Abstract
Peptidoglycan polymerization complexes contain multimodular penicillin-binding proteins (PBP) of classes A and B that associate a conserved C-terminal transpeptidase module to an N-terminal glycosyltransferase or morphogenesis module, respectively. In Enterococcus faecalis, class B PBP5 mediates intrinsic resistance to the cephalosporin class of β-lactam antibiotics, such as ceftriaxone. To identify the glycosyltransferase partner(s) of PBP5, combinations of deletions were introduced in all three class A PBP genes of E. faecalis JH2-2 (ponA, pbpF, and pbpZ). Among mutants with single or double deletions, only JH2-2 ΔponA ΔpbpF was susceptible to ceftriaxone. Ceftriaxone resistance was restored by heterologous expression of pbpF from Enterococcus faecium but not by mgt encoding the monofunctional glycosyltransferase of Staphylococcus aureus. Thus, PBP5 partners essential for peptidoglycan polymerization in the presence of β-lactams formed a subset of the class A PBPs of E. faecalis, and heterospecific complementation was observed with an ortholog from E. faecium. Site-directed mutagenesis of pbpF confirmed that the catalytic serine residue of the transpeptidase module was not required for resistance. None of the three class A PBP genes was essential for viability, although deletion of the three genes led to an increase in the generation time and to a decrease in peptidoglycan cross-linking. As the E. faecalis chromosome does not contain any additional glycosyltransferase-related genes, these observations indicate that glycan chain polymerization in the triple mutant is performed by a novel type of glycosyltransferase. The latter enzyme was not inhibited by moenomycin, since deletion of the three class A PBP genes led to high-level resistance to this glycosyltransferase inhibitor.
High-molecular-weight penicillin-binding proteins (PBPs) are the main determinants of clinically relevant β-lactam resistance phenotypes in streptococci, staphylococci, and enterococci, although the genetic basis for resistance differs in these bacteria. Resistant isolates of Streptococcus pneumoniae harbor multiple mosaic PBP genes generated by recombination with gene fragments acquired from related streptococci by natural transformation (19). Methicillin-resistant Staphylococcus aureus (MRSA) strains have acquired an additional PBP gene (mecA) encoding PBP2a, which is sufficient for high-level resistance to virtually all β-lactams in the absence of modification of other PBPs (34). Since MRSA strains grow in the presence of β-lactam antibiotics at concentrations sufficient to rapidly saturate all PBPs except PBP2a, it is assumed that the latter enzyme is sufficient for peptidoglycan cross-linking (26). Similarly, Enterococcus faecalis and Enterococcus faecium strains produce PBP5 that appears sufficient for transpeptidation in the presence of β-lactams (7, 29, 30). Resistance is an intrinsic property of these species, as virtually all isolates are resistant to moderate (e.g., ampicillin) or high (e.g., ceftriaxone) levels of β-lactams and produce a species-specific PBP5 (24). Acquisition of high-level resistance to ampicillin, mainly in clinical isolates of E. faecium, results from overproduction of PBP5 (12, 13, 29) and from amino acid substitutions that decrease the interaction of PBP5 with the drug (27, 35). Alteration of other as-yet-unknown accessory factors is also involved (18, 29).
The high-molecular-weight PBPs fall into two classes based on the association of the conserved C-terminal transpeptidase module with an N-terminal glycosyltransferase module (class A) or a morphogenesis module (class B) devoid of any known catalytic activity (14). As the S. aureus PBP2a and the enterococcal PBP5 are class B PBPs, peptidoglycan polymerization in the presence of high concentrations of β-lactams is thought to require cooperation between the d,d-transpeptidase module of these PBPs and the glycosyltransferase module of class A PBPs. Evidence for such cooperation has been obtained in MRSA strains based on selective inactivation by site-directed mutagenesis of the glycosyltransferase activity of PBP2, which led to a viable mutant susceptible to methicillin (26). Polymerization of the glycan chains in the mutant was presumably catalyzed by a monofunctional glycosyltransferase (Mgt), as PBP2 is the sole class A PBP produced by S. aureus (26, 32). The sets of peptidoglycan-polymerizing enzymes of S. aureus and enterococci are significantly different, since the genome of E. faecalis harbors three putative class A PBP genes, no homologue of mgt, and three putative class B PBP genes (Table 1). To evaluate the role of the three class A PBP genes of E. faecalis in intrinsic β-lactam resistance, we have developed a method to construct multiple deletions in the chromosome of this bacterium. We report the deletion of all combinations of one to three class A PBP genes and their impact on bacterial growth, peptidoglycan cross-linking, and susceptibility to cell wall synthesis inhibitors.
TABLE 1.
Multimodular PBPs of E. faecalis and putative orthologs from various species
| Species | PBP of class and subclass (calculated mass in kDa)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| A
|
B
|
|||||||
| A3 | A4 | A5 | A6 | B1 | B4 | B5 | B3 | |
| E. faecalisb | ponA (85.4) | pbpF (79.5) | pbpZ (88.5) | NP | pbp5 (74.0) | pbpB (81.7) | pbpA (77.9) | NP |
| E. faeciumb | ponA (86.6) | pbpF (79.9) | pbpZ (86.7) | NP | pbp5 (73.7) | pbpB (81.0) | pbpA (78.9) | NP |
| S. aureusc | PBP2 (79.3) | NP | NP | NP | PBP2a (76.3) | PBP1 (82.7) | PBP3 (77.2) | NP |
| S. pneumoniae | PBP1a (79.8) | PBP2a (80.8) | PBP1b (89.5) | NP | NP | PBP2x (82.3) | PBP2b (73.9) | NP |
| B. subtilis | PBP1 (99.6) | PBP2c (79.3) | PBP4 (70.6) | PBP2d (71.8) | PBP3 (74.4) | PBP2b (79.3) | PBP2a (80.1) | PBPVD (71.3) |
Classification is that of Goffin and Ghuysen and is based on amino acid sequence comparison (14). NP, not present.
PBPs of E. faecalis and E. faecium are identified by the gene designation used in the text.
MRSA.
MATERIALS AND METHODS
Growth conditions.
Bacterial strains were grown in brain heart infusion (BHI) broth or agar (Becton Dickinson, le Pont de Claix, France) at 37°C. MICs of ampicillin (Bristol-Myers, Paris, France), ceftriaxone (Laboratoires Roche, Neuilly, France), and moenomycin (Hoechst, Mainz, Germany) were determined with 105 CFU per spot on BHI agar after 48 h of incubation.
Replication and transfer properties of suicide vector pHS1.
The vector pHS1 was constructed to introduce serial deletions in the chromosome of E. faecalis JH2-2 (17) by homologous recombination. This vector (Fig. 1A) is composed of (i) the origin of replication and the repA(Ts) gene encoding the thermosensitive replication protein of plasmid pGhost4 (20), (ii) the aph3′-aac6" bifunctional gene of plasmid pAT392 (2) conferring resistance to all aminoglycosides, including gentamicin, and (iii) the origin of transfer of transposon Tn916 (oriTTn916) allowing conjugal transfer between gram-positive bacteria provided that the donor harbors Tn916 (8). The vector pHS1 and derivatives were propagated at 37°C in Escherichia coli EC101 (5) with selection for gentamicin resistance (16 μg/ml). The plasmids were introduced into E. faecalis JH2Sm::Tn916 (8) by electroporation (10) and maintained in this host at permissive temperature for replication (28°C) in media containing gentamicin (128 μg/ml). Plasmid pHS1 and derivatives were transferred by mating from JH2Sm::Tn916 (resistant to tetracycline and streptomycin) to JH2-2 (resistant to rifampin and fusidic acid). Transconjugants were selected on BHI agar containing rifampin (40 μg/ml), fusidic acid (20 μg/ml), and gentamicin (128 μg/ml). Typical transfer frequencies were in the order of 10−4 per donor.
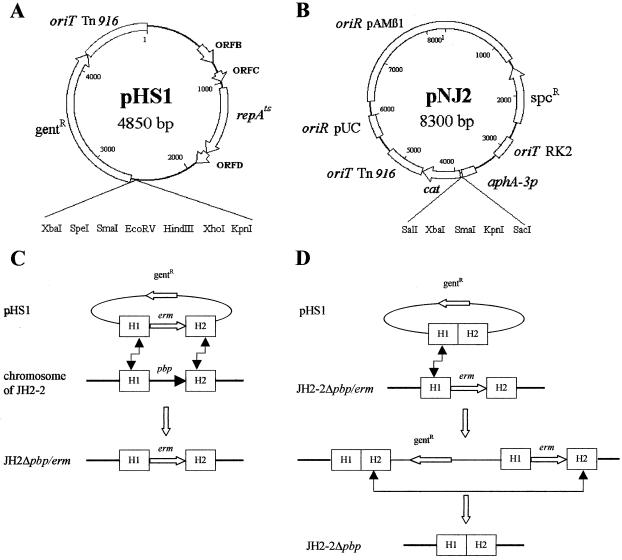
FIG. 1.
Schematic representation of vectors and approach used to generate chromosomal deletions. The maps of suicide vector pHS1 (A) and shuttle vector pNJ2 (B) show unique restriction sites used for cloning. The plasmids confer resistance to gentamicin (gentR) and spectinomycin (spcR), respectively. (C) Replacement of the pbp genes by the erm erythromycin resistance cassette was generated by a double crossover, as indicated by broken arrows. (D) The erm cassette was removed from the chromosome in two steps. In the first step, integration of plasmid pHS1ΩH1-H2 by a single crossover involving H1 (as represented) or H2 was selected at 42°C on agar containing gentamicin. Integration generated a partial duplication of the locus, since the sequence of the pHS1 vector was flanked by the H1-H2 and H1-erm-H2 alleles. Serial subcultures at the permissive (28°C) and nonpermissive (42°C) temperatures in the absence of antibiotic were used to stimulate the excision and loss of pHS1ΩH1-erm-H2, leaving the H1-H2 allele in the chromosome.
Recombination events generating chromosomal deletions.
DNA fragments (ca. 500 bp) flanking the sequence targeted for deletion, designated H1 and H2, were independently amplified by PCR. The H1 and H2 fragments, separated by an erythromycin resistance cassette (erm), were cloned into pHS1. The resulting pHS1 derivatives carrying the H1-erm-H2 insertions were introduced into E. faecalis JH2-2 as described above. Replacement of the sequence targeted for deletion by the erm cassette by a double crossover (Fig. 1C) was selected at the nonpermissive temperature for plasmid replication (42°C) on media containing erythromycin (10 μg/ml).
The erm resistance cassette was removed from the chromosomal pbp loci by using derivatives of pHS1 carrying H1 directly fused to H2 (Fig. 1D). Chromosomal integration of the plasmids by a single crossover was obtained by selecting for gentamicin resistance (128 μg/ml) at nonpermissive temperature for plasmid replication. Plasmid excision was obtained by subculturing clones in the absence of antibiotic at 28°C, as the activity of the Rep(Ts) protein at permissive temperature was reported to stimulate homologous recombination (5). The excised plasmids were cured at 42°C, and clones susceptible to gentamicin and erythromycin were screened by replica plating.
Amplification of H1 and H2 sequences.
The following pairs of oligonucleotides were used to amplify by PCR the H1 and H2 sequences flanking ponA, pbpF, pbpZ, and pbp5: ponA H1, 5′-TTATCCCAAACGAAGTG-3′ and 5′-AGATCTGTGTTGGATGCATGTCT-3′; ponA H2, 5′-AGATCTGCAACCACCTGAAAGTAG-3′ and 5′-TTGTGGGCTTAGAAGATG-3′; pbpF H1, 5′-TTAAGGTGACACAATCG-3′ and 5′-AGATCTTTGTCCATAGTACTCCC-3′; pbpF H2, 5′-AGATCTTGGGACAAATTAAAGACG-3′ and 5′-TATCACGCACAGGAGTC-3′; pbpZ H1, 5′-TGGATCACCAATCATGC-3′ and 5′-AGATCTCAAAAGCTTCACCTCA-3′; pbpZ H2, 5′-AGATCTATTACGCTTCTTACTGG-3′ and 5′-GTTGGTGTGGTATTATC-3′. BglII restriction sites (underlined) were used to ligate the H1, H2, and erm fragments as shown in Fig. 1C and D.
The H1-H2 DNA fragment used for deletion of pbp5 was constructed by two sequential amplifications with partially complementary primers as previous described (1). In the first step, the H1 and H2 fragments were separately amplified (primers H1F, 5′-AGAATCATTTTTGACTG-3′, and H1R, 5′-CAAATGGTTCGCTGGGTTTCAATAATCCCCTAAC-3′, for H1; primers H2Fb, 5′-ACCCAGCGAACCATTTGAAAAGAGAAAATGAACG-3′, and H2R, 5′-AGGGAAATATGTTGGTC-3′, for H2). Seventeen bases of primers H1R and H2Fb were complementary (underlined). In the second step, the H1 and H2 fragments were denatured, annealed, and coamplified with primers H1F and H2R (see above). The same method was used to obtain the H1-erm-H2 fragment with primers H1F, H1R, H2R, and H2F. Primers H1R (see above) and H2F (5′-GTAAGTTAAGGGACTGCAAAAGAGAAAATGAACG-3′) contained sequences complementary to the erm resistance cassette (italicized).
Analysis of the structure of the deletions.
Genomic DNA was prepared (Wizard genomic DNA purification kit; Promega, Madison, Wis.), separately digested with AccI and XmnI, except for pbp5 (digested with PstI and SspI), and analyzed by Southern blot hybridization. The probes were obtained by labeling DNA of derivatives of plasmid pCRblunt (Invitrogen, Carlsbad, Calif.) harboring the H1 and H2 sequences with [α-32P]dCTP (Megaprime DNA labeling system; Amersham Biosciences, Little Chalfont, England). For each of the four PBP genes (ponA, pbpF, pbpZ, and pbp5), the hybridization patterns corresponded to the predicted map of the wild-type locus, pbp replaced by erm, and deletion of the gene (data not shown). In addition, PCR was performed with oligonucleotides adjacent to the H1 and H2 sequences, to confirm the reduction in size of the PCR products resulting from gene replacement and deletion. Finally, the presence of the fused H1 and H2 fragments in the chromosome of the mutants was verified by direct sequencing of the PCR products.
Properties of expression vector pNJ2.
Plasmid pNJ2 was constructed to obtain expression of cloned genes under the control of the heterologous aphA-3p promoter, which is active in E. faecalis (3). This vector (Fig. 1B) is composed of (i) plasmid pAT28 that replicates both in E. coli (oriR pUC) and gram-positive bacteria (oriR pAMβ1), and confers resistance to spectinomycin (31), (ii) the chloramphenicol acetyltransferase gene (cat) and the aphA-3p promoter of plasmid pAT79 (3), and (iii) the origin of transfer of transposon Tn916 (oriTTn916) (8). The vector pNJ2 and its derivatives were introduced into E. faecalis JH2Sm::Tn916 by electroporation and transferred by conjugation to derivatives of E. faecalis JH2-2 (frequency of ca. 10−4 per donor). The plasmids were selected with spectinomycin (60 μg/ml) and chloramphenicol (20 μg/ml). Rifampin (40 μg/ml) and fusidic acid (20 μg/ml) were added to the media for selection of the E. faecalis JH2-2 recipients.
Shuttle plasmids for pbp and mgt expression in E. faecalis.
The pbp5 open reading frame and ribosome binding site of E. faecalis JH2-2 (pbp5fs) was amplified with primers n-PB1 (5′-ATAGGTGAAACACAAGC-3′) and PB2 (5′-ACAGAAACCTGTTTCG-3′) and cloned under the control of the aphA-3p promoter to generate pNJ2Ωpbp5fs. The same approach was used to express the pbpF genes from E. faecalis JH2-2 (pbpFfs) and E. faecium D344S (22) (pbpFfm) and the mgt gene of S. aureus NCTC 8325 (mgtSa). The following primers containing SacI and XbaI sites (underlined) were used for amplification: pbpFfs, primers pbpF1S (5′-GGTGGTGAGCTCTAGACTTAGCCAAGAAACG-3′) and PBPF4S (5′-GGTGGTCTGCAGTCTAGACAACTAATTTCCTAATAAG-3′); pbpFfm, primers D344-F-1 (5′-TTGAGCTCACTACAACTTAAGCAGGA-3′) and D344-F-2 (5′-TTTCTAGAGTAGTTACTCTCTATTGT-3′); mgtSa,primers MGT1 (5′-TTGAGCTCAAGGTATATACTAAGTGAG-3′) andMGT2 (5′-TTTCTAGAGCAAGTATTTAACGATTTAA-3′). DNA sequencing was performed for all recombinant plasmids used in this study to confirm the accuracy of the PCR.
Site-directed mutagenesis of E. faecalis pbpF.
The codon specifying the catalytic serine residue (TCG, Ser402) was replaced by a GGA glycine codon. The 5′ portion of pbpFfs was amplified with pbpF1S (see above) and PBPF2S (5′-GGTGGTGGATCCGCCTGGTGAACGTTTTGTT-3′) to introduce the GGA codon (bold) and a BamHI site (underlined). The 3′ portion of pbpFfs was amplified with PBPF4S (see above) and PBPF3S (5′-GGTGGTGGATCCTTAAAACCAATTTCTG-3′). The PCR fragments were digested with BamHI, ligated, and introduced into pNJ2 to generate plasmid pNJ2ΩpbpFfs(S402-G).
Peptidoglycan structure analysis.
Bacteria were grown at 37°C to an optical density of 0.8 in BHI broth. Peptidoglycan was extracted with 4% sodium dodecyl sulfate at 100°C, treated with pronase (200 μg/ml) and trypsin (200 μg/ml), and digested with lysozyme (200 μg/ml) and mutanolysin (200 μg/ml). Muropeptides were reduced with sodium borohydride and separated by reverse-phase high-performance liquid chromatography on a C18 column (3 μm, 4.6 by 250 mm; Interchrom, Montluçon, France) at a flow rate of 0.5 ml/min with a 0 to 20% gradient applied between 10 and 90 min (buffer A, 0.05% [vol/vol] trifluoroacetic acid in water; buffer B, 0.035% [vol/vol] trifluoroacetic acid in acetonitrile). The relative abundance of muropeptides was estimated by the percentage of the integrated area of peaks detected by the absorbance at 210 nm. Mass spectral data were collected with an electrospray time-of-flight mass spectrometer operating in the positive mode (Qstar Pulsar I; Applied Biosystems, Courtaboeuf, France) directly connected to the C18 column (flow rate, 0.5 ml/min). The data were acquired with a capillary voltage of 5,200 V and a declustering potential of 20 V. The mass scan range was from m/z 400 to 2,500, and the scan cycle was 1 s. Structure assignment of muropeptides based on mass determination was performed as previously described (6).
Analysis of PBPs.
The technique used for the analysis of PBPs of the different strains was as previously described (33), except that labeling was performed with 40 μg of benzyl[14C]penicillin potassium (2.11 GBq/mmol; Amersham Pharmacia Biotech)/ml.
RESULTS
Deletion of the pbp5 gene from the chromosome of E. faecalis JH2-2.
The vector pHS1 was constructed to delete chromosomal PBP genes by homologous recombination as outlined in Fig. 1. E. faecalis JH2-2 was highly resistant to the expanded-spectrum cephalosporin ceftriaxone, and deletion of pbp5 led to a 4,000-fold reduction in the MIC of this drug (Table 2). The strain was less resistant to ampicillin, and deletion of pbp5 produced only a fourfold reduction in the MIC of this antibiotic. The pbp5 gene of E. faecalis JH2-2 cloned under the control of the aphA-3p promoter of the shuttle vector pNJ2 (Fig. 1B) restored wild-type β-lactam resistance in JH2-2 Δpbp5.
TABLE 2.
Susceptibility of E. faecalis strains to inhibitors of peptidoglycan synthesis
| Strain | MIC (μg/ml) of:
|
||
|---|---|---|---|
| Ceftriax- one | Ampi- cillin | Moeno- mycin | |
| JH2-2 | 1,000 | 2 | 0.25 |
| JH2-2 Δpbp5 | 0.25 | 0.5 | 0.25 |
| JH2-2/pNJ2Ωpbp5fs | 1,000 | 2 | 0.25 |
| JH2-2 Δpbp5/pNJ2Ωpbp5fs | 1,000 | 2 | 0.25 |
| JH2-2 ΔponA | 1,000 | 2 | 0.25 |
| JH2-2 ΔpbpF | 1,000 | 2 | 0.25 |
| JH2-2 ΔpbpZ | 1,000 | 2 | 0.25 |
| JH2-2 ΔponA ΔpbpF | 1 | 1 | >128 |
| JH2-2 ΔponA ΔpbpZ | 512 | 2 | 0.25 |
| JH2-2 ΔpbpF ΔpbpZ | 1,000 | 2 | 0.5 |
| JH2-2 ΔponA ΔpbpF ΔpbpZ | 1 | 1 | >128 |
| JH2-2 ΔponA ΔpbpF/pNJ2Ωpbp5fs | 1 | 1 | >128 |
| JH2-2 ΔponA ΔpbpF ΔpbpZ/pNJ2Ωpbp5fs | 1 | 1 | >128 |
Identification of class A PBP genes essential for PBP5-mediated β-lactam resistance.
Single deletion of any of the three class A PBP genes of E. faecalis (ponA, pbpF, or pbpZ) had no impact on the MICs of β-lactam antibiotics (Table 2). Among the three combinations of double deletions, only the deletion of ponA and pbpF resulted in a large decrease (1,000-fold) in ceftriaxone resistance. Thus, either ponA or pbpF was required for intrinsic ceftriaxone resistance mediated by PBP5. Overexpression of the pbp5 gene under the control of the aphA-3p promoter of pNJ2 did not restore ceftriaxone resistance in JH2-2 ΔponA ΔpbpF (Table 2).
Deletion of the three class A PBP genes.
Previous analyses showed that the self-transferable plasmid pIP964(hly) can mobilize chromosomal genes between E. faecalis strains by conjugation (4). To determine whether this system could be used to generate combinations of chromosomal deletions, E. faecalis JH2-2 ΔpbpZ erm/pIP964(hly), obtained by replacing pbpZ with erm, was mated with JH2Sm. Transfer of the hemolysin marker (hly) of pIP964 occurred at a frequency of ca. 10−1, as determined on blood agar indicator plates. The chromosomal erythromycin resistance cassette located at the pbpZ locus was also transferable but at a lower frequency (ca. 10−8). An E. faecalis JH2Sm transconjugant harboring the ΔpbpZ erm allele and pIP964(hly) was in turn used as a donor in mating experiments, with E. faecalis JH2-2 ΔponA ΔpbpF as a recipient. The ponA, pbpF, and pbpZ loci of five transconjugants obtained on selective media containing erythromycin, rifampin, and fusidic acid were analyzed by PCR. As expected, all of them had received the ΔpbpZ erm allele. Cotransfers were observed in two transconjugants that received pbpF alone or ponA and pbpF in addition to ΔpbpZ erm. The remaining three transconjugants acquired the ΔpbpZ erm allele and retained the ΔponA and ΔpbpF alleles of the recipient. The erm cassette was removed from the chromosome of one of these transconjugants to generate JH2-2 ΔponA ΔpbpF ΔpbpZ. Deletion of the three PBP genes was confirmed by Southern blot and PCR analyses (data not shown). This result indicates that none of the three class A PBPs is essential for viability.
Growth rate.
Deletion of the three class A PBP genes led to an increase of the generation time (70.0 ± 4.6 min for JH2-2 ΔponA ΔpbpF ΔpbpZ versus 37.7 ± 1.0 min for JH2-2). The increase in the generation time was marginal (<12%) for the other mutants lacking one or two class A PBP genes.
Susceptibility to moenomycin.
The impact of the deletion of class A PBP genes on the activity of the glycosyltransferase inhibitor moenomycin was studied by the agar dilution method (Table 2). Deletion of ponA and pbpF or of all three class A PBP genes resulted in high-level resistance to moenomycin. This surprising observation indicates that the antibacterial activity of moenomycin requires ponA, pbpF, or both genes.
Analysis of PBP patterns labeled with benzyl[14C]penicillin.
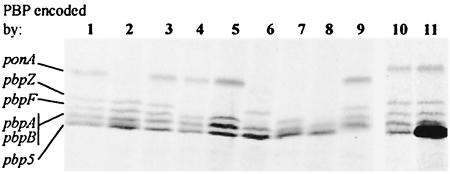
The chromosome of E. faecalis harbors six genes encoding putative multimodular PBPs belonging to class A (ponA, pbpF, and pbpZ) and class B (pbp5, pbpA, and pbpB) (Table 1). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis resolved five high-molecular penicillin-labeled protein bands in membrane extracts from E. faecalis JH2-2 (Fig. 2, lane 1) in addition to the low-molecular weight d,d-carboxypeptidase DacA (data not shown). Based on the analysis of mutants constructed in the present study, three of the five penicillin-labeled protein bands could be assigned to class A PBPs. The band with the lower electrophoretic mobility should correspond to the ponA gene product, since it was absent from JH2-2 ΔponA (lane 2), JH2-2 ΔponA ΔpbpF (lane 6), JH2-2 ΔponA ΔpbpZ (lane 7), and JH2-2 ΔponA ΔpbpF ΔpbpZ (lane 8). The PBP encoded by ponA had a much lower electrophoretic mobility than expected from its calculated molecular mass (Table 1), as previously shown for putative orthologs from other gram-positive bacteria (25). The second protein band by order of electrophoretic mobility should be the pbpZ gene product, as it was absent from JH2-2 ΔpbpZ (lane 4), JH2-2 ΔpbpF ΔpbpZ (lane 5), JH2-2 ΔponA ΔpbpZ (lane 7), and JH2-2 ΔponA ΔpbpF ΔpbpZ (lane 8). The third protein band disappeared totally in the triple mutant JH2-2 ΔponA ΔpbpF ΔpbpZ (lane 8) and may therefore contain the pbpF gene product. However, this band cannot solely correspond to the PBP encoded by pbpF, since the band was present in JH2-2 ΔpbpF (lane 3), JH2-2 ΔpbpF ΔpbpZ (lane 5), JH2-2 ΔponA ΔpbpF (lane 6), and JH2-2 ΔponA ΔpbpZ (lane 7). These observations may imply that the third band contained truncated forms of PBPs encoded by ponA and pbpZ, in addition to the PBP encoded by pbpF. Deletion of pbp5 was associated with loss of the fifth PBP band (lane 9). This band was more intense in a JH2-2 derivative containing a copy of pbp5 cloned into pNJ2 (Fig. 2, lane 11), confirming that it corresponded to PBP5. The remaining PBP band (fourth band) may contain the putative class B PBPs encoded by pbpA and pbpB, since it was unaffected by the ponA, pbpF, pbpZ, and pbp5 deletions.
FIG. 2.
PBP profiles of single, double, and triple PBP mutants. Lanes: 1 and 10, JH2-2; 2, JH2-2 ΔponA; 3, JH2-2 ΔpbpF; 4, JH2-2 ΔpbpZ; 5, JH2-2 ΔpbpF ΔpbpZ; 6, JH2-2 ΔponA ΔpbpF; 7, JH2-2 ΔponA ΔpbpZ; 8, JH2-2 ΔponA ΔpbpF ΔpbpZ; 9, JH2-2 Δpbp5; 11, JH2-2/pNJ2Ωpbp5fs.
Complementation of class A PBP gene deletions.
The pbpF gene of E. faecalis JH2-2 (pbpFfs) was cloned under the control of the aphA-3p promoter of the shuttle plasmid pNJ2 and introduced into JH2-2 ΔponA ΔpbpF and JH2-2 ΔponA ΔpbpF ΔpbpZ strains. As expected, the resulting plasmid restored ceftriaxone resistance in these mutants (MIC of 1,000 μg/ml for both hosts). The active-site serine of the transpeptidase module encoded by pbpFfs was replaced by a glycine by site-directed mutagenesis. Expression of the resulting gene (pbpFfsS402-G) cloned into pNJ2 also restored a wild-type level of ceftriaxone resistance in the same hosts (MIC of 1,000 μg/ml). Thus, the glycosyltransferase module of the PBP encoded by pbpFfs was sufficient for ceftriaxone resistance in the absence of a functional C-terminal d,d-transpeptidase module.
Heterologous expression of the pbpF ortholog of E. faecium (pbpFfm) in JH2-2 ΔponA ΔpbpF and JH2-2 ΔponA ΔpbpF ΔpbpZ led to high-level resistance to ceftriaxone (MIC of 1,000 μg/ml for both hosts), indicating that the glycosyltransferase module of the E. faecium PBP is functional when expressed in E. faecalis. In contrast, expression of mgtSa encoding the monofunctional glycosyltransferase of S. aureus had no effect on the MIC of the antibiotic.
Peptidoglycan structure.
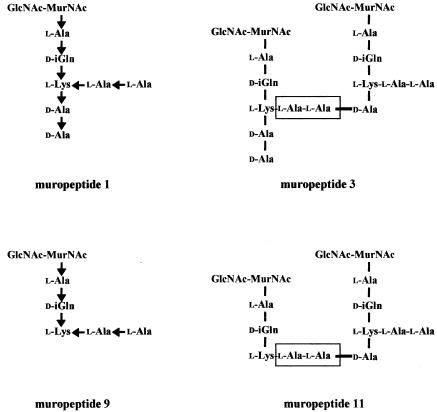
In E. faecalis, the peptidoglycan is polymerized from a subunit consisting of two sugars (GlcNAc and MurNAc), a linear pentapeptide stem (l-Ala1-d-isoglutamine [iGln]2-l-Lys3-d-Ala4-d-Ala5) linked to MurNAc, and a side chain (l-Ala-l-Ala) branched to the ɛ-amino group of l-Lys (Fig. 3) (6, 28). The d,d-transpeptidase activity of PBPs catalyzes formation of Lys3-(l-Ala-l-Ala)-d-Ala4 cross bridges by cleavage of the d-Ala4-d-Ala5 bond of a donor stem peptide and linkage of d-Ala4 to the extremity of the l-Ala-l-Ala side chain of an acceptor stem peptide.
FIG. 3.
Structure of E. faecalis muropeptide monomers and dimers. The most abundant muropeptides (e.g., muropeptides 1 and 3) contained two d-Ala residues at the free C-terminal end and two l-Ala residues both at the free N-terminal end and in the cross bridge (boxed). Less-abundant muropeptides (e.g., 9 and 11) contained a tripeptide stem lacking the two C-terminal d-Ala residues. The orientations of the CO→NH peptide bonds are indicated by arrows.
The diversity of peptidoglycan fragments (muropeptides) obtained by digestion of the peptidoglycan of E. faecalis JH2-2 by muramidases had three main origins (Table 3), as previously described (6). First, the muropeptides differed by the number of disaccharide peptide subunits linked together by the transpeptidases (from 1 to 4 for the monomers and tetramers, respectively). Second, the most abundant muropeptides contained a pentapeptide stem (l-Ala-d-iGln-l-Lys-d-Ala-d-Ala), whereas less-abundant forms contained tripeptide stems (l-Ala-d-iGln-l-Lys) lacking the d-Ala residues at the free C-terminal end. The latter muropeptides may be generated by hydrolysis of the l-Lys3-d-Ala4 peptide bond by l,d-carboxypeptidases (6). Third, a fraction of the muropeptides contained O-acetylated sugars.
TABLE 3.
Muropeptide composition of peptidoglycan from E. faecalis JH2-2 and derivatives
| Muropeptide no.a | Form | Massb | Relative abundance (%) in:
|
||
|---|---|---|---|---|---|
| JH2-2c | JH2-2 ΔponA ΔpbpFd | JH2-2 ΔponA ΔpbpF ΔpbpZe | |||
| 1 | Monomer | 1,109.6 | 20.3 | 29.4 | 31.9 |
| 2 | O-Acetylated monomer | 1,151.6 | 3.6 | 3.9 | 4.3 |
| 3 | Dimer | 2,130.1 | 25.2 | 28.4 | 21.2 |
| 4 | O-Acetylated dimer | 2,172.1 | 8.4 | 6.7 | 9.1 |
| 5 | Trimer | 3,150.5 | 12.9 | 6.4 | 3.5 |
| 6 | O-Acetylated trimer | 3,192.6 | 3.6 | 1.2 | 1.3 |
| 7 | Tetramer | 4,171.0 | 4.6 | 0.8 | 0.5 |
| 8 | O-Acetylated tetramer | 4,213.1 | 1.4 | 0.1 | 0.1 |
| 9 | Monomer | 967.5 | 4.2 | 6.8 | 9.3 |
| 10 | O-Acetylated monomer | 1,009.5 | 0.9 | 1.5 | 3.9 |
| 11 | Dimer | 1,988.0 | 6.5 | 9.3 | 7.6 |
| 12 | O-Acetylated dimer | 2,030.0 | 3.2 | 3.1 | 4.9 |
| 13 | Trimer | 3,008.5 | 3.0 | 1.9 | 1.4 |
| 14 | O-Acetylated trimer | 3,050.5 | 1.3 | 0.5 | 0.7 |
| 15 | Tetramer | 4,029.0 | 0.8 | 0.1 | 0.1 |
| 16 | O-Acetylated tetramer | 4,071.0 | 0.3 | 0.0 | 0.0 |
Muropeptides are separated into two subgroups depending upon the presence (major forms, muropeptides 1 to 8, containing a pentapeptide stem [l-Ala-d-iGln-l-Lys-d-Ala-d-Ala]) or absence (forms of lesser abundance, muropeptides 9 to 16, containing a tripeptide stem [l-Ala-d-iGln-l-Lys]) of the d-Ala-d-Ala residues at the free C-terminal end of the peptide stem. For each of these subgroups, the relative abundance is given for the individual monomer, dimer, trimer, and tetramer and for the derivatives resulting from sugar O-acetylation.
The monoisotopic masses of the proposed structures differed from those deduced from m/z determinations by less than 0.3 atomic mass units.
The relative abundances for total monomers, dimers, trimers, and tetramers were 28.9, 43.3, 20.8, and 7.0%, respectively.
The relative abundances for total monomers, dimers, trimers, and tetramers were 41.6, 47.5, 10.0, and 1.0%, respectively.
The relative abundances for total monomers, dimers, trimers, and tetramers were 49.5, 42.8, 6.9, and 0.8%, respectively.
To analyze the impact of the deletion of class A PBP genes on cell wall cross-linking, the muropeptides of strains JH2-2, JH2-2 ΔponA ΔpbpF, and JH2-2 ΔponA ΔpbpF ΔpbpZ were compared (Table 3). The double and triple deletions were associated with a decrease in the relative abundance of the trimers and tetramers. These results indicate that class A PBPs contribute to peptidoglycan cross-linking in JH2-2. The relative abundance of muropeptides with O-acetylated sugars and incomplete tripeptide stems was not altered.
DISCUSSION
The role of PBP5 in intrinsic β-lactam resistance of enterococci has been well established by the analysis of various spontaneous mutants of E. faecium obtained in the laboratory or isolated from patients, as susceptibility testing provided a powerful screen for monitoring alterations of pbp5 expression (7, 18, 29, 30). In contrast, the role of other PBPs remained uncharacterized due to insufficiently developed genetic tools. In particular, electroporation is inefficient and certain natural isolates and mutants of E. faecium and E. faecalis are refractory to transformation by this method. For this reason, we have constructed the expression vector pNJ2 and the suicide plasmid pHS1 (Fig. 1), which were mobilizable between enterococcal strains by Tn916 at a frequency of ca. 10−4 per donor. Derivatives of the thermosensitive replicon pHS1 were designed to serially introduce deletions into the chromosome of E. faecalis by homologous recombination (Fig. 1C and D). The deletions obtained by this approach are precise, as they are delineated by the oligonucleotides used to amplify the H1 and H2 sequences flanking the chromosomal region targeted for deletion. In this study, the mutants contained deletions removing at least 95% of the open reading frames, and two of them were in-frame deletions to minimize the risk of polar effects on expression of downstream genes after deletion of the erm cassette. The transfer properties of pHS1 combined to direct mobilization of chromosomal markers by plasmid pIP964 are powerful tools to generate combinations of chromosomal deletions in E. faecalis.
Deletion of pbp5 led to a 4,000-fold reduction in the MIC of ceftriaxone for E. faecalis JH2-2, whereas the MIC of ampicillin was only reduced 4-fold (Table 2). A spontaneous deletion of pbp5 in E. faecium led to a larger decrease in the MIC of ampicillin (800-fold) in addition to the loss of resistance to cephalosporins (29). Therefore, the contribution of PBP5 to intrinsic ampicillin resistance appears smaller in E. faecalis than in E. faecium. This difference is worth noting, since the emergence of high-level ampicillin resistance by modification of PBP5 has been mostly, if not exclusively, reported for clinical isolates of E. faecium (24).
The bacterial cell wall is polymerized by large complexes that include glycosyltransferases and transpeptidases for insertion of new material in the murein layer (15). In the presence of high concentrations of ceftriaxone, the transpeptidase module of all PBPs, except that of PBP5, is thought to be inactivated by the antibiotic (7, 29, 30). Resistance to ceftriaxone was used as a screen to identify the glycosyltransferases that cooperate with the transpeptidase module of PBP5 for peptidoglycan polymerization in the presence of the drug (Table 2). The screen identified the class A PBPs encoded by ponA and pbpF as essential partners of PBP5. Site-directed mutagenesis of pbpF confirmed that the catalytic activity of the transpeptidase module of the PBP did not play a role in resistance. S. aureus produces a single class A PBP (PBP2) that is similarly essential for β-lactam resistance mediated in this organism by PBP2a (26). The ponA gene of E. faecalis and the gene encoding PBP2 in S. aureus are putative orthologs, as are the genes encoding low-affinity class B PBP5 and PBP2a (Table 1) (14). The same functional interactions might therefore occur between subclasses of A- and B-type PBPs in different bacteria. In agreement, expression of the putative pbpF ortholog from E. faecium restored ceftriaxone resistance in the ΔponA ΔpbpF mutant, whereas pbpZ of E. faecalis and mgt of S. aureus had no effect. The peptidoglycan precursors of E. faecalis and E. faecium contain side chains consisting of the sequence l-alanyl-l-alanine and a single γ-d-asparaginyl or γ-d-aspartyl residue, respectively (28). In spite of this difference, the glycosyltransferase module of the E. faecium PBP was functional in the heterologous host, indicating for the first time that heterospecific complementation can be used to get insight into the function of class A PBPs.
Deletion of ponA, pbpF, and pbpZ led to a viable mutant, indicating that the class A PBPs are unessential. The chromosome of E. faecalis does not encode a monofunctional glycosyltransferase (MGT) or any additional protein displaying similarity to the glycosyltransferase module of class A PBPs. In the triple mutant, transglycosylation is therefore performed by a distinct class of proteins that do not display similarity with known glycosyltransferases. A similar observation was recently reported for a mutant of Bacillus subtilis lacking all four genes encoding class A PBPs in this organism (23). In contrast, at least one class A PBP is required for viability in E. coli (PBP1a or PBP1b) (11) and S. pneumoniae (PBP1a or PBP2a) (16, 25). Although unessential, the class A PBPs contribute to peptidoglycan cross-linking in E. faecalis, since deletion of the three class A PBP genes led to an increase in the proportion of monomers to the detriment of trimers and tetramers (Table 3). Except for this difference, the mode of cross-linking and structure of the muropeptides were essentially unaltered in comparison to the parental strain JH2-2. These observations indicate that the transpeptidation reaction catalyzed by the entire set of PBPs, or solely by the class B PBPs, may involve the same precursors with respect to the presence of tripeptide or pentapeptide at the free C terminus of the acceptor stems.
Moenomycin was recently shown to inhibit the glycosyltransferase activity of purified PBP1b of E. coli in vitro, although the drug was not competitive with respect to the lipid II substrate (9). The mutants lacking ponA and pbpF or the three class A PBP genes were resistant to moenomycin, whereas the parental strain and all other single and double deletion mutants were susceptible to this antibiotic (Table 2). Thus, susceptibility to moenomycin in E. faecalis depends upon production of at least one of the class A PBPs encoded by ponA and pbpF. This observation implies that binding of moenomycin to its targets has a toxic effect despite the fact that the PBPs encoded by ponA and pbpF are not essential for viability. The antibacterial activity of moenomycin appears, therefore, to result from poisoning of the polymerization complexes containing class A PBPs rather than simply inhibiting their glycosyltransferase active site. This complex mode of action has important implications for the discovery of new drugs targeting the transglycosylation reaction and the improvement of existing molecules, such as biphenyl derivatives of vancomycin and moenomycin (9). In particular, in vitro inhibition of the transglycosylase activity of purified class A PBPs is not expected to strictly correlate with antibacterial activity, since poisoning of the peptidoglycan polymerization complexes and inhibition of enzyme activity may occur independently. Moreover, class A PBPs and the related monofunctional glycosyltransferases may no longer be considered essential targets in human gram-positive pathogens, since the E. faecalis JH2-2 ΔponA ΔpbpF ΔpbpZ null mutant was viable.
In conclusion, neither the d,d-transpeptidase nor the glycosyltransferase activity of E. faecalis class A PBPs is essential for peptidoglycan synthesis. Complete bypass of the d,d-transpeptidase activity of the PBPs by an l,d-transpeptidase insensitive to β-lactam inhibition has been recently reported for E. faecium (21, 22). The l,d transpeptidase is responsible for the synthesis of new peptidoglycan cross bridges (l-Lys3→d-Asx-l-Lys3) that replace the cross bridges formed by the d,d-transpeptidases (d-Ala4→d-Asx-l-Lys3). These complementary observations indicate that peptidoglycan polymerization in the total absence of multimodular PBPs is theoretically possible in enterococci.
Acknowledgments
This work was supported by Wyeth Research, by the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires (MENRT), and by the Fondation pour la Recherche Médicale. A.A. was the recipient of a fellowship from the Gobierno Vasco.
E. faecalis genome sequence data were kindly provided by The Institute for Genomic Research, as publicly released at http://www.tigr.org.
REFERENCES
- 1.Arthur, M., F. Depardieu, and P. Courvalin. 1999. Regulated interactions between partner and non-partner sensors and response regulators that control glycopeptide resistance gene expression in enterococci. Microbiology 145:1849-1858. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, M., F. Depardieu, H. A. Snaith, P. E. Reynolds, and P. Courvalin. 1994. Contribution of VanY d,d-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob. Agents Chemother. 38:1899-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur, M., C. Molinas, and P. Courvalin. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baptista, M., F. Depardieu, P. Reynolds, P. Courvalin, and M. Arthur. 1997. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol. Microbiol. 25:93-105. [DOI] [PubMed] [Google Scholar]
- 5.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouhss, A., N. Josseaume, A. Severin, K. Tabei, J. E. Hugonnet, D. Shlaes, D. Mengin-Lecreulx, J. Van Heijenoort, and M. Arthur. 2002. Synthesis of the L-alanyl-L-alanine cross-bridge of Enterococcus faecalis peptidoglycan. J. Biol. Chem. 277:45935-45941. [DOI] [PubMed] [Google Scholar]
- 7.Canepari, P., M. del mar Lleo, G. Cornaglia, R. Fontana, and G. Satta. 1986. In Streptococcus faecium penicillin-binding protein 5 alone is sufficient for growth at sub-maximal but not at maximal rate. J. Gen. Microbiol. 132:625-631. [DOI] [PubMed] [Google Scholar]
- 8.Celli, J., and P. Trieu-Cuot. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103-117. [DOI] [PubMed] [Google Scholar]
- 9.Chen, L., D. Walker, B. Sun, Y. Hu, S. Walker, and D. Kahne. 2003. Vancomycin analogues active against vanA-resistant strains inhibit bacterial transglycosylase without binding substrate. Proc. Natl. Acad. Sci. USA 100:5658-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz-Rodz, A. L., and M. S. Gilmore. 1990. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224:152-154. [DOI] [PubMed] [Google Scholar]
- 11.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duez, C., W. Zorzi, F. Sapunaric, A. Amoroso, I. Thamm, and J. Coyette. 2001. The penicillin resistance of Enterococcus faecalis JH2-2r results from an overproduction of the low-affinity penicillin-binding protein PBP4 and does not involve a psr-like gene. Microbiology 147:2561-2569. [DOI] [PubMed] [Google Scholar]
- 13.Fontana, R., M. Aldegheri, M. Ligozzi, H. Lopez, A. Sucari, and G. Satta. 1994. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 38:1980-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Höltje, J. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoskins, J., P. Matsushima, D. L. Mullen, J. Tang, G. Zhao, T. I. Meier, T. I. Nicas, and S. R. Jaskunas. 1999. Gene disruption studies of penicillin-binding proteins 1a, 1b, and 2a in Streptococcus pneumoniae. J. Bacteriol. 181:6552-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klare, I., A. C. Rodloff, J. Wagner, W. Witte, and R. Hakenbeck. 1992. Overproduction of a penicillin-binding protein is not the only mechanism of penicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 36:783-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laible, G., B. G. Spratt, and R. Hakenbeck. 1991. Interspecies recombinational events during the evolution of altered PBP2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 5:1993-2002. [DOI] [PubMed] [Google Scholar]
- 20.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mainardi, J. L., V. Morel, M. Fourgeaud, J. Cremniter, D. Blanot, R. Legrand, C. Frehel, M. Arthur, J. Van Heijenoort, and L. Gutmann. 2002. Balance between two transpeptidation mechanisms determines the expression of beta-lactam resistance in Enterococcus faecium. J. Biol. Chem. 277:35801-35807. [DOI] [PubMed] [Google Scholar]
- 22.Mainardi, J.-L., R. Legrand, M. Arthur, B. Schoot, J. van Heijenoort, and L. Gutmann. 2000. Novel mechanism of β-lactam resistance due to by-pass of DD-transpeptidation in Enterococcus faecium. J. Biol. Chem. 275:16490-16496. [DOI] [PubMed] [Google Scholar]
- 23.McPherson, D. C., and D. L. Popham. 2003. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J. Bacteriol. 185:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray, B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paik, J., I. Kern, R. Lurz, and R. Hakenbeck. 1999. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J. Bacteriol. 181:3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinho, M. G., H. de Lencastre, and A. Tomasz. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. USA 98:10886-10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rybkine, T., J. L. Mainardi, W. Sougakoff, E. Collatz, and L. Gutmann. 1998. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of beta-lactam resistance. J. Infect. Dis. 178:159-163. [DOI] [PubMed] [Google Scholar]
- 28.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sifaoui, F., M. Arthur, L. Rice, and L. Gutmann. 2001. Role of penicillin-binding protein 5 in expression of ampicillin resistance and peptidoglycan structure in Enterococcus faecium. Antimicrob. Agents Chemother. 45:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Signoretto, C., M. Boaretti, and P. Canepari. 1994. Cloning, sequencing and expression in Escherichia coli of the low-affinity penicillin binding protein of Enterococcus faecalis. FEMS Microbiol. Lett. 123:99-106. [DOI] [PubMed] [Google Scholar]
- 31.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZα gene for conjugal transfer of DNA from Escherichia coli to Gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Q. M., R. B. Peery, R. B. Johnson, W. E. Alborn, W. K. Yeh, and P. L. Skatrud. 2001. Identification and characterization of a monofunctional glycosyltransferase from Staphylococcus aureus. J. Bacteriol. 183:4779-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williamson, R., C. le Bouguenec, L. Gutmann, and T. Horaud. 1985. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J. Gen. Microbiol. 131:1933-1940. [DOI] [PubMed] [Google Scholar]
- 34.Wu, S. W., H. de Lencastre, and A. Tomasz. 2001. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J. Bacteriol. 183:2417-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zorzi, W., X. Y. Zhou, O. Dardenne, J. Lamotte, D. Raze, J. Pierre, L. Gutmann, and J. Coyette. 1996. Structure of the low-affinity penicillin-binding protein 5 PBP5fm in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J. Bacteriol. 178:4948-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]