Abstract
The trace metal copper (Cu) plays an essential role in biology as a cofactor for many enzymes that include Cu, Zn superoxide dismutase, cytochrome oxidase, ceruloplasmin, lysyl oxidase, and dopamine β-hydroxylase. Consequently, Cu transport at the cell surface and the delivery of Cu to intracellular compartments are critical events for a wide variety of biological processes. The components that orchestrate intracellular Cu trafficking and their roles in Cu homeostasis have been elucidated by the studies of model microorganisms and by the characterizations of molecular basis of Cu-related genetic diseases, including Menkes disease and Wilson disease. However, little is known about the mechanisms for Cu uptake at the plasma membrane and the consequences of defects in this process in mammals. Here, we show that the mouse Ctr1 gene encodes a component of the Cu transport machinery and that mice heterozygous for Ctr1 exhibit tissue-specific defects in copper accumulation and in the activities of copper-dependent enzymes. Mice completely deficient for Ctr1 exhibit profound growth and developmental defects and die in utero in mid-gestation. These results demonstrate a crucial role for Cu acquisition through the Ctr1 transporter for mammalian Cu homeostasis and embryonic development.
Copper (Cu) is a micronutrient that plays an essential role in biology, serving as a cofactor for enzymes that modify neuropeptides, generate cellular energy, detoxify oxygen-derived radicals, mobilize iron, coagulate blood, and cross-link connective tissue (1, 2). Human and animal genetic diseases including Menkes disease and Wilson disease underscore critical roles for Cu absorption and distribution (3, 4). The entrapment of Cu in intestinal cells in Menkes disease patients leads to Cu deficiency as ascertained by defects in the activities of Cu-containing enzymes. Patients with Wilson disease accumulate Cu in the liver, resulting in liver cirrhosis and neurodegeneration.
Recent studies in microorganisms and the characterization of the molecular basis of Cu-related genetic diseases have elucidated many of the components that orchestrate intracellular Cu metabolism (2–7). Mutations in one of two human genes encoding Cu transporting P-type ATPases that reside in the secretory compartment cause Menkes and Wilson disease, which result in a defect in intestinal Cu absorption and Cu maldistribution in the liver, respectively (3, 4). Cu is carried to subcellular compartments or Cu-dependent enzymes through the action of target-specific Cu chaperone proteins that include Atx1/Atox1, Cox17, and CCS (5–9). The Atx1/Atox1 Cu chaperone directly interacts with the cytosolic Cu-binding domains of the Cu-transporting P-type ATPases to provide Cu to the secretory compartment, where it is incorporated into iron homeostasis proteins such as Fet3 in yeast and ceruloplasmin in mammals (9–11). CCS directly interacts with apo-Cu, Zn superoxide dismutase (SOD) for the incorporation of Cu (12). Mitochondrial Sco1 and Sco2 protein are involved in the incorporation of Cu delivered by Cox17 into cytochrome oxidase (COX) subunits (13). Consistent with the role of Sco1 and Sco2 in Cu incorporation into COX, mutations in Sco1 or Sco2 genes have been identified from the characterization of the patients with COX deficiency (14, 15).
Studies in yeast cells first identified genes encoding high-affinity Cu ion transport proteins in the plasma membrane. Either before or concomitant with high-affinity uptake, Cu(II) is reduced to Cu(I) by one or more metalloreductases encoded by the Fre1 through Fre7 genes (16, 17). Cu(I) is thought to be delivered across the plasma membrane by the high-affinity transporters Ctr1 and Ctr3 in Saccharomyces cerevisiae and Ctr4 in Schizosaccharomyces pombe, with Km values in the low micromolar range (18–20). Yeast cells lacking high-affinity Cu transporters exhibit striking defects in Cu and Fe uptake, mitochondrial respiration, and Cu, Zn SOD activity (18–20). Recently, candidates for human and murine plasma membrane Cu transporters have been isolated by complementation of yeast Ctr mutants and by database homology searches (21, 22). Mammalian Ctr1 mRNA is expressed in all tissues examined, with higher expression levels in liver and kidney and lower levels in brain and spleen (21, 22). Murine Ctr1 displays 92% sequence identity to human Ctr1 and maps to a syntenic locus in the mouse genome, 4C1–2 (22). Whereas mammalian Ctr1 functionally replaces yeast high-affinity Cu transporters and there are structural similarities among Cu transporters in yeast and mammals, little is known about the function of the putative Ctr1 Cu transporter in mammals and the consequences of inactivation of the mammalian Ctr1 gene.
We have examined the role of mouse Ctr1 protein in Cu transport by ectopic expression in human cells and by the generation and characterization of Ctr1 gene knock-out mice. We observed that (i) the mouse Ctr1 gene encodes a component of the Cu transport machinery, (ii) the mice heterozygous for Ctr1 exhibit tissue-specific defects in copper accumulation and in the activities of copper-dependent enzymes, and (iii) mice completely deficient for Ctr1 exhibit profound growth and developmental defects and die in utero in mid-gestation. These results demonstrate a crucial role for Cu acquisition through the Ctr1 transporter for mammalian Cu homeostasis and embryonic development.
Materials and Methods
64Cu Uptake Assay in Hek293 Cells.
Human embryonic kidney cells (Hek293) were cultured in DMEM (GIBCO) with 10% FBS under 5% CO2. Cells were transfected with the pcDNA3.1 vector (Invitrogen) or pcDNA3.1 expressing the mouse Ctr1. 64Cu (10 μM as CuCl2) was added to culture medium 2 days after transfection and incubated for different time points. Parallel experiments were conducted at 4°C for cell-surface binding values, which were subtracted from the values obtained at 37°C to obtain net copper uptake. Cu uptake was quenched by adding ice-cold EDTA (10 mM final concentration); cells were washed three times with ice-cold PBS, resuspended in SDS/Triton X-100 PBS buffer for lysis, and aliquots of cell lysate were counted by using a γ-counter (Packard Cobra II). Copper uptake was calculated by using a standard curve and normalized to protein concentrations of cell lysates.
Targeted Disruption of the Ctr1 Gene.
BAC clones containing mouse Ctr1 sequences were isolated from a 129/SvJ genomic library (22). The targeting vector contains a neomycin (neo) gene expression cassette driven by the PGK promoter (23) and ≈2.0 kb and 3.5 kb of genomic sequences that flank Ctr1 coding exons 5′ and 3′, respectively. The linearized targeting vector was introduced into the 129Sv mouse-derived R1 embryonic stem (ES) cell line (24) by electroporation. The thymidine kinase (tk) gene and neomycin resistance (neo) gene were used to select ES cells in which the targeting construct had integrated into the genome. Four clones carrying the targeted Ctr1 allele were injected into blastocysts collected from C57BL/6 mice following standard procedure (25). The Ctr1+/− mice were generated by breeding between male chimeric mice and female C57BL/6 mice. Mice were genotyped by Southern blotting and PCR analysis with primer sets specific to the Ctr1 gene and the neo gene.
RNA Analysis.
Total RNA was isolated from organs and tissues with standard methods (26). Twenty micrograms total RNA was separated by electrophoreses on a 1% formaldehyde–agarose gel and transferred to a nylon membrane; Ctr1 and Gapdh mRNA levels were detected by using radiolabeled mouse Ctr1 and rat Gapdh coding sequences as probes.
Morphological Studies.
Embryos from Ctr1+/− intercrosses were dissected from the decidua with the removal of Reichert's membrane and ectoplacental cone tissue, and photographed. Approximate gestational stages were estimated from dates of the appearance of vaginal plugs. Embryos were genotyped by PCR or Southern blotting analysis.
Histological Studies.
Mouse embryos were fixed with 4% paraformaldehyde overnight at 4°C, then dehydrated with graded ethanol. Embryos were embedded in paraffin, sectioned (5 μm), and stained with hematoxylin and eosin. Genotypes of the embryos were determined by the Southern blotting analysis with genomic DNA extracted from remaining embryo tissue used for histological studies.
Cu Supplementation.
Drinking water containing 2 mM or 6 mM CuCl2 was provided to female Ctr1+/− mice from 3 weeks before intercrossing through gestation and lactation. By providing copper-supplemented drinking water, these mice ingested ≈50–100 times more copper compared with mice reared under normal dietary conditions.
Measurement of Cu Levels and Enzyme Activities.
Organs were wet-digested with concentrated HNO3 and then analyzed for copper and iron levels by flame atomic absorption spectroscopy (27). Activities of COX and Cu, Zn SOD were determined spectrophotometrically on whole homogenate as described previously (28).
Results
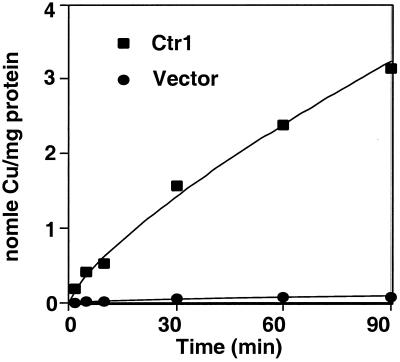
Mouse and human cDNAs have been isolated that encode proteins with significant sequence identity to yeast high-affinity Cu transporters and that complement phenotypes of yeast cells defective in the high-affinity Cu transport machinery (21, 22). To ascertain whether the encoded mammalian Ctr1 proteins function in Cu uptake, a mouse Ctr1 cDNA was placed under the control of the human CMV promoter and was expressed in Hek293 cells by transient transfection. The accumulation of 64Cu was measured and compared between cells expressing mouse Ctr1 or the empty vector control. The data in Fig. 1 demonstrate that expression of mouse Ctr1 dramatically stimulates 64Cu uptake (≈30-fold) in a time-dependent manner. This observation, together with demonstrations that human and mouse Ctr1 complement Cu uptake defects associated with S. cerevisiae cells lacking high-affinity Cu transporters (21, 22), that human Ctr1 transports Cu with high affinity, and that Ctr1 is localized to the plasma membrane (J.L. and D.J.T., unpublished results), strongly suggests that Ctr1 functions as a Cu transporter in mammals.
Figure 1.
Expression of mouse Ctr1 stimulates 64Cu uptake. Human embryonic kidney cells (HEK293) were transfected with vector (●) or the mouse Ctr1 cDNA under control of the CMV promoter (■). Cu64 (10 μM) was added to the culture medium. 64Cu uptake was calculated at different times by using a standard curve and normalized to protein concentration of total cell lysates.
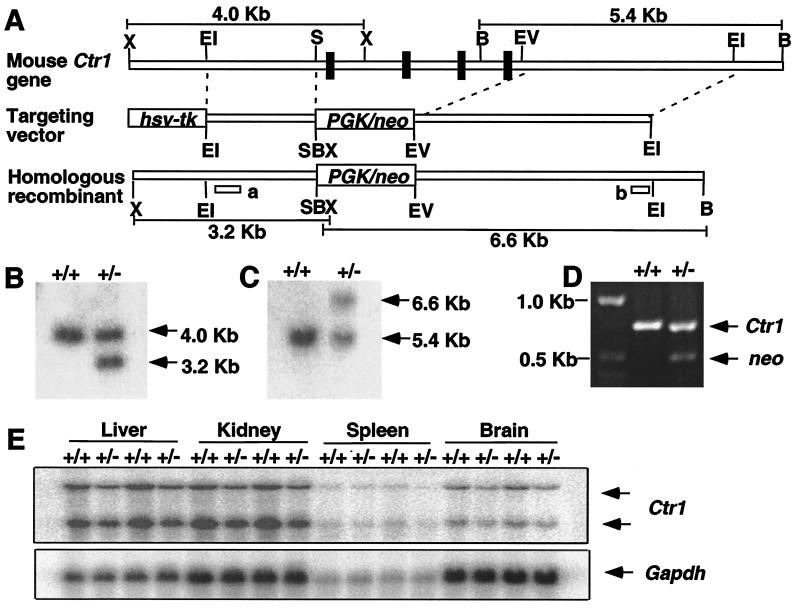
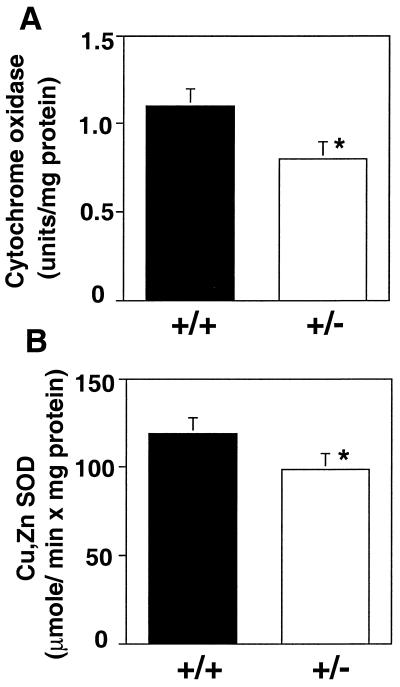
To investigate the role of the Ctr1 Cu transporter in mammals, one copy of the Ctr1 chromosomal locus was insertionally inactivated in mouse ES cells. All four Ctr1 coding exons were replaced by a PGK/neo cassette (Fig. 2A); homologous recombinants were selected on the basis of neomycin resistance and screened by Southern blotting and PCR to identify ES cell clones heterozygous for the Ctr1 deletion allele (Fig. 2 B–D). The Ctr1+/− ES cells were used to generate Ctr1+/− mice by standard techniques. RNA blotting experiments demonstrated that Ctr1+/− mice expressed ≈50% the levels of the two Ctr1 mRNA species as compared with wild-type littermates in liver, kidney, spleen, and brain (Fig. 2E). The presence of two distinct Ctr1 transcripts in mammals (21, 22), both of which are reduced to half wild-type levels in Ctr1+/− mice (Fig. 1E), demonstrate that both transcripts are derived from a single mouse Ctr1 gene. Interestingly, although there are overall reductions of Ctr1 mRNA in all organs examined, atomic absorption spectroscopy demonstrates an ≈50% reduction in total Cu levels in brain and spleen with no significant changes in liver or kidney (Table 1). Because iron homeostasis through ceruloplasmin and hephaestin is Cu-dependent (29, 30), we also measured iron levels in Ctr1+/− mice and found that its levels in all four tissues were slightly decreased with a significant difference in the liver (Table 1). Furthermore, as shown in Fig. 3, both Cu, Zn SOD activity and COX activity are reduced by ≈20% in total brain homogenates from Ctr1 heterozygotes as compared with wild-type littermates. Therefore, perturbations in the levels of Ctr1 expression, through the inactivation of one Ctr1 allele, give more pronounced affects in brain and spleen than in the other tissues examined. Taken together, characterization of Ctr1+/− mice demonstrates that Ctr1 plays an important physiological role in Cu acquisition.
Figure 2.
Targeted disruption of the mouse Ctr1 gene by homologous recombination. (A) A restriction map of the genomic locus encompassing the mouse Ctr1 coding exons is diagrammed (X, XbaI; EI, EcoRI; EV, EcoRV; S, SacI; B, BamBI). All Ctr1 coding exons were replaced by a neomycin (neo) gene expression cassette driven by the PGK promoter. The predicted locus derived from homologous recombination is shown. Hybridization probes “a” and “b” used to detect homologous recombinant allele by Southern blotting analysis are indicated. The DNA fragment sizes derived from the wild-type and recombinant Ctr1 locus, by cleavage with XbaI and BamHI, are indicated. Genomic DNA isolated from ES cells or mouse tail biopsies was genotyped by Southern blotting after restriction enzyme digestion with XbaI and hybridized with probe “a” (B) or by restriction enzyme digestion with BamHI and hybridized with probe “b” (C). (D) Primer sets specific to the Ctr1 and neo genes were used to screen Ctr1+/+ and Ctr1+/− mice by PCR analysis. Both Ctr1 and neo gene-specific DNA fragments were amplified from the genomic DNA isolated from Ctr1+/− mice, but only a Ctr1-specific product was amplified from genomic DNA isolated from Ctr1+/+ mice. (E) Ctr1 mRNA blot analysis from organs derived from wild-type and Ctr1+/− mice. Gapdh mRNA levels were used as a loading control.
Table 1.
Copper and iron levels in wild-type and Ctr1+/− mice
| Liver
|
Kidney
|
Spleen
|
Brain
|
|||||
|---|---|---|---|---|---|---|---|---|
| +/+ | +/− | +/+ | +/− | +/+ | +/− | +/+ | +/− | |
| Copper | 6.1 ± 1.0 | 6.3 ± 0.8 | 4.6 ± 0.1 | 4.6 ± 0.3 | 1.0 ± 0.4 | 0.6 ± 0.1* | 3.2 ± 0.1 | 1.7 ± 0.2** |
| Iron | 81.2 ± 10.6 | 67.8 ± 9.0* | 52.1 ± 9.6 | 45.4 ± 6.7 | 175.5 ± 55.9 | 143.1 ± 32.6 | 17.7 ± 1.0 | 15.8 ± 2.4 |
Copper and iron levels (μg/g of tissue) were analyzed for 5- to 6-week-old wild-type (+/+) and Ctr1+/− (+/−) littermates from Ctr1+/− intercrosses. Means of 5–7 mice were compared by Student's t test. *, P < 0.05; **, P < 0.01.
Figure 3.
Brain cupro-enzyme activities in wild-type and Ctr1+/− mice. Activities of cytochrome oxidase (Cox) (μmol/min × mg protein) and Cu, Zn SOD (units/mg protein) from whole brain homogenates were analyzed for 5–6-week-old wild-type (+/+) and Ctr1+/− (+/−) littermates. Means of five to seven mice were compared by Student's t test. *, P < 0.01.
Several independent crosses between Ctr1+/− male and female mice were established to generate Ctr1−/− mice, and pups derived from these crosses were genotyped for Ctr1 status by Southern blotting and PCR. Although the expected Mendelian ratio of Ctr1+/+ and Ctr1+/− pups was generated from these crosses, of 378 pups analyzed no Ctr1−/− offspring were identified (Table 2). Dietary Cu supplementation (see Materials and Methods) of pregnant mice did not rescue Ctr1−/− offspring, suggesting that Ctr1−/− embryos cannot acquire Cu because of the lack of the plasma membrane Ctr1 transporter and that there is no alternate system for Cu uptake that can compensate for loss of Ctr1. However, the Ctr1−/− mice could be rescued by expression of the Ctr1 ORF with a regulatable promoter by using standard transgenic approaches (J.L. and D.J.T., unpublished observations). These observations demonstrated that the presence of at least one functional copy of the Ctr1 Cu transporter gene is essential for normal embryonic development through parturition, and the embryonic lethal phenotype of Ctr1−/− mice is Ctr1 deletion specific. To investigate this in more detail, embryos were isolated from Ctr1+/− crosses at days 8.5, 10.5, and 12.5 postfertilization and Ctr1 allele status genotyped by PCR. Although the expected ratio of Ctr1−/− embryos was identified through embryonic day 10.5, no Ctr1−/− embryos were recovered 12.5 days postfertilization (Table 2). Furthermore, evidence for resorption of 29% of the embryos at E12.5, as indicated by the presence of a residual placenta with a shriveled embryo attached, revealed that all Ctr1−/− embryos were resorbed into the uterine wall around this point of gestation. Consistent with the essential role of Ctr1 in embryo development as demonstrated by Ctr1−/− mouse, Ctr1 is expressed both in embryonic stem cells (data not shown) and in early mouse embryos (22).
Table 2.
Embryonic lethality of homozygous Ctr1 null mice
| Embryo or pups
|
Genotype
(%)
|
Resorbed (%) | |||
|---|---|---|---|---|---|
| Day | Number | +/+ | +/− | −/− | |
| 3 weeks | 378 | 140 (37%) | 238 (63%) | 0 (0%) | ND |
| E 12.5 | 132 | 30 (23%) | 63 (48%) | 0 (0%) | 39 (29%) |
| E 10.5 | 169 | 44 (26%) | 79 (47%) | 41 (24%) | 5 (3%) |
| E 8.5 | 138 | 33 (24%) | 70 (51%) | 28 (20%) | 7 (5%) |
Genotypes of offspring from Ctr1+/− intercrosses were determined by PCR and/or Southern blotting analysis of genomic DNA isolated from 3-week-old mice or embryos at the indicated stages post fertilization. ND, not determined.
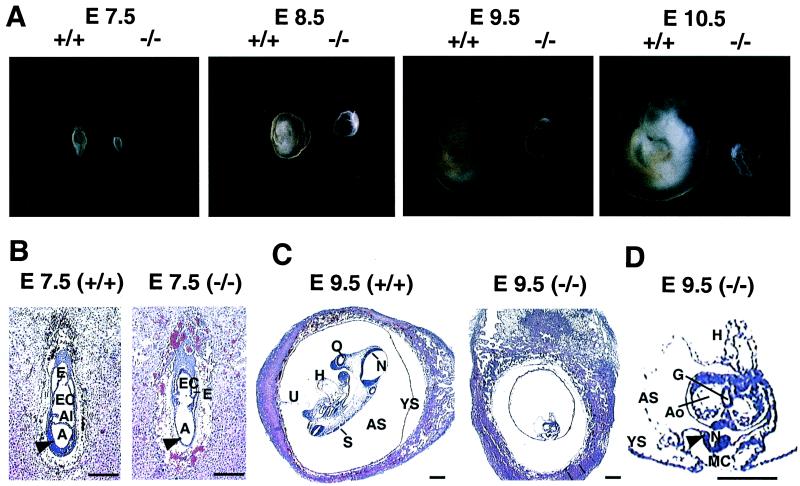
Morphological and histological evaluation of wild-type and Ctr1−/− embryos demonstrated severe growth and developmental consequences arising from complete loss of the Ctr1 gene. As shown in Fig. 4A, Ctr1−/− embryos exhibited a dramatic reduction in size at E7.5, which is exacerbated as compared with wild-type embryos as in utero development progressed through day E10.5. Histological evaluation of Ctr1−/− embryos demonstrated that although the fundamental mouse embryonic structures were conserved at E7.5, many structures including the neural ectoderm and mesoderm cell layers were poorly developed (Fig. 4B, arrowhead). Furthermore, at E9.5, closure of the neural tube was impaired (Fig. 4D, arrowhead), and there was a severe diminution in mesenchyme cell formation (Fig. 4D, MC). Moreover, this analysis revealed that the process of turning, which is essential to achieve the characteristic fetal position, had not initiated at E9.5 in Ctr1−/− embryos (Fig. 4D). The devastating morphological and histological abnormalities that are manifest approximately mid-way through gestation suggest that one or more Cu-dependent enzymes that rely on Ctr1 as a source of Cu may be critical for mouse embryonic development.
Figure 4.
Morphological and histological analysis of littermate embryos from Ctr1+/− intercrosses. (A) Morphology of Ctr1+/+ and Ctr1−/− embryos are compared. Embryos were isolated with yolk sac at E7.5, E8.5, E9.5, and E10.5 days postfertilization. Genotypes (+/+ or −/−) of Ctr1 alleles are indicated. (B and C) Embryos with decidua were dissected from the uterus at the indicated days postfertilization, sectioned, and stained with hematoxylin and eosin. (D) Four-fold magnification of the E9.5 Ctr1−/− embryo in C for detail. A, amnionic cavity; Al, allantois; Ao, aorta; AS, amnionic sac; E, ectoplacental cavity; EC, ecocoelomic cavity; G, gut; H, heart; N, neuroepithelium; O, otic vesicle; S, somite; U, umbilical cord; YS, yolk sac; arrowhead in B, ectoderm and mesoderm layers; arrowhead in D, defective neural tube closure; MC, mesenchyme cells. (Bar = 250 μm.)
Discussion
Cu uptake and its distribution at the cellular and organismal level are critical processes to provide adequate Cu to a number of Cu-containing enzymes and proteins. In this study, we have clearly demonstrated that the Ctr1 protein is able to stimulate Cu uptake and act as a gateway for Cu acquisition. Normal levels of Ctr1 expression are critical to maintain Cu levels in organs and tissues and for embryo development. Furthermore, characterization of Ctr1 heterozygous and homozygous deletion mice address important issues regarding Cu metabolism and its implication in development.
Although mammalian Ctr1 protein functionally complements yeast high-affinity Cu transporters (21, 22), the biological function of Ctr1 in mammals has not been investigated. It was recently reported that human Ctr1 expression under the control of a strong promoter stimulated Cu uptake in transfected cells (31). Our data demonstrate that expression of mouse Ctr1 stimulates Cu uptake in cultured cells and that mice heterozygous for Ctr1 display a Cu accumulation defect, further supporting the notion that mammalian Ctr1 is a component of the Cu transport machinery and a limiting molecule for the Cu uptake system. Furthermore, reduced Cu accumulation in both brain and spleen from Ctr1+/− mice underscores the notion that both Ctr1 alleles are critical for normal overall Cu homeostasis. The reason for the tissue-specific defect in Cu accumulation in Ctr1 heterozygous mice (Table 1) is not clearly understood yet. This could reflect low levels of Ctr1 expression in brain and spleen that may be just sufficient for normal Cu homeostasis in these organs (Fig. 2E) (21, 22) or tissue-specific compensatory mechanisms in other tissues that have not yet been elucidated. Although brain and spleen Cu levels and Cu-dependent enzyme activities are reduced in Ctr1+/− mice, no obvious abnormalities in gross anatomy or fertility have been observed thus far. However, it will be interesting to test whether alteration of Cu levels and Cu-dependent enzyme activities in the brains of Ctr1+/− mice alter the function of neuronal cells and the nervous system.
The iron levels in several tissues examined are slightly reduced in the Ctr1+/− mice with a small but statistically significant difference in the liver (Table 1), consistent with the previously established roles for Cu in iron metabolism. The yeast Ctr1 high-affinity Cu transporter was initially identified through a screen of mutants defective in Fe transport (18), providing a mechanistic explanation for the observed link between Cu and iron metabolism (32). In the absence of functional yeast high-affinity Cu transporters, the inactive Fet3 multicopper ferroxidase fails to assemble with the Ftr1 iron permease as an iron–transporter complex (33). In mammals, it has been known that ceruloplasmin and hephaestin, multicopper ferroxidases in serum and in the intestine, respectively, have critical roles for iron homeostasis. Individuals with a mutation in the ceruloplasmin gene cannot mobilize iron from tissues (29). Mice with sex-linked anemia that accumulate iron in intestinal epithelial cells have been shown to be defective in the hephaestin protein thought to be essential for intestinal iron transport into the portal circulation (30). Although we have not examined Cu levels in enterocytes, one possibility for the reduced iron levels in the liver observed in Ctr1+/− mice is that hephaestin function is partially defected in the Ctr1+/− mice because of Cu limitation.
The baker's yeast S. cerevisiae expresses two high-affinity Cu transporters, Ctr1 and Ctr3, at the plasma membrane (18, 19). Ctr3 was identified as a suppressor of the Cu starvation phenotypes associated with a Ctr1-deleted strain (19), although Ctr3 gene expression in many laboratory strains of S. cerevisiae is silenced by the insertion of a Ty2 retroviral-type transposable element (19). Recent studies suggest that Cu ion uptake in S. cerevisiae can also occur through low-affinity Cu transporters encoded by the Fet4 (34), Ctr2 (35), and Smf1/2 (36) genes. A putative low-affinity mammalian Cu transporter, Ctr2, was also identified by the sequence homology with human Ctr1 protein (21), and rat intestinal DCT1 (Nramp2, DMT1) has been suggested to transport Cu (37). However, our results demonstrate that other potential Cu transporters cannot complement Ctr1 Cu transporter defects, including embryonic lethality (Table 2) and reduced Cu levels in the Ctr1+/− mice (Table 1).
Our results obtained from the characterization of theCtr1+/− and Ctr1−/− mice underscore a crucial requirement for the Ctr1 Cu transporter in mammalian Cu homeostasis and development. The critical roles for Cu in embryonic development demonstrated in the Ctr1−/− mice are consistent with the developmental abnormalities observed in Menkes disease patients and animal models (3, 4, 38) and Atox1 Cu chaperone-deficient mice (39). However, individual deficiency of any Cu-containing enzyme or protein examined does not manifest such a severe developmental defect as observed from mice lacking the Ctr1 Cu transporter. For example, mice deficient in Cu, Zn SOD, tyrosinase, ceruloplasmin, dopamine β-hydroxylase, or coagulation factor V do not demonstrate complete embryonic lethality (40–46). It is more likely that defects in multiple Cu proteins rather than loss of one specific protein function are responsible for the early embryonic lethal phenotype of the Ctr1−/− mice. Interestingly, insertional inactivation of the Drosophila PHM gene encoding a subunit of the Cu-dependent peptidylglycyl α-amidating enzyme is embryonically lethal (47), although the role of this enzyme in mammalian development has not been determined yet. Therefore, whether the embryonic lethal phenotype observed in Ctr1-deficient mice is because of the defective activity of a protein that is critical for early embryo development or a cumulative effect of an overall reduction in the activities of many Cu-dependent proteins remains to be elucidated. Furthermore, given the role of Ctr1 in maintaining normal Cu levels, it will be important to determine whether Cu acquisition through Ctr1 plays a role in the initiation or progression of Cu-related diseases including Alzheimer's disease, amyotrophic lateral sclerosis, or transmissible spongiform encephalopathies (48–51).
Acknowledgments
We thank Jonathan Gitlin and colleagues for sharing information before publication and for insightful discussions; Linda Samuelson, Sally Camper, Thom Saunders, and Patrick Gillespie of the University of Michigan Transgenic Animal Facility for invaluable advice; Sue O'Shea for advice on mouse histological analysis; the Mallinckrodt Institute of Radiology at Washington University for 64Cu; and Miranda Lau, Chen Kuang, and the Histology Core Facility at the University of Michigan School of Dentistry for excellent technical assistance. We gratefully acknowledge Andras Nagy for the R1 embryonic stem cell line and Richard Mulligan for the plasmid pPNT. This work was supported in part by National Institutes of Health Grant GM62555, a grant from the International Copper Association (to D.J.T.), and American Heart Association Postdoctoral Fellowship 9920536 (to J.L.).
Abbreviations
- ES
embryonic stem
- SOD
superoxide dismutase
- COX
cytochrome oxidase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 6543.
References
- 1.Linder M C. Biochemistry of Copper. New York: Plenum; 1991. [Google Scholar]
- 2.Pena M M O, Lee J, Thiele D J. J Nutr. 1999;129:1251–1260. doi: 10.1093/jn/129.7.1251. [DOI] [PubMed] [Google Scholar]
- 3.DiDonato M, Sarkar B. Biochim Biophys Acta. 1997;1360:3–16. doi: 10.1016/s0925-4439(96)00064-6. [DOI] [PubMed] [Google Scholar]
- 4.Schaeffr M, Gitlin J D. Am J Physiol. 1999;276:G311–G314. doi: 10.1152/ajpgi.1999.276.2.G311. [DOI] [PubMed] [Google Scholar]
- 5.Valentine J S, Gralla E B. Science. 1997;278:817–818. doi: 10.1126/science.278.5339.817. [DOI] [PubMed] [Google Scholar]
- 6.Askwith C, Kaplan J. Trends Biochem Sci. 1998;23:135–138. doi: 10.1016/s0968-0004(98)01192-x. [DOI] [PubMed] [Google Scholar]
- 7.Harrison M D, Jones C E, Solioz M, Dameron C T. Trends Biochem Sci. 2000;25:29–32. doi: 10.1016/s0968-0004(99)01492-9. [DOI] [PubMed] [Google Scholar]
- 8.Culotta V C, Lin S J, Schmidt P, Klomp L W, Casareno R L, Gitlin J. Adv Exp Med Biol. 1999;448:247–254. doi: 10.1007/978-1-4615-4859-1_22. [DOI] [PubMed] [Google Scholar]
- 9.Pufahl R A, Singer C P, Peariso K L, Lin S-J, Schmidt P J, Fahrni C J, Culotta V C, Penner-Haha J E, O'Halloran T V. Science. 1997;278:853–856. doi: 10.1126/science.278.5339.853. [DOI] [PubMed] [Google Scholar]
- 10.Hamza I, Schaefer M, Klomp L W, Gitlin J D. Proc Natl Acad Sci USA. 1999;96:13363–13368. doi: 10.1073/pnas.96.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Askwith C, Eide D, Ho A V, Bernard P S, Li L, Davis-Kaplan S, Sipe D M, Kaplan J. Cell. 1994;76:403–410. doi: 10.1016/0092-8674(94)90346-8. [DOI] [PubMed] [Google Scholar]
- 12.Casareno R L B, Waggoner D, Gitlin J D. J Biol Chem. 1998;273:23625–23628. doi: 10.1074/jbc.273.37.23625. [DOI] [PubMed] [Google Scholar]
- 13.Glerum D M, Shtanko A, Tzagoloff A. J Biol Chem. 1996;271:20531–20535. doi: 10.1074/jbc.271.34.20531. [DOI] [PubMed] [Google Scholar]
- 14.Papadopoulou L C, Sue C M, Davidson M M, Tanji K, Nishino I, Sadlock J E, Krishna S, Walker W, Selby J, Glerum D M, et al. Nat Genet. 1999;23:333–337. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 15.Valnot I, Osmond S, Gigarel N, Mehaye B, Amiel J, Cormier-Daire V, Munnich A, Bonnefont J-P, Rustin P, Rotig A. Am J Hum Genet. 2000;67:1104–1109. doi: 10.1016/s0002-9297(07)62940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hassett R, Kosman D J. J Biol Chem. 1995;270:128–134. doi: 10.1074/jbc.270.1.128. [DOI] [PubMed] [Google Scholar]
- 17.Martins L J, Jensen L T, Simons J R, Keller G L, Winge D R. J Biol Chem. 1998;273:23716–23721. doi: 10.1074/jbc.273.37.23716. [DOI] [PubMed] [Google Scholar]
- 18.Dancis A, Yuan D S, Haile D, Askwith C, Eide D, Moehle C, Kaplan J, Klausner R D. Cell. 1994;76:393–402. doi: 10.1016/0092-8674(94)90345-x. [DOI] [PubMed] [Google Scholar]
- 19.Knight S A B, Labbe S, Kwon L F, Kosman D J, Thiele D J. Genes Dev. 1996;10:1917–1929. doi: 10.1101/gad.10.15.1917. [DOI] [PubMed] [Google Scholar]
- 20.Labbe S, Pena M M O, Fernandes A R, Thiele D J. J Biol Chem. 1999;274:36252–36260. doi: 10.1074/jbc.274.51.36252. [DOI] [PubMed] [Google Scholar]
- 21.Zhou B, Gitschier J. Proc Natl Acad Sci USA. 1997;94:7481–7486. doi: 10.1073/pnas.94.14.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Prohaska J R, Dagenais S L, Glover T W, Thiele D J. Gene. 2000;254:87–96. doi: 10.1016/s0378-1119(00)00287-0. [DOI] [PubMed] [Google Scholar]
- 23.Tybulewicz V L J, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 24.Nagy A, Rossant J, Nagy R, Abranmow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 26.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 27.Prohaska J R, Bailey W R. J Nutr. 1993;123:1226–1234. doi: 10.1093/jn/123.7.1226. [DOI] [PubMed] [Google Scholar]
- 28.Prohaska J R. J Nutr. 1991;121:355–363. doi: 10.1093/jn/121.3.355. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan J, O'Halloran T V. Science. 1996;271:1510–1512. doi: 10.1126/science.271.5255.1510. [DOI] [PubMed] [Google Scholar]
- 30.Vulpe C D, Kuo Y-M, Murphy T L, Cowley L, Askwith C, Libina N, Gitschier J, Anderson G J. Nat Genet. 1999;21:195–199. doi: 10.1038/5979. [DOI] [PubMed] [Google Scholar]
- 31.Moller L B, Petersen C, Lund C, Horn N. Gene. 2000;257:13–22. doi: 10.1016/s0378-1119(00)00394-2. [DOI] [PubMed] [Google Scholar]
- 32.Lee G R, Nacht S, Lukens J N, Cartwright G E. J Clin Invest. 1968;47:2058–2069. doi: 10.1172/JCI105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stearman R, Yuan D S, Yamaguchi-Iwai Y, Klausner R D, Dancis A. Science. 1996;271:1552–1557. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- 34.Hassett R, Dix D R, Eide D J, Kosman D J. Biochem J. 2000;351:477–484. [PMC free article] [PubMed] [Google Scholar]
- 35.Kampfenkel K, Kushnir S, Babiychuk E, Inze D, Montagu M V. J Biol Chem. 1995;270:28479–28486. doi: 10.1074/jbc.270.47.28479. [DOI] [PubMed] [Google Scholar]
- 36.Liu X F, Supek F, Nelson N, Culotta V C. J Biol Chem. 1997;272:11763–11769. doi: 10.1074/jbc.272.18.11763. [DOI] [PubMed] [Google Scholar]
- 37.Gunshin H, Mackenzie B, Berger U V, Gunshin Y, Romero M F, Boron W F, Nussberger S, Gollan J L, Hediger M A. Nature (London) 1997;383:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 38.Hunt D M. Nature (London) 1974;249:852–854. doi: 10.1038/249852a0. [DOI] [PubMed] [Google Scholar]
- 39.Hamza I, Faisst A, Prohaska J, Chen J, Gruss P, Gitlin J D. Proc Natl Acad Sci USA. 2001;98:6848–6852. doi: 10.1073/pnas.111058498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas S A, Matsumoto A M, Palmiter R D. Nature (London) 1995;374:643–646. doi: 10.1038/374643a0. [DOI] [PubMed] [Google Scholar]
- 41.Ho Y-S, Gargano M, Cao J, Bronson R T, Heimler I, Hutz R J. J Biol Chem. 1998;273:7765–7769. doi: 10.1074/jbc.273.13.7765. [DOI] [PubMed] [Google Scholar]
- 42.Harris J L, Durley A P, Man T K, Gitlin D J. Proc Natl Acad Sci USA. 1999;96:10812–10817. doi: 10.1073/pnas.96.19.10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui J, O'Shea K S, Purkayastha A, Saunders T L, Ginsburg D. Nature (London) 1996;384:66–68. doi: 10.1038/384066a0. [DOI] [PubMed] [Google Scholar]
- 44.Buiakova O I, Xu J, Lutsenko S, Zeitlin S, Das K, Das S, Ross B M, Mekios C, Scheinberg H, Gilliam T C. Hum Mol Genet. 1999;8:1665–1671. doi: 10.1093/hmg/8.9.1665. [DOI] [PubMed] [Google Scholar]
- 45.Theophilos M B, Cox D W, Mercer J F. Hum Mol Genet. 1996;5:1619–1624. doi: 10.1093/hmg/5.10.1619. [DOI] [PubMed] [Google Scholar]
- 46.Wong P C, Waggoner D, Subramaniam J R, Tessarollo L, Bartnikas T B, Cullotta V C, Price D L, Rothstein J, Gitlin J D. Proc Natl Acad Sci USA. 2000;97:2886–2891. doi: 10.1073/pnas.040461197. . (First Published August 15, 2000; 10.1073/pnas.170280397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolhekar K S, Roberts M S, Jiang N, Johnson R C, Mains R E, Eipper B A, Taghert P H. J Neurosci. 1997;17:1363–1376. doi: 10.1523/JNEUROSCI.17-04-01363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Multhaup G, Schlicksupp A, Hesse L, Beher D, Ruppert T, Masters C L, Beyreuther K. Science. 1996;271:1406–1409. doi: 10.1126/science.271.5254.1406. [DOI] [PubMed] [Google Scholar]
- 49.Wiedau-Pazos M, Goto J J, Rabizadeh S, Gralla E B, Roe J A, Lee M K, Valentine J S, Bredesen D E. Science. 1996;271:515–518. doi: 10.1126/science.271.5248.515. [DOI] [PubMed] [Google Scholar]
- 50.Brown D R, Qin K, Herms J W, Madlung A, Manson J, Strome R, Fraser P E, Kruck T, Bohlen A V, Schulz-Schaeffer W, et al. Nature (London) 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 51.Bush A I. Cur Opin Chem Biol. 2000;4:184–191. doi: 10.1016/s1367-5931(99)00073-3. [DOI] [PubMed] [Google Scholar]