 10.1084/jem.172.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse BE, Frayo RS, Schwartz MW, Cummings DE. Reversal of cancer anorexia by blockade of central melanocortin receptors in rats. Endocrinology. 2001;142:3292–3301. doi: 10.1210/endo.142.8.8324. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zheng H, Zhou Y, Tang X, Yu B, Li J. Association of IL-1beta gene polymorphism with cachexia from locally advanced gastric cancer. BMC Cancer. 2007;7:45. doi: 10.1186/1471-2407-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.