Abstract
The nuclear receptor peroxisome proliferator-activated receptor gamma (PPARγ) is required for differentiation and function of mature adipocytes. Its expression is induced during adipogenesis where it plays a key role in establishing the transcriptome of terminally differentiated white fat cells. Here, we review findings indicating that PPARγ expression and activity are intricately regulated through control of chromatin structure. Hierarchical and combinatorial activation of transcription factors, noncoding RNAs, and chromatin remodelers allows for temporally controlled expression of PPARγ and its target genes through sequential chromatin remodelling. In obesity, these regulatory pathways may be altered and lead to modified PPARγ activity.
1. Introduction
Peroxisome proliferator-activated receptor gamma (PPARγ) is a transcription factor (TF) belonging to the superfamily of nuclear receptors. PPARγ has a well-established central role in differentiation and function of mature adipocytes [1–5]. This nuclear receptor is activated by endogenous ligands such as nitrated linoleic acid or oxidized fatty acids (9- and 13-HODE), prostaglandin J2 as well as by various synthetic ligands, including the insulin sensitizers pioglitazone and rosiglitazone [6]. Indeed, activation of PPARγ by the thiazolidinediones (TZDs) pioglitazone or rosiglitazone improves insulin resistance associated with obesity and diabetes [7]. Recent studies suggest that this effect is mainly mediated through activation of PPARγ in adipocytes [4, 8], although studies also suggest that macrophage [9] and brain PPARγ [10] contributes to this therapeutic effect.
Transcriptional regulation in mammalian cells is intimately linked to the genomic organisation of the DNA in a highly dynamic chromatin structure. Indeed, chromatin intrinsically impedes transcription factor access to the DNA. This is illustrated by the finding that TFs bind to a highly limited number of potential response elements within the genome of eukaryotic cells [11]. Typically, chromatin regions bound by TFs are devoid of nucleosomes since they can mask their DNA recognition motifs [12]. An additional layer of regulation is provided by epigenomic signatures including histone variant incorporation, histone posttranslational modifications as well as DNA (hydroxy)methylation. Transcriptional regulators and epigenetic modifications mutually regulate each other in order to achieve proper cell type and environment-specific usage of all functional DNA sites embedded within the genome [13–15]. Hence, transcriptional regulatory regions are characterized by different epigenetic signatures when inactive, poised, or active [16, 17].
In this context, results from several biochemical and genetic studies led to the concept that many transcription factors, including nuclear receptors, require presetting of the chromatin for binding to their response elements in the DNA sequence [13, 15]. This consists of preliminary remodelling of the chromatin landscape allowing subsequent TF binding. This process involves so-called pioneer factors that can be recruited to nucleosomal DNA. Such pioneer factors would be required to initiate chromatin remodelling and competency of enhancers that are subsequently used by TFs to mediate transcriptional regulatory signals. Strikingly, some TFs bearing these functions can remain associated with mitotic chromatin suggesting they could bookmark regulatory sites and represent “epigenetic cues” [13, 15].
During adipocyte differentiation, chromatin remodelling events take place to allow proper PPARγ expression and activity. We review here our current knowledge of an integrated control of PPARγ expression and function in adipocytes emphasizing the central role of chromatin remodelling.
2. Regulation of PPARγ Gene Expression in Adipocytes
2.1. Transcriptional Regulation of PPARγ
The study of adipogenesis and adipocyte functions has benefited from cellular models which can be easily manipulated in vitro [18, 19]. Numerous studies made use of the mouse preadipocyte 3T3-L1 cell-line, which can adopt an adipocyte-like phenotype with lipid droplet accumulation upon stimulation with a cocktail of adipogenic inducers (isobutylmethylxanthine (IBMX), dexamethasone, and insulin). During this process, PPARγ expression is strongly induced. Two isoforms of PPARγ are encoded from alternative promoters in the mouse, namely, PPARγ1 and PPARγ2. While PPARγ1 is found in numerous tissues, PPARγ2 expression is mostly restricted to white and brown adipose tissues [20]. PPARγ2 possesses 30 additional aminoacids, which renders the PPARγ aminoterminal transactivation domain more active [21, 22]. Thus, while both PPARγ isoforms can induce adipogenesis, PPARγ2 is thought to play a dominant role in this process [23, 24].
Very early during adipogenesis, the expression of ecotropic viral integration site 1 (Evi1), CCAAT/enhancer binding protein (C/EBP) β and C/EBPδ is induced [25, 26]. This results in expression of low levels of the two PPARγ isoforms and of C/EBPα [25, 26] maintained in a repressed state in preadipocytes by the transcriptional corepressor SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) [27]. PPARγ and C/EBPα can then induce each other's expression in a positive feedback loop promoting and maintaining the differentiated state of the adipocyte [3]. Interestingly, genomic profiling of PPARγ binding sites in adipocytes has revealed that it is present both at the PPARγ2 promoter and at potential enhancers in the vicinity or within its own gene [28, 29]. Expression of PPARγ also requires the activity of the krüppel-like factors 5 and 15 (KLF5 and KLF15) secondarily to their induction by C/EBP transcription factors [30, 31]. Additionally, the transcription factors nuclear family I (NFI) and nuclear factor E2-related factor 2 (Nrf2) regulate both C/EBPα and PPARγ during adipogenesis most probably through direct binding to the PPARγ1 and PPARγ2 promoter, respectively [32, 33]. Additionally, NFI could exert its activities through binding to enhancers within or near both genes [33].
Gene expression is induced by TFs and their cofactors through chromatin remodelling events triggered by cofactor enzymatic activities catalyzing histone and DNA modifications [13–15]. Indeed, the transcriptional activation of PPARγ during adipogenesis correlates with an epigenetic switch at the PPARγ gene. For instance, adipocyte differentiation is associated with a strong increase in levels of histone activation marks at the two PPARγ promoters. This includes acetylation of histone H3 lysine 27 (H3K27ac) and methylation of H3K4 (H3K4me2/3) and H4K20 (H4K20me1) [29, 34]. H3K27ac, which is catalyzed by the transcriptional coactivators CREB-binding protein (CBP) and p300 [35] and typically found at active transcriptional regulatory regions, also increases at enhancers within or near the PPARγ gene [28]. Activation of the PPARγ promoters is also associated with the removal of repressive marks including H3K9me2 and H3K27me3 [17, 34, 36, 37]. The switch from methylation to acetylation at H3K27 could therefore represent a point of integration between activating and repressing signals. Concomitantly, demethylation of the PPARγ2 promoter, which leads to the release of the transcriptional inhibitor methyl CpG-binding protein 2 (MeCP2), occurs gradually during differentiation paralleling the continuing rise in PPARγ2 mRNA expression [34, 38].
These epigenetic changes create an environment competent for gene induction. However, additional remodelling is required. Indeed, PPARγ promoters also undergo chromatin reconfiguration through the binding of the nucleosome-remodeling complex switch/sucrose non-fermentable (SWI/SNF) [39]. SWI/SNF is required neither for epigenetic changes nor for general TF recruitment, but promotes transcription elongation [39]. Hence, PPARγ induction is a multistep process in which sequential chromatin remodelling events eventually lead to the release of stalled RNA polymerase II. Control of transcription elongation through modulation of RNA polymerase II release from promoters has recently emerged as a central mechanism governing developmental gene expression [40]. Like developmental gene promoters in pluripotent cells [41], the PPARγ1 promoter bears H3K4me3 in preadipocytes [28], which could facilitate its induction during differentiation [41]. RNA polymerase II stalling is not a mere consequence of transcription regulation but is by itself an integral part of gene regulation by competing with nucleosomes at promoters and therefore setting the ground for induction [40, 42]. Finally, release of stalled RNA polymerase II results in trimethylation of H3K36 within the PPARγ gene, a feature of actively transcribed regions (Figure 1) [28, 43].
Figure 1.
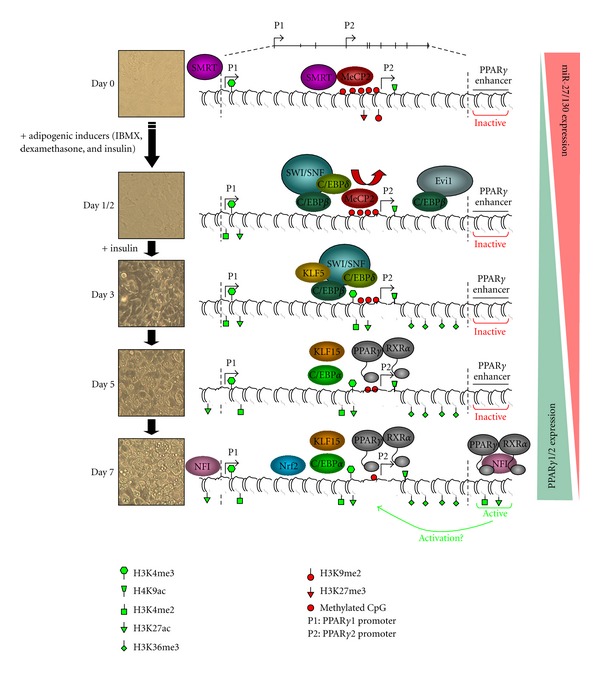
Chromatin-based regulation of PPARγ expression in adipogenesis. Schematic describing epigenomic events as well as sequential transcription factor binding involved in the regulation of PPARγ1/2 expression during adipogenesis. Expression of miR-27/130 targeting PPARγ is also indicated.
The function of adipose tissues is severely altered in obesity [44]. However, this does not stem from a reduced expression of PPARγ, which remains unchanged or increased in adipose tissues from obese rats, mice, and humans [45–48]. Accordingly, knock-out of Nrf2 decreases PPARγ expression, impairs adipogenesis, and protects mice from obesity [32]. Sustained expression of PPARγ in WAT of obese mice may involve a decrease in levels of the orphan nuclear receptor chicken ovalbumin upstream promoter transcription factor II (COUP-TFII), which represses transcription of the PPARγ gene by bringing SMRT and decreasing histone acetylation levels at its promoters [49]. Note however that COUP-TFII role in adipogenesis is still to be clarified since contradictory results have been reported [50, 51]. In this context, perturbation of adipocyte functions in obesity might be linked essentially to modified rather than deficient PPARγ transcriptional regulatory activities as discussed hereafter. Alternatively, altered adipogenesis might be linked to the concomitant perturbed expression of other genes controlling adipocyte differentiation and functions such as genes of the Wnt, Notch, and Sonic Hedgehog signaling pathways [47]. On the contrary to what was observed in obesity, PPARγ expression is decreased in visceral adipose tissues of mouse models of diabetes (db/db), which may directly affect adipocyte differentiation and/or function. The authors showed that this decrease in PPARγ expression is linked to DNA methylation of its promoter [34].Interestingly, recent findings have highlighted a link between epigenetics and metabolism [52] showing that altered metabolism can lead to changes in activity of chromatin-modifying enzymes [53, 54]. Whether and how this could participate in PPARγ abnormal expression in adipocytes remain to be investigated.
2.2. Posttranscriptional Regulation of PPARγ Gene Expression in Adipocytes: Role of miRNA
Epigenomic transitions during adipogenesis often occur at regions distinct from promoters of annotated coding genes [28, 33, 55]. While some of these regions have been defined as enhancers regulating these genes, others are most probably linked to modulation of noncoding RNA (ncRNA) expression. A growing body of evidence points to a major role for ncRNAs in the control of cellular differentiation. Among those are microRNAs (miRNAs), which are short (~22 nucleotides) ncRNAs that posttranscriptionally repress gene expression [56]. By pairing to partially complementary sites in target mRNAs, miRNAs trigger their degradation and/or repress their translation [57]. Several miRNAs play key roles in the control of adipogenesis and adipocyte functions acting as pro- or antiadipogenic factors including miR-30 [58], miR-21 [59], and miR-637 [60] (for review [61, 62]).
Among those, miR-27a/b [63–65] and miR-130a/b [66] are negative regulators of terminal adipocyte differentiation. This inhibition of adipogenesis stems, at least in part, from their ability to prevent the transcriptional induction of PPARγ in preadipocytes. In line, expression of these miRNAs is downregulated during adipogenesis. Both miR-27a/b and miR-130a/b directly target the 3′-untranslated region (3′-UTR) of PPARγ [63, 64, 66]. Additionally, miR-130a/b could also recognize a sequence within the coding region of PPARγ [66].
Interestingly, in agreement with their negative effect on PPARγ expression observed in vitro, miR-130a/b expression correlates inversely with PPARγ expression and BMI (body mass index) in abdominal fat depots of female subjects [66]. On the other hand, in contrast to their opposite expression observed during adipogenesis in vitro, miR-27a/b and PPARγ are both increased in epididymal fat pads from obese mice (ob/ob) [65]. Therefore, while these studies demonstrate that miR-130a/b play a key role in post-transcriptional regulation of PPARγ expression in adipogenesis and obesity, additional work is required to clarify the role of miR-27a/b in these processes.
The 5′- and 3′-UTRs of PPARγ mRNA are relatively short (173 and 211 nucleotides long, resp.), which may exclude interaction with a large number of miRNAs [67, 68]. However, since miRNAs can simultaneously target several mRNAs within defined gene networks [69], it would be interesting to analyze whether some additional miRNAs, among those regulating adipogenesis [62], also target PPARγ. Additionally, some miRNAs controlling adipogenesis indirectly regulate PPARγ expression. For instance, miR-31 and miR-155 negatively impact on adipogenesis by directly targeting C/EBPα and C/EBPβ mRNA, respectively, which is secondarily associated with a decrease in PPARγ expression levels [70, 71].
3. Chromatin-Based Regulation of PPARγ Activity in Adipocytes
3.1. PPARγ Transcriptional Activities Require Chromatin Presetting
Recent insights in our understanding of the transcriptional mechanisms controlling adipogenesis indicate that, reminiscent of other nuclear receptors, PPARγ activities require chromatin presetting. For instance, C/EBPβ can bind to condensed chromatin in preadipocytes and trigger interdependent recruitment of additional TFs including the glucocorticoid receptor (GR), signal transducers and activators of transcription 5 (STAT5), retinoid-X-receptor (RXR) and C/EBPδ to alleviate the repression exerted by SMRT [27, 55, 72]. Altogether, these factors are thought to induce early chromatin opening at enhancers allowing their replacement by PPARγ (and C/EBPα) in more mature adipocytes [55, 73, 74]. Consequently, PPARγ binds to enhancers characterized by early nucleosome depletion and presence of histone posttranslational modifications typical of competent/active sites (methylation of histone H3 lysine 4 (H3K4me) and acetylation of histone H3 lysine 9 (H3K9ac)) [28, 33]. These chromatin-based regulatory mechanisms are most probably involved in defining the adipocyte-specific PPARγ transcriptional activities. Indeed, cell type-specific PPARγ binding to chromatin and gene expression regulatory activities depend on different cell-specific collaborating TFs. For example, PPARγ is recruited to enhancers that bind the pioneer factor PU.1 in macrophages [75].
In addition to enhancers whose chromatin is preset early, PPARγ also binds to many enhancers where chromatin remodelling occurs during adipocyte differentiation [55]. In this case, how PPARγ is directed to these regulatory regions and whether chromatin modifications precede or correlate with its recruitment are not clear yet.
3.2. Control of Chromatin Structure by PPARγ
Even though recent studies highlight the need for chromatin presetting in the regulation of PPARγ transcriptional activities, PPARγ activation in turn also leads to additional posttranslational histone modifications. Indeed, PPARγ activation triggers an exchange of interacting cofactors from corepressors to coactivators. These complexes bear enzymatic activities targeting histone acetylation and methylation. For instance, adipogenic differentiation is linked to a shift from complexes containing histone deacetylase (HDAC) to complexes containing hisone acetyltransferase (HAT) activities [76]. This exchange is observed during adipogenesis as well as upon activation of PPARγ with synthetic agonists leading, for example, to increased acetylation of H3K9 at enhancers [75]. It emerges therefore that the activity of PPARγ-dependent enhancers is controlled through sequential stages of chromatin remodelling linked to the hierarchical binding of TFs and cofactors. Chromatin remodelling at these enhancers also involves hydroxymethylation of cytosines through mechanisms that remain to be elucidated [77].
Unlike enhancers whose accessibility is highly variable and cell-type specific, promoters generally lie in open and nucleosome free chromatin regardless of the cell type [78, 79]. However, promoters are controlled by epigenetic modifications that are partly different from those operating at enhancers [79]. For instance, in addition to chromatin remodelling taking place at enhancers, gene activation during adipogenesis also involves modifications at their promoters. This includes methylation of H4K20 by the SET domain containing lysine methyltransferase 8 (Setd8), an enzyme whose expression is induced by PPARγ resulting in the activation of target gene promoters [29]. PPARγ-mediated gene activation during adipogenesis also requires the mediator complex [80]. This complex not only serves as a platform for recruitment of general TFs and RNA polymerase II but can also recruit chromatin remodelers such as chromodomain helicase DNA-binding protein 1 (CHD1) [81]. Altogether, these studies indicate that activation of PPARγ target genes involves a coordinated remodelling of chromatin at both enhancers and promoters. In this context, PPARγ-mediated regulation could involve a defined three-dimensional organisation of chromatin allowing enhancers and promoters to interact, reminiscent of gene activation by other nuclear receptors such as the estrogen receptor α [82, 83]. Importantly, PPARγ-bound enhancers could regulate the expression of coding genes important for adipocyte functions both directly and/or indirectly by modulating the levels of miRNAs that control adipogenesis including miR-103 [84, 85]. Obesity leads to altered gene expression profiles in adipose tissue [86, 87]. PPARγ transcriptional activity is exacerbated in obese compared to lean visceral WAT [46]. PPARγ binds to DNA as a heterodimer with RXR, a nuclear receptor subfamily consisting of the isotypes RXRα, β, and γ. Interestingly, RXRα protein expression levels are specifically downregulated in visceral white adipose tissue of obese mice and humans through proteasomal degradation. This leads to reduced proportions of the RXRα-PPARγ heterodimer and, as a result, to an increased sensitivity to PPARγ agonists, since the SMRT corepressor is more readily dismissed from the remaining RXRβ-PPARγ complex. Even though not formally demonstrated, the effect of RXRα on the interaction with SMRT most probably influences the chromatin structure resulting in a blunted response to PPARγ agonists due to enhanced interaction with HDACs [46]. In addition, the expression of Setd8, which is also increased in white adipose tissue of obese mice [29], may also participate in the strong PPARγ transcriptional response of adipocytes from obese subjects through increased histone methylation.
4. An Integrated View of PPARγ Regulation in Adipocytes
Taken as a whole, regulation of PPARγ expression and activity is a highly integrative process defining an adipogenic transcriptional network involving key cross-regulatory loops between its members (Figure 2). Epigenomic transitions during adipocyte differentiation allow for both temporallycontrolled induction of PPARγ expression and subsequent regulation of its target genes. Chromatin presetting is observed both at PPARγ and its target genes in preadipocytes and exploited to implement the adipogenic transcriptional program. This process is regulated by an intricate network of hierarchical and combinatorial transcriptional regulatory events. For instance, the transcription factor C/EBPβ plays a pioneer role early in the course of adipogenesis to induce expression of PPARγ, C/EBPα, and KLF5 and 15, which subsequently collaborate to maintain their own expression and activate adipogenic genes. Importantly, this activation requires presetting of chromatin at enhancers by C/EBPβ [55]. Adipogenesis also involves inactivation of repressors, including miR-27a/b, which target both C/EBPα and PPARγ in preadipocytes [63–65]. Among PPARγ target genes are chromatin remodelers including the histone methyltransferase Setd8 [29] and transducin-like enhancer of split 3 (TLE3) [88]. Both factors are involved in a positive autoregulatory loop with PPARγ simultaneously maintaining its high level of expression and induction of target gene expression. While TLE factors are known to have chromatin remodelling activities [89, 90], how TLE3 induces gene expression in adipocytes has not yet been characterized. Overall, PPARγ in adipocytes is therefore controlled by an intricate network of transcriptional regulators that notably license the chromatin structure to allow for appropriate expression and activity of this nuclear receptor.
Figure 2.
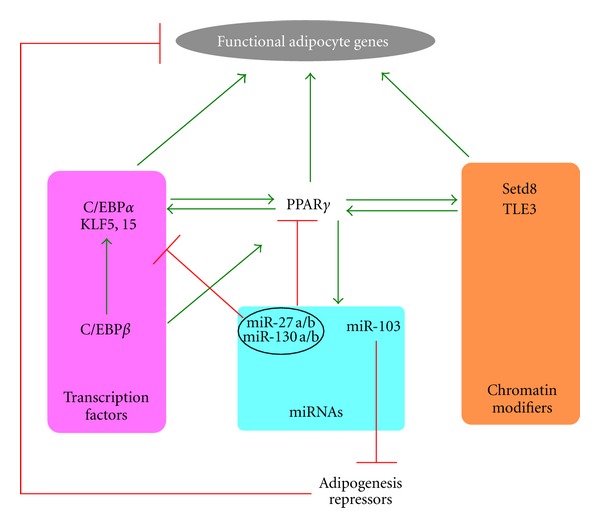
Integrated regulation of PPARγ expression and activity in adipocytes. Schematic showing transcriptional regulators involved in control of PPARγ expression and activity in adipocytes. Green arrows, positive regulation; red bars, negative regulation. See text for further details.
5. Concluding Remarks and Perspectives
We envision that future studies will extend the adipogenic transcriptional network by identifying additional transcriptional regulators and chromatin-associated events controlling PPARγ activities. In addition to a better definition of epigenetic marks involved in this process, studies aimed at identifying mechanisms required for long-range activities of PPARγ-bound enhancers are awaited. This will include a thorough description of the three-dimensional organisation of the chromatin during adipogenesis. Chromosome conformation capture (3C) and its derivatives are recent approaches that have refined our view of the genome spatial organisation and that may prove useful [91]. Taking into account the large diversity of newly discovered ncRNA species, the role of these RNAs in adipogenesis and control of PPARγ is likely at its infancy. Another major question regarding the control of PPARγ activity during adipogenesis relates to the identification of physiologically relevant endogenous ligands [6]. This also requires a better understanding of regulation of PPARγ activities by alternative mechanisms including notably posttranslational modifications [92–95]. Finally, how these regulatory pathways are affected in pathophysiological conditions, such as obesity, will also deserve to be further addressed in order to improve PPARγ targeting strategies. In this context, we will need to better understand the consequences of metabolic perturbations on the enzymatic activities of chromatin modifiers and their consequences for gene transcriptional regulation [52].
Authors' Contribution
J. Eeckhoute and F. Oger contributed equally as first co-authors.
Acknowledgments
This work was supported by grants from “European Genomic Institute for Diabetes” (E.G.I.D., ANR-10-LABX-46). F. Oger was supported by OSEO-ANVAR (IT-DIAB). B. Staels is a member of the Institut Universitaire de France.
References
- 1.Barak Y, Nelson MC, Ong ES, et al. PPARγ is required for placental, cardiac, and adipose tissue development. Molecular Cell. 1999;4(4):585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 2.Imai T, Takakuwa R, Marchand S, et al. Peroxisome proliferator-activated receptor γ is required in mature white and brown adipocytes for their survival in the mouse. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(13):4543–4547. doi: 10.1073/pnas.0400356101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosen ED, Hsu CH, Wang X, et al. C/EBPα induces adipogenesis through PPARγ: a unified pathway. Genes and Development. 2002;16(1):22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugii S, Olson P, Sears DD, et al. PPARγ activation in adipocytes is sufficient for systemic insulin sensitization. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(52):22504–22509. doi: 10.1073/pnas.0912487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 6.Schupp M, Lazar MA. Endogenous ligands for nuclear receptors: digging deeper. The Journal of Biological Chemistry. 2010;285(52):40409–40415. doi: 10.1074/jbc.R110.182451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARγ . Annual Review of Biochemistry. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 8.Li P, Fan W, Xu J, et al. Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPARγ activity and insulin sensitivity. Cell. 2011;147(4):815–826. doi: 10.1016/j.cell.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hevener AL, Olefsky JM, Reichart D, et al. Macrophage PPARγ is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. The Journal of Clinical Investigation. 2007;117(6):1658–1669. doi: 10.1172/JCI31561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu M, Sarruf DA, Talukdar S, et al. Brain PPAR-γ promotes obesity and is required for the insuling-sensitizing effect of thiazolidinediones. Nature Medicine. 2011;17(5):618–622. doi: 10.1038/nm.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eeckhoute J, Métivier R, Salbert G. Defining specificity of transcription factor regulatory activities. Journal of Cell Science. 2009;122(22):4027–4034. doi: 10.1242/jcs.054916. [DOI] [PubMed] [Google Scholar]
- 12.Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474(7352):516–522. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magnani L, Eeckhoute J, Lupien M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends in Genetics. 2011;27(11):465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Minard ME, Jain AK, Barton MC. Analysis of epigenetic alterations to chromatin during development. Genesis. 2009;47(8):559–572. doi: 10.1002/dvg.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes and Development. 2011;25(21):2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature Genetics. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nature Reviews Molecular Cell Biology. 2011;12(11):722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poulos SP, Dodson MV, Hausman GJ. Cell line models for differentiation: preadipocytes and adipocytes. Experimental Biology and Medicine. 2010;235(10):1185–1193. doi: 10.1258/ebm.2010.010063. [DOI] [PubMed] [Google Scholar]
- 20.Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142(10):4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- 21.Fajas L, Auboeuf D, Raspé E, et al. The organization, promoter analysis, and expression of the human PPARγ gene. The Journal of Biological Chemistry. 1997;272(30):18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 22.Werman A, Hollenberg A, Solanes G, Bjørbæk C, Vidal-Puig AJ, Flier JS. Ligand-independent activation domain in the N terminus of peroxisome proliferator-activated receptor γ (PPARγ). Differential activity of PPARγ1 and -2 isoforms and influence of insulin. The Journal of Biological Chemistry. 1997;272(32):20230–20235. doi: 10.1074/jbc.272.32.20230. [DOI] [PubMed] [Google Scholar]
- 23.Mueller E, ori SD, Aiyer A, et al. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor γ isoforms. The Journal of Biological Chemistry. 2002;277(44):41925–41930. doi: 10.1074/jbc.M206950200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Fu M, Cui T, et al. Selective disruption of PPARγ2 impairs the development of adipose tissue and insulin sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(29):10703–10708. doi: 10.1073/pnas.0403652101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishibashi J, Firtina Z, Rajakumari S, et al. An Evi1-C/EBPβ complex controls peroxisome proliferator-activated receptor γ2 gene expression to initiate white fat cell differentiation. Molecular and Cellular Biology. 2012;32(12):2289–2299. doi: 10.1128/MCB.06529-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh WC, Cao Z, Classon M, McKnight SL. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes and Development. 1995;9(2):168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- 27.Raghav S, Waszak S, Krier I, et al. Integrative genomics identifies the corepressor SMRT as a gatekeeper of adipogenesis through the transcription factors C/EBPβ and KAISO. Molecular Cell. 2012;46(3):335–350. doi: 10.1016/j.molcel.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 28.Mikkelsen TS, Xu Z, Zhang X, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143(1):156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakabayashi KI, Okamura M, Tsutsumi S, et al. The peroxisome proliferator-activated receptor γ/retinoid X receptor α heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Molecular and Cellular Biology. 2009;29(13):3544–3555. doi: 10.1128/MCB.01856-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori T, Sakaue H, Iguchi H, et al. Role of krüppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. The Journal of Biological Chemistry. 2005;280(13):12867–12875. doi: 10.1074/jbc.M410515200. [DOI] [PubMed] [Google Scholar]
- 31.Oishi Y, Manabe I, Tobe K, et al. Krüppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metabolism. 2005;1(1):27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Pi J, Leung L, Xue P, et al. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. The Journal of Biological Chemistry. 2010;285(12):9292–9300. doi: 10.1074/jbc.M109.093955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waki H, Nakamura M, Yamauchi T, et al. Global mapping of cell type-specific open chromatin by FAIRE-seq reveals the regulatory role of the NFI family in adipocyte differentiation. PLoS Genetics. 2011;7(10) doi: 10.1371/journal.pgen.1002311.e1002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujiki K, Kano F, Shiota K, Murata M. Expression of the peroxisome proliferator activated receptor γ gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biology. 2009;7, article 38 doi: 10.1186/1741-7007-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tie F, Banerjee R, Stratton CA, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136(18):3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eeckhoute J, Lupien M, Meyer CA, et al. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Research. 2009;19(3):372–380. doi: 10.1101/gr.084582.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noer A, Lindeman LC, Collas P. Histone H3 modifications associated with differentiation and long-term culture of mesenchymal adipose stem cells. Stem Cells and Development. 2009;18(5):725–736. doi: 10.1089/scd.2008.0189. [DOI] [PubMed] [Google Scholar]
- 38.Sasai N, Defossez PA. Many paths to one goal? The proteins that recognize methylated DNA in eukaryotes. International Journal of Developmental Biology. 2009;53(2-3):323–334. doi: 10.1387/ijdb.082652ns. [DOI] [PubMed] [Google Scholar]
- 39.Salma N, Xiao H, Mueller E, Imbalzano AN. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor γ nuclear hormone receptor. Molecular and Cellular Biology. 2004;24(11):4651–4663. doi: 10.1128/MCB.24.11.4651-4663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145(4):502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 42.Gilchrist DA, Dos Santos G, Fargo DC, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143(4):540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao B, Shibata Y, Strahl BD, Lieb JD. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Molecular and Cellular Biology. 2005;25(21):9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun K, Kusminski CM, Scherer PE. Adipose tissue remodeling and obesity. The Journal of Clinical Investigation. 2011;121(6):2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorla-Bajszczak A, Siegrist-Kaiser C, Boss O, Burger AG, Meier CA. Expression of peroxisome proliferator-activated receptors in lean and obese Zucker rats. European Journal of Endocrinology. 2000;142(1):71–78. doi: 10.1530/eje.0.1420071. [DOI] [PubMed] [Google Scholar]
- 46.Lefebvre B, Benomar Y, Guédin A, et al. Proteasomal degradation of retinoid X receptor α reprograms transcriptional activity of PPARγ in obese mice and humans. The Journal of Clinical Investigation. 2010;120(5):1454–1468. doi: 10.1172/JCI38606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roldan M, Macias-Gonzalez M, Garcia R, Tinahones FJ, Martin M. Obesity short-circuits stemness gene network in human adipose multipotent stem cells. The FASEB Journal. 2011;25(12):4111–4126. doi: 10.1096/fj.10-171439. [DOI] [PubMed] [Google Scholar]
- 48.Vidal-Puig AJ, Considine RV, Jimenez-Liñan M, et al. Peroxisome proliferator-activated receptor gene expression in human tissues: effects of obesity, weight loss, and regulation by insulin and glucocorticoids. The Journal of Clinical Investigation. 1997;99(10):2416–2422. doi: 10.1172/JCI119424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okamura M, Kudo H, Wakabayashi KI, et al. COUP-TFII acts downstream of Wnt/β-catenin signal to silence PPARγ gene expression and repress adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(14):5819–5824. doi: 10.1073/pnas.0901676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, Xie X, Qin J, et al. The nuclear orphan receptor COUP-TFII plays an essential role in adipogenesis, glucose homeostasis, and energy metabolism. Cell Metabolism. 2009;9(1):77–87. doi: 10.1016/j.cmet.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Z, Yu S, Hsu CH, Eguchi J, Rosen ED. The orphan nuclear receptor chicken ovalbumin upstream promoter-transcription factor II is a critical regulator of adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(7):2421–2426. doi: 10.1073/pnas.0707082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148(1-2):24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Pirola L, Balcerczyk A, Tothill RW, et al. Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells. Genome Research. 2011;21(10):1601–1615. doi: 10.1101/gr.116095.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao M, Yang H, Xu W, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes and Development. 2012;26(12):1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siersbfk R, Nielsen R, John S, et al. Extensive chromatin remodelling and establishment of transcription factor hotspots during early adipogenesis. The EMBO Journal. 2011;30(8):1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaikkonen MU, Lam MTY, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovascular Research. 2011;90(3):430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaragosi LE, Wdziekonski B, Le Brigand K, et al. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biology. 2011;12, article R64 doi: 10.1186/gb-2011-12-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim YJ, Hwang SH, Cho HH, Shin KK, Bae YC, Jung JS. MicroRNA 21 regulates the proliferation of human adipose tissue-derived mesenchymal stem cells and high-fat diet-induced obesity alters microRNA 21 expression in white adipose tissues. Journal of Cellular Physiology. 2012;227(1):183–193. doi: 10.1002/jcp.22716. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J-F, Fu W-M, He M-L, et al. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Molecular Biology of the Cell. 2011;22(21):3955–3961. doi: 10.1091/mbc.E11-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hilton C, Neville MJ, Karpe F. MicroRNAs in adipose tissue: their role in adipogenesis and obesity. doi: 10.1038/ijo.2012.59. International Journal of Obesity. In press. [DOI] [PubMed] [Google Scholar]
- 62.Romao JM, Jin W, Dodson MV, Hausman GJ, Moore SS, le Guan L. MicroRNA regulation in mammalian adipogenesis. Experimental Biology and Medicine. 2011;236(9):997–1004. doi: 10.1258/ebm.2011.011101. [DOI] [PubMed] [Google Scholar]
- 63.Karbiener M, Fischer C, Nowitsch S, et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARγ . Biochemical and Biophysical Research Communications. 2009;390(2):247–251. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- 64.Kim SY, Kim AY, Lee HW, et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARγ expression. Biochemical and Biophysical Research Communications. 2010;392(3):323–328. doi: 10.1016/j.bbrc.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 65.Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. A role of miR-27 in the regulation of adipogenesis. FEBS Journal. 2009;276(8):2348–2358. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee EK, Lee MJ, Abdelmohsen K, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor γ expression. Molecular and Cellular Biology. 2011;31(4):626–638. doi: 10.1128/MCB.00894-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng C, Bhardwaj N, Gerstein M. The relationship between the evolution of microRNA targets and the length of their UTRs. BMC Genomics. 2009;10, article 431 doi: 10.1186/1471-2164-10-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang C, Tian R, Zhao Q, et al. Computational inference of mRNA stability from histone modification and transcriptome profiles. Nucleic Acids Research. 2012;40(14):6414–6423. doi: 10.1093/nar/gks304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsu JB, Chiu CM, Hsu SD, et al. miRTar: an integrated system for identifying miRNA-target interactions in Human. BMC Bioinformatics. 2011;12, article 300 doi: 10.1186/1471-2105-12-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu S, Yang Y, Wu J. TNFα-induced up-regulation of miR-155 inhibits adipogenesis by down-regulating early adipogenic transcription factors. Biochemical and Biophysical Research Communications. 2011;414(3):618–624. doi: 10.1016/j.bbrc.2011.09.131. [DOI] [PubMed] [Google Scholar]
- 71.Sun F, Wang J, Pan Q, et al. Characterization of function and regulation of miR-24-1 and miR-31. Biochemical and Biophysical Research Communications. 2009;380(3):660–665. doi: 10.1016/j.bbrc.2009.01.161. [DOI] [PubMed] [Google Scholar]
- 72.Steger DJ, Grant GR, Schupp M, et al. Propagation of adipogenic signals through an epigenomic transition state. Genes and Development. 2010;24(10):1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lefterova MI, Zhang Y, Steger DJ, et al. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes and Development. 2008;22(21):2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nielsen R, Pedersen TÅ, Hagenbeek D, et al. Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes and Development. 2008;22(21):2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lefterova MI, Steger DJ, Zhuo D, et al. Cell-specific determinants of peroxisome proliferator-activated receptor γ function in adipocytes and macrophages. Molecular and Cellular Biology. 2010;30(9):2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sugii S, Evans RM. Epigenetic codes of PPARγ in metabolic disease. FEBS Letters. 2011;585(13):2121–2128. [Google Scholar]
- 77.Sérandour A, Avner S, Oger F, et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. doi: 10.1093/nar/gks595. Nucleic Acids Research. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144(3):327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nature Reviews Genetics. 2011;12(4):283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grøntved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S. MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor γ and is required for full transcriptional activity and adipogenesis. Molecular and Cellular Biology. 2010;30(9):2155–2169. doi: 10.1128/MCB.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin JJ, Lehmann LW, Bonora G, et al. Mediator coordinates PIC assembly with recruitment of CHD1. Genes and Development. 2011;25(20):2198–2209. doi: 10.1101/gad.17554711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fullwood MJ, Liu MH, Pan YF, et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Splinter E, De Laat W. The complex transcription regulatory landscape of our genome: control in three dimensions. The EMBO Journal. 2011;30(21):4345–4355. doi: 10.1038/emboj.2011.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.John E, Wienecke-Baldacchino A, Liivrand M, Heinäniemi M, Carlberg C, Sinkkonen L. Dataset integration identifies transcriptional regulation of microRNA genes by PPARγ in differentiating mouse 3T3-L1 adipocytes. Nucleic Acids Research. 2012;40(10):4446–4460. doi: 10.1093/nar/gks025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58(5):1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guan H, Arany E, Van Beek JP, et al. Adipose tissue gene expression profiling reveals distinct molecular pathways that define visceral adiposity in offspring of maternal protein-restricted rats. American Journal of Physiology. 2005;288(4):E663–E673. doi: 10.1152/ajpendo.00461.2004. [DOI] [PubMed] [Google Scholar]
- 87.Vohl MC, Sladek R, Robitaille J, et al. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obesity Research. 2004;12(8):1217–1222. doi: 10.1038/oby.2004.153. [DOI] [PubMed] [Google Scholar]
- 88.Villanueva CJ, Waki H, Godio C, et al. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metabolism. 2011;13(4):413–427. doi: 10.1016/j.cmet.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Holmes KA, Hurtado A, Brown GD, et al. Transducin-like enhancer protein 1 mediates estrogen receptor binding and transcriptional activity in breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(8):2748–2753. doi: 10.1073/pnas.1018863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jennings BH, Ish-Horowicz D. The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biology. 2008;9(1, article 205) doi: 10.1186/gb-2008-9-1-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes and Development. 2012;26(1):11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Berrabah W, Aumercier P, Lefebvre P, Staels B. Control of nuclear receptor activities in metabolism by post-translational modifications. FEBS Letters. 2011;585(11):1640–1650. doi: 10.1016/j.febslet.2011.03.066. [DOI] [PubMed] [Google Scholar]
- 93.Choi JH, Banks AS, Kamenecka TM, et al. Antidiabetic actions of a non-agonist PPARγ ligand blocking Cdk5-mediated phosphorylation. Nature. 2011;477(7365):477–481. doi: 10.1038/nature10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dutchak PA, Katafuchi T, Bookout AL, et al. Fibroblast growth factor-21 regulates PPARγ activity and the antidiabetic actions of thiazolidinediones. Cell. 2012;148(3):556–567. doi: 10.1016/j.cell.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beekum OV, Fleskens V, Kalkhoven E. Posttranslational modifications of PPAR-γ: fine-tuning the metabolic master regulator. Obesity. 2009;17(2):213–219. doi: 10.1038/oby.2008.473. [DOI] [PubMed] [Google Scholar]


