Abstract
Type IV secretion systems mediate conjugative plasmid transfer as well as the translocation of virulence factors from various gram-negative pathogens to eukaryotic host cells. The translocation apparatus consists of 9 to 12 components, and the components from different organisms are believed to have similar functions. However, orthologs to proteins of the prototypical type IV system, VirB of Agrobacterium tumefaciens, typically share only 15 to 30% identical amino acids, and functional complementation between components of different type IV secretion systems has not been achieved. We here report a heterologous complementation in the case of A. tumefaciens virB1 defects with its orthologs from Brucella suis (VirB1s) and the IncN plasmid pKM101 (TraL). In contrast, expression of the genes encoding the VirB1 orthologs from the IncF plasmid (open reading frame 169) and from the Helicobacter pylori cag pathogenicity island (HP0523) did not complement VirB1 functions. The complementation of VirB1 activity was assessed by T-pilus formation, by tumor formation on wounded plants, by IncQ plasmid transfer, and by IncQ plasmid recipient assay. Replacement of the key active-site Glu residue by Ala abolished the complementation by VirB1 from B. suis and by TraL, demonstrating that heterologous complementation requires an intact lytic transglycosylase active site. In contrast, the VirB1 active-site mutant from A. tumefaciens retained considerable residual activity in various activity assays, implying that this protein exerts additional effects during the type IV secretion process.
Type IV secretion systems (T4SS) mediate the translocation of macromolecules across the envelope of gram-negative bacteria. These systems were initially discovered to be essential for the conjugative transfer of plasmids of different incompatibility groups (F, H, N, P, and W) (22, 56). In addition to the plasmid DNA, proteins are translocated, suggesting that those may carry the recognition signals for T4SS translocation (34, 36). This notion was further supported by the discovery that the VirB/D4 system from Agrobacterium tumefaciens, which shares significant sequence similarities with plasmid-encoded T4SS, translocates several proteins in addition to its T-DNA substrate (52). Subsequently, T4SS were found to mediate the translocation of proteinaceous effector molecules from several bacterial pathogens (3, 11, 12, 20, 36).
Compared to other T4SS, the T-DNA translocation system from A. tumefaciens has been studied in the greatest detail (3, 11). Its 12 components (VirB1 to VirB11 and VirD4) localize to the inner and the outer membrane, and different lines of evidence suggest that this complex spans the periplasm. Biochemical experiments suggested that VirB7, VirB8, VirB9, and VirB10, which are mostly exposed to the periplasm but anchored to the membranes, constitute the core complex required for the stabilization of many other membrane-bound VirB proteins (1, 4, 7, 14, 18, 19, 29). Protein-protein interactions between VirB7 to VirB10 were also detected with the yeast two-hybrid system (13, 14). VirB8 and VirB10 contain N-terminal hydrophobic domains and are likely anchored to the inner membrane. In contrast, the lipoprotein VirB7 predominantly localizes to the outer membrane together with covalently linked VirB9. The T4SS core likely links the surface-exposed T-pilus components VirB2 and VirB5 to the cell, and VirB6 and VirB7 play a key role during that process (17, 25, 27, 29, 32, 45). The mechanism of T4SS assembly is unknown, and energy provided by one or more of the nucleotide triphosphatases VirB4, VirB11, and VirD4 is believed to energize this process (30, 43).
The murein layer imposes a barrier to the assembly of trans-envelope structures, and VirB1 likely exerts its effect here (5, 15, 16, 28). VirB1 is required for efficient T4SS function, but virB1 deletion strains of A. tumefaciens were reported to retain between 1 and 10% residual virulence (8, 39). Similarly, in the absence of VirB1 ortholog P19, plasmid R1 conjugation and R17 phage infection were reduced to 10% of the wild-type level and traL mutants of pKM101 had 5% residual activity (5, 54). The finding that VirB1 is not essential was attributed to its putative function as a lytic transglycosylase, which may facilitate T4SS assembly by localized lysis of the peptidoglycan (39). The relatively thin murein layer of gram-negative bacteria may permit a reduced level of T4SS assembly even in the absence of VirB1 orthologs (28). Alternatively, cellular enzymes may partly substitute for its function (26). Direct evidence for the transglycosylase activity has not been presented, but VirB1 derivatives with alterations in the putative active sites had strongly reduced virulence (39). Overproduction of the VirB1 ortholog P19 from plasmid R1 led to cell lysis, implying a role in murein metabolism (6). In addition to its suggested function as a transglycosylase, there may be an additional contribution of VirB1 to T4SS functionality. VirB1 undergoes C-terminal processing after export into the periplasm, resulting in an N-terminal transglycosylase domain and the 14-kDa C-terminal processing product VirB1*, which is partly secreted across the outer membrane (2). Both domains partly complemented a virB1 deletion mutant, implying that they independently contribute to virulence (37). Cross-linking experiments showed that VirB1* localizes in close proximity to VirB9, and this interaction may be required for T4SS function (2). A role of VirB1 in T4SS assembly via interactions with other proteins was further supported by the results of two-hybrid experiments and by its impact on the localization of VirB10 in the cell (35, 53).
We present here a detailed analysis of the consequences of a mutational alteration of the active site of VirB1, showing that it contributes to efficient gene transfer from A. tumefaciens but it is not required for processing of VirB1 and secretion of the C-terminal VirB1* product. In addition, our data strongly suggest that the protein contributes a transglycosylase-independent function to the type IV secretion process. Functional complementation with VirB1 orthologs from some other T4SS was achieved, suggesting that the requirement for transglycosylase activity is conserved among T4SS.
MATERIALS AND METHODS
Bacteria, plasmid constructions, and mutagenesis.
Strains and plasmids used in this study are given in Table 1. Cultures of Escherichia coli for cloning experiments were grown at 37°C in Luria-Bertani medium (1% peptone, 0.5% yeast extract, 0.5% NaCl), and DNA manipulations followed standard procedures (38). Antibiotics were added for plasmid propagation as appropriate (spectinomycin [SPC], 50 μg/ml; streptomycin, 50 μg/ml; carbenicillin [CAR], 100 μg/ml).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JM109 | endA1 gyr96 thi hsdR71 supE44 recA1 relA1 Δ(lac-proAB) (F′ traD36 proAB+lacIqlacZΔM15) | 55 |
| A. tumefaciens A348 | Wild type, pTiA6NC | 23 |
| A. tumefaciens PC1001 | pTiA6NC carrying an in-frame deletion of virB1 | 8 |
| A. tumefaciens UIA143(pTiA6) | C58 derivative; recA ery140 pTiA6 Eryr | 9 |
| Plasmids | ||
| pTrc200 | Strr, Spcr, pVS1 origin, lacIq, trc promoter expression vector | 46 |
| pTrcB1 | pTrc200, carrying 759-bp NcoI/BglII virB1 fragment from A. tumefaciens C58 | 45 |
| pTrcB1E→A | pTrcB1, mutated A. tumefaciens virB1 gene, active-site Glu60 changed to Ala | This work |
| pTrcB1s | pTrc200, carrying 717-bp NcoI/ScaI virB1 fragment from B. suis | This work |
| pTrcB1sE→A | pTrcB1s, mutated B. suis virB1 gene, active-site Glu27 changed to Ala | This work |
| pTrcTraL | pTrc200, carrying 735-bp NcoI/ScaI traL fragment from pKM101 | This work |
| pTrcTraLE→A | pTrcTraL, mutated pKM101 traL gene, active-site Glu53 changed to Ala | This work |
| pTrc169 | pTrc200, carrying 507-bp NcoI/ScaI fragment from the F plasmid | This work |
| pTrc523 | pTrc200, carrying 507-bp NcoI/ScaI fragment from H. pylori chromosomal DNA | This work |
| pBAD18 | Carr, cloning vector | 24 |
| pT7-7 | Carr, cloning vector | 48 |
| pLS1 | Carr, IncQ plasmid for VirB/D4-mediated transfer experiments | 47 |
| pKM101 | Carr, mucA, mucB, IncN broad-host-range plasmid | 54 |
| pUCvirB | Carr, virB region from B. suis 1330 | 40 |
For complementation analysis, vector pTrc200 was cleaved with NcoI and SmaI treated with alkaline phosphatase followed by ligation with PCR fragments of genes encoding VirB1 orthologs as follows. Similar PCR conditions were used in all cases (1 ng of plasmid template; cycle conditions: 2 min at 95°C for denaturation (one time); 44°C for 1 min, 72°C for 2 min, and 95°C for 30 s for cycling (30 times); 44°C for 1 min and 72°C for 5 min for strand completion (one time); and termination at 4°C).
Brucella suis virB1 was amplified from pUCvirB (oligonucleotides BsB1-5′, 5′-GGACCATGGTGCCATTCCTTGTCCTCGCGC-3′, and BsB1-3′, 5′-GCTAGTACTTAGAAAACAACTACGCCGTCCGTATTATCCTTC-3′). TraL was amplified from pKM101 (oligonucleotides TraL5, 5′-GGGGCCATGGGTAAACATCCAAAACTCC-3′, and TraL3′, 5′-GAAAGTACTCATTCCCCCTTCG-3′) (41). Open reading frame (ORF) 169 was amplified from the F plasmid (oligonucleotides 169-N1, 5′-GGGGCCATGGCAAAATGGATGTTAGCCATCTGCCT-3′, and 169-C, 5′-CCCAGTACTTAATTGTTCTGCACGCTGTTAATTTC-3′) (22). The reading frame of HP0523 was amplified from Helicobacter pylori 26695 chromosomal DNA (oligonucleotides Cag4-5, 5′-GGGGCCATGGTCGAGAAATGGATTGGTCT-3′, and Cag4-3, 5′-CCCAGTACTACTCGTTATATCGCACTTG-3′) (49). The fragments were cleaved with NcoI and ScaI (underlined in oligonucleotide sequences above) followed by ligation to similarly cut pTrc200 and sequencing with an ABI 310 sequencer, resulting in pTrcB1s, pTrcTraL, pTrc169, and pTrc523.
The codons determining active-site Glu residues were changed to Ala in pTrcB1 (Glu60), pTrcB1s (Glu27), and pTrcTraL (Glu53) by site-directed mutagenesis by using the Gene editor in vitro site-directed mutagenesis system (Promega) according to the supplier's description. The genes encoding VirB1 and its orthologs were subcloned and subjected to mutagenesis as follows. The virB1 gene from pTrcB1 was excised with Eco32I/HindIII and ligated into similarly cut pBAD18 followed by mutagenesis with oligonucleotide B1M5 (GCAGCGATCGCTCAGGTCGCTAGCCGCTTTGATCCGCTTGCT). The virB1 gene from pTrcB1s was excised with NcoI/HindIII and ligated into similarly cut pT7-7 followed by mutagenesis with oligonucleotide B1suisM5 (GCAGCAATCGTGCAGGTCGCTAGCGGCTTCAATCCTTATGCA). The traL gene from pTrcTraL was excised with NcoI/HindIII and ligated into similarly cut pT7-7 followed by mutagenesis with oligonucleotide TraLM5 (GCGTACATCGTCGGCCATGCTAGCTCAAATGGACCGTACAGG). Mutations were identified by restriction analysis with NheI (underlined above) and confirmed by sequencing.
Cultivation of A. tumefaciens and analysis of exocellular fractions.
Overnight cultures of A. tumefaciens were grown in YEB medium (0.5% beef extract, 0.5% peptone, 0.1% yeast extract, 0.5% sucrose, 2 mM MgSO4) in the absence of antibiotics (wild-type strains) or with SPC (300 μg/ml) and streptomycin (100 μg/ml) for the propagation of pTrc200 derivatives. For propagation of pLS1, CAR was added at 150 μg/ml, and for the selection of UIA143(pTiA6) pLS1 transconjugants, erythromycin (ERY) was added at 150 μg/ml in addition to CAR. Further, the cells were inoculated to an optical density at 600 nm of 0.1 in liquid AB medium (10 g of glucose/liter, 4 g of MES [morpholineethanesulfonic acid]/liter, 0.3 g of MgSO4 · 7 H2O/liter, 0.15 g of KCl/liter, 0.01 g of CaCl2/liter, 0.0025 g of FeSO4 · 7H2O/liter, and 1 mM potassium phosphate [pH 5.5]) and grown for 5 h at 20°C followed by specific procedures depending on the assay used. First, for the analysis of VirB1* secretion in liquid cultures, cells were grown at 20°C for 18 h with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for the induction of the trc promoter with or without 200 μM acetosyringone (AS) for virulence gene induction. VirB1* was precipitated from cell-free culture supernatants with acetone as described followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis (2). Second, for the isolation of surface-exposed T-pili, 1 ml of cells was plated per large (15 cm2) AB medium plate (20 g of agar/liter) with or without 200 μM AS and the plates were further incubated at 20°C for 4 days. T-pili were isolated from the cells grown on 4 plates each with and without AS by shearing and ultracentrifugation as described previously (45). Third, for plant infection experiments, leaf squares (4 by 4 mm) of Nicotiana tabacum cv. Havana 425 were cocultivated with the bacteria in the presence of 0.5 mM IPTG and 300 μM AS on hormone-free Murashige-Skoog medium for 2 days at 25°C, rinsed with Luria-Bertani plus 200 μg of vancomycin/ml and 200 μg of timentin/ml, and then cultivated on hormone-free Murashige-Skoog medium plus 200 μg of vancomycin/ml and 200 μg of timentin/ml at 25°C in the dark (35). Tumor formation was scored 12 days after the start of the cocultivation. Fourth, for the analysis of pLS1 donor activity, A348 and PC1001 carrying pTrc200 with and without VirB1 ortholog-encoding genes were cocultivated with UIA143(pTiA6) recipient cells in a 1:5 ratio for 3 days on AB minimal medium agar containing 500 μM AS and 500 μM IPTG followed by plating on selective agar media (CAR, 150 μg/ml; ERY, 150 μg/ml) and quantitation of recipient and donor cells as described previously (9). Fifth, for the analysis of pLS1 recipient activity, donor A348 pLS1 cells were cocultivated with PC1001 recipient cells carrying pTrc200 with and without VirB1 ortholog-encoding genes in a 5:1 ratio for 2 days on AB induction medium plus 500 μM AS and 500 μg of IPTG/ml followed by plating on Luria-Bertani agar plus antibiotics (150 μg of CAR/ml and/or 100 μg of SPC/ml) as necessary for quantitation of donors, recipients, and transconjugants as described previously (35).
SDS-PAGE and Western blotting.
Cells and exocellular fractions were incubated in Laemmli sample buffer for 5 min at 100°C followed by SDS-PAGE according to the method of Laemmli (VirB1 and VirB8) (31) or Schägger and von Jagow (VirB2) (44), and Western blotting was performed in a tank blot apparatus. Detection was performed with a chemiluminescence-based system (NEN) with A. tumefaciens strain C58 VirB protein-specific antisera.
RESULTS
Choice and cloning of VirB1 orthologs.
We assessed the hypothesis that VirB1 orthologs may be exchangeable between different T4SS. Complementation of the virB1 deletion mutant A. tumefaciens strain A348 (PC1001) was performed with VirB1 from A. tumefaciens strain C58 and its orthologs from B. suis (VirB1s) and pKM101 (TraL). Similar to other T4SS components, these proteins share relatively limited sequence similarities. They were identified based on their location in virB-like operons and their putative active-site signatures (Fig. 1). The sequence similarity to the ORF169 from the F plasmid protein is even smaller, but different lines of evidence suggest that this protein may be a lytic transglycosylase (5, 6). In addition, we analyzed the HP0523 (cag4) gene product from H. pylori strain 26692. This protein had not been identified as a VirB1 ortholog initially (49), but a recent publication shows suggestive evidence for a role in T4SS complex assembly (42). The transglycosylase active-site signature sequence was identified by visual inspection of gene products from the cag pathogenicity island. Similar to ORF169, the similarity to VirB1 is limited, but the putative active-site residues are well conserved (Fig. 1B) and the gene is necessary for virulence (21). The genes encoding the different VirB1 orthologs were PCR amplified, cloned into broad-host-range vector pTrc200, and transformed into strain PC1001 (ΔvirB1) for analysis of their biological activities. In addition, the genes encoding VirB1, VirB1s, and TraL were modified by site-directed mutagenesis to change the essential active-site Glu residues of their respective products.
FIG. 1.
VirB1 orthologs and active-site mutants used in this study. (A) Schematic representation of the analyzed proteins and their amino acid sequence identities (percentages noted on the right) to A. tumefaciens C58 VirB1. Amino acid residues in boxes A, B, and C (black boxes) are implicated in the enzymatic activity of lytic transglycosylases. They were assigned in the different orthologs according to the method of Bayer et al. (5). Signal peptides (grey boxes) were assigned by the SignalP V1.1 algorithm (http://www.cbs.dtu.dk/services/SignalP). The C-terminal processing site of VirB1 (A173QQ) is shown as a hatched box. (B) Alignment of VirB1 orthologs investigated in this study; residues identical to those of VirB1 are shaded. Conserved residues are indicated by black boxes A, B, and C, and the putative active-site Glu residues are labeled with a star. Active-site residues corresponding to Glu60 from A. tumefaciens VirB1 were changed to Ala in VirB1s and TraL in the course of this study. The alignment was generated with the MegAlign program (DNA Star).
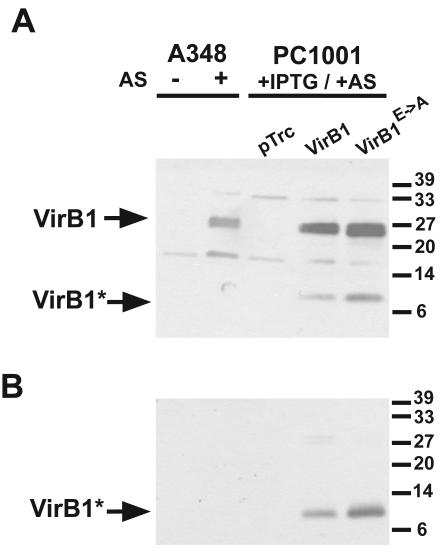
The VirB1 content of cell lysates and of culture supernatants was monitored by SDS-PAGE and Western blotting to assess the impact of the active-site mutation. Equal levels of the full-length protein and of VirB1* were detected in samples from PC1001 carrying pTrcB1 and pTrcB1E→A (Fig. 2), demonstrating that the putative active site is not required for stability, processing, and secretion. Different assays were conducted to assess the complementation of the A. tumefaciens virB1 defect.
FIG. 2.
Processing of VirB1 and secretion of VirB1*. Strains A348 and PC1001 (ΔvirB1) carrying pTrc200 (pTrc), pTrcB1 (VirB1), or pTrcB1E→A (VirB1E→A) were cultivated under virulence gene-inducing (+AS) or noninducing conditions (−AS). IPTG was added to cultures carrying pTrc200 constructs for induction of gene expression from the trc promoter. Cell lysates (A) and secreted proteins precipitated from the supernatant (B) were separated by SDS-PAGE followed by Western blotting with A. tumefaciens C58 VirB1-specific antiserum. Arrows indicate VirB1 and its C-terminal processing product VirB1*. A VirB1* processing product of strain A348 VirB1 was not detected with strain C58 VirB1-specific antiserum. Numbers on the right indicate reference proteins.
Complementation of T-pilus formation.
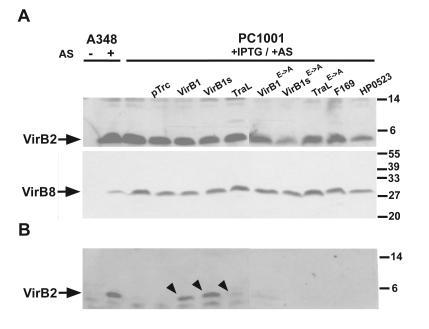
VirB1 was previously shown to be required for T-pilus formation in A. tumefaciens strain A348, demonstrating that it is required for T4SS assembly (33, 45). To test the complementation, the A348 wild type and strain PC1001 (ΔvirB1) carrying pTrc200 with or without virB1 ortholog-carrying genes were grown on AB agar plates under virulence gene-inducing conditions followed by subcellular fractionation and analysis of VirB protein content. Analysis of cell lysates with VirB2- and VirB8-specific antisera showed that the accumulation of the major T-pilus component as well as of the T4SS core component VirB8 were not affected by the absence of VirB1 (Fig. 3A). Surface-exposed high-molecular-mass structures were removed from the cells by shearing and ultracentrifugation, and as previously described, significant amounts of VirB2 were not detected in exocellular fractions from strain PC1001 (Fig. 3B). The introduction of pTrcB1 and of pTrcB1s restored T-pilus formation almost to wild-type levels. In two of three experiments, pTrcTraL showed weak complementing activity, but pTrc169 and pTrc523 never restored T-pilus formation. Similarly, plasmids directing the production of active site variants of VirB1, VirB1s, and TraL did not reproducibly complement T-pilus formation, demonstrating that the putative active site is required. Since T-pilus formation it is not readily amenable to quantitation, low degrees of complementation are difficult to monitor and it does not permit conclusions on substrate translocation.
FIG. 3.
Complementation of T-pilus formation by strain PC1001. Strains A348 and PC1001 (ΔvirB1) carrying pTrc200 and virB1 ortholog-encoding constructs were cultivated under virulence gene-inducing (+AS) or noninducing conditions (−AS). IPTG was added to cultures carrying pTrc200 constructs for induction of gene expression from the trc promoter. Cell lysates (A) and exocellular high-molecular-mass T-pilus fractions (B) were separated by SDS-PAGE followed by Western blotting with VirB2- or VirB8-specific antisera. The strains used are as follows: A348 and PC1001 carrying pTrc200 (pTrc), pTrcB1 (VirB1), pTrcB1s (VirB1s), pTraL (TraL), pTrcB1E→A (VirB1E→A), pTrcB1sE→A (VirB1sE→A), pTrcTraLE→A (TraLE→A), pTrc169 (F169), or pTrc523 (HP0523). The experiments were performed three times, and the arrows indicate detection of T-pilus component VirB2 in exocellular fractions in at least two of those experiments. Numbers on the right indicate reference proteins.
Complementation of tumor formation.
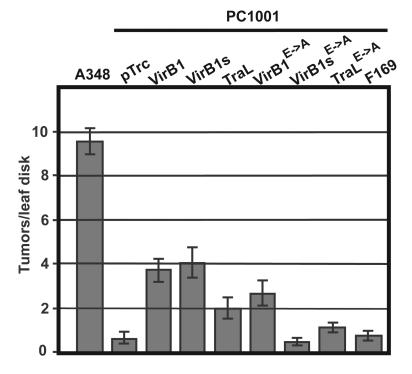
To test the efficiency of substrate translocation, we next assessed the efficiency of tumor formation by strain A348 and by PC1001 carrying pTrc200 with and without virB1 orthologous genes. Freshly cut N. tabacum leaf squares were dipped into bacterial cultures, cocultivated for 2 days, and then incubated on plant medium plus antibiotics to eliminate the bacteria. These assays (Fig. 4) demonstrated that strain PC1001 initiated 5 to 10% of tumors compared to A348, similar to previous observations (39). The introduction of pTrcB1 into PC1001 restored tumor formation to 40% of the level of the wild type, and similar results were obtained by the introduction of pTrcB1s (Fig. 4). The introduction of pTrcTraL led to a more modest increase of tumor formation to 20% of the level of the wild type, whereas pTrc169 did not significantly increase the ability of PC1001 to incite tumors. Surprisingly, the introduction of pTrcB1E→A, which directs the production of an active-site variant of VirB1, significantly increased bacterial virulence to 30% of the level of the wild type, whereas the introduction of pTrcB1sE→A and pTrcTraLE→A did not cause significant effects. The results of the tumor formation assays are similar to those of the pilus formation assay, but the strong effect of pTrcTraL and the partial restoration of tumor formation by an active-site variant of VirB1 constitute a marked difference. However, the relatively low complementation by pTrcB1, which could be caused by inefficient promoter induction by IPTG in the wound, complicates the interpretation. We therefore next chose IncQ plasmid transfer as a more quantitative assay of substrate translocation.
FIG. 4.
Complementation of PC1001 (ΔvirB1) tumor formation on N. tabacum leaf disks. Strains A348 and PC1001 (ΔvirB1) carrying pTrc200 and virB1 ortholog-carrying constructs were cocultivated with N. tabacum leaf disks for 2 days followed by elimination of the bacteria and quantitation of tumor formation at the wound sites. The strains used are as follows: A348 and PC1001 carrying pTrc200 (pTrc), pTrcB1 (VirB1), pTrcB1s (VirB1s), pTraL (TraL), pTrcB1E→A (VirB1E→A), pTrcB1sE→A (VirB1sE→A), pTrcTraLE→A (TraLE→A), or pTrc169 (F169). Error bars indicate standard errors (n = 14).
Complementation of IncQ plasmid transfer.
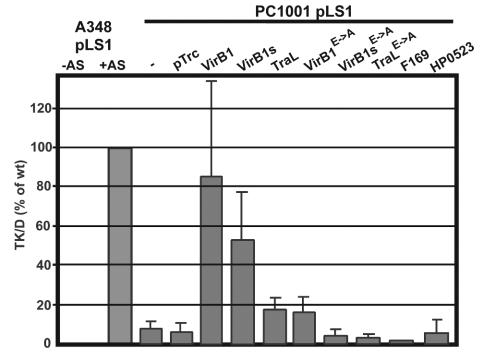
VirB/D4-mediated translocation of IncQ plasmids relies on the recognition of relaxosome complexes by the A. tumefaciens T4SS (47). The transfer of IncQ plasmids between agrobacteria can be readily quantitated, and since the mating is being performed on agar media, the induction of pTrc200-borne genes with IPTG can be easily controlled. The IncQ plasmid pLS1 (Carr) was introduced into strains A348 and PC1001 carrying pTrc200 with and without virB1 orthologous genes. The strains were mated with the recipient UIA143(pTiA6) (Eryr), followed by the quantitation of plasmid transfer on selective media (ERY and CAR). Similar to previous observations (9), the plasmid transfer ability of PC1001 was reduced to about 10% of the wild-type level. The introduction of pTrcB1 conferred almost full complementation, and pTrcB1s increased pLS1 transfer to 50% of the wild-type level (Fig. 5). The presence of pTrcTraL led to a smaller increase of pLS1 transfer efficiency (20% of the wild-type level), whereas the presence of pTrc169 and pTrc523 did not significantly increase transfer. Among the plasmids encoding active-site derivatives, only pTrcB1E→A increased pLS1 transfer, and the stimulation was modest but significant (20% of the wild-type level, comparable to TraL). Those results are qualitatively similar to those obtained in tumor formation assays.
FIG. 5.
Complementation of pLS1 donor activity of strain PC1001. Donor strains A348(pLS1) and PC1001 (ΔvirB1)(pLS1) carrying pTrc200 and virB1 ortholog-carrying constructs were cocultivated with recipient strain A. tumefaciens UIA143(pTiA6) for 3 days in the presence of IPTG under virulence gene-inducing (+AS) or noninducing (−AS) conditions followed by the quantitation of pLS1-carrying recipients (TK/D = transconjugants per donor). The strains used are as follows: A348(pLS1) and PC1001(pLS1) carrying pTrc200 (pTrc), pTrcB1 (VirB1), pTrcB1s (VirB1s), pTraL (TraL), pTrcB1E→A (VirB1E→A), pTrcB1sE→A (VirB1sE→A), pTrcTraLE→A (TraLE→A), pTrc169 (F169), or pTrc523 (HP0523). Error bars indicate standard deviations derived from the results from three independent experiments.
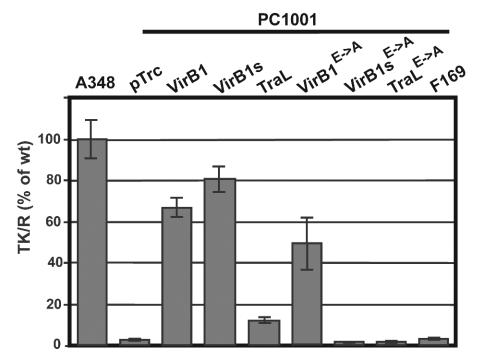
The production of VirB proteins on the recipient side also stimulated conjugative transfer of IncQ plasmids (9, 35). The mechanism is unknown, but VirB1 is a key factor because its deletion has stronger negative impacts than on the donor side (9). Compared to the wild type, the recipient activity of PC1001 was reduced to 3% and the introduction of pTrcB1 restored plasmid transfer to 67% of that of strain A348 (Fig. 6). The introduction of pTrcB1s led to an even stronger increase than that of pTrcB1 (80% of the wild-type level), whereas the introduction of pTrcTraL only modestly increased plasmid recipient activity to 10% and pTrc169 showed no effect. Surprisingly, PC1001 producing the active-site mutant of VirB1 showed 45% of the IncQ plasmid recipient activity compared to that of the wild type, whereas the active-site mutants of VirB1s and TraL had no stimulating effect (Fig. 6). These results are in accord with those of the tumor formation and pLS1 donor assays, suggesting that in addition to its likely function as transglycosylase, VirB1 contributes a second function to T4SS secretion.
FIG. 6.
Complementation of pLS1 recipient activity of strain PC1001 (ΔvirB1). Donor strain A348(pLS1) was cocultivated with recipient strains A348(pTrc200) and PC1001 carrying pTrc200 and/or virB1 ortholog-carrying constructs for 3 days in the presence of IPTG under virulence gene-inducing conditions followed by the quantitation of pLS1-carrying recipients (TK/R = transconjugants per recipient). The strains used are as follows: A348(pTrc200) and PC1001(ΔvirB1) carrying pTrc200 (pTrc), pTrcB1 (VirB1), pTrcB1s (VirB1s), pTraL (TraL), pTrcB1E→A (VirB1E→A), pTrcB1sE→A (VirB1sE→A), pTrcTraLE→A (TraLE→A), or pTrc169 (F169). Error bars indicate standard deviations derived from the results from three independent conjugations.
DISCUSSION
Lytic transglycosylases play a role in murein metabolism, and the active-site residues were defined based on the crystal structure of the soluble lytic transglycosylase Slt70 from E. coli (16). Analysis of the crystal structures of the soluble lytic transglycosylases Slt35 and Slt70 in complexes with their substrates further substantiated the picture of the active-site requirements (50, 51). The existence of a clearly defined active-site signature with three characteristic regions led to the identification of putative transglycosylases in a variety of macromolecular secretion systems (5, 28). Direct evidence for transglycosylase activity is still missing, but different types of evidence indicate a function of these proteins as murein-metabolizing enzymes. The most direct observation was that overproduction of P19, the putative lytic transglycosylase from the IncP plasmid R1, led to perforation of the bacterial cell envelope (6). In addition, mutational alterations of the active-site Glu residues of P19 and VirB1 essentially abolished their abilities to complement the defects in their respective T4SS (6, 39). Finally, in contrast to all other T4SS components, VirB1 orthologs are generally not essential, and the reason may be that other murein-metabolizing enzymes partly substitute for their function.
This led to the hypothesis that in contrast to other T4SS components (10, 25, 46), exchange between different VirB1 orthologs may be possible (28). We directly tested this hypothesis here and found that VirB1s from B. suis and TraL from pKM101 partly complemented the virB1 defect of A. tumefaciens strain PC1001. A variety of different assay systems were employed to substantiate this finding, such as T-pilus formation, tumor formation, and IncQ plasmid donor and IncQ plasmid recipient activity, and the results of all assays were qualitatively similar. In spite of the limited degree of sequence similarity, VirB1s complemented the virB1 defect to a similar level as VirB1 from A. tumefaciens. In contrast, the complementation by TraL, which has approximately the same sequence similarity to VirB1 as VirB1s, was reduced but still significant. In addition to the previously assigned VirB1 orthologs, we identified the H. pylori HP0523 gene product as a putative transglycosylase. HP0523 and the F plasmid-encoded ORF169 gene product are significantly smaller than VirB1, VirB1s, and TraL, and the overall sequence identities to VirB1 are below 20%. These proteins essentially comprise the N-terminal domain of VirB1, which partly complements virB1 defects (37). The putative active-site residues are well conserved, suggesting a function of these proteins as lytic transglycosylases. The facts that the HP0523 gene product is essential for the virulence of H. pylori (21, 42) and that P19 of plasmid R1, which is almost identical to ORF169, is required for efficient conjugative transfer (5), support a role for these proteins in T4SS assembly. While we did not observe complementation of the A. tumefaciens virB1 defect, which may be due to low protein levels in the cell, the results of an independent study support a role for HP0523 in T4SS assembly in the cell envelope (42).
An obvious difference between the virB1 complementing and noncomplementing orthologs is the length of the proteins. VirB1, VirB1s, and TraL share a C-terminal extension, which is processed and secreted in the case of VirB1. Production of the C-terminal processing product VirB1* alone partly complemented virB1 defects of A. tumefaciens, implying that this part of the protein may exert an effect during the type IV secretion process independent of the N-terminal domain (37). VirB1* was cross-linked to VirB9 in vivo (2), and mutational alterations of different residues N- and C-terminal to the processing site negatively affected processing and complementation (C. Höppner and C. Baron, unpublished data). Thus, the interactions of the C-terminal processing product with other components of the T4SS machinery may determine the outcome of complementation experiments, and this may explain our inability to achieve complementation with the ORF169 and HP0523 gene products. One possibility to test this hypothesis would be to study the complementation by fusion proteins of ORF169 and HP0523 with the VirB1* domain.
Based on results obtained with the yeast two-hybrid system, it was recently suggested that VirB1 may interact with several VirB proteins, such as VirB4, VirB8, VirB9, VirB10, and VirB11 (53). Supporting this hypothesis are recent data demonstrating that VirB1, VirB2, and VirB3 impact the subcellular localization of VirB10 (35). To distinguish between the contribution of the transglycosylase and other activities of the proteins, we investigated the complementation by active-site mutants of VirB1, VirB1s, and TraL. Exchange of the active-site Glu residue by Ala did not affect the processing of VirB1 and the secretion of VirB1*, showing that these processes do not require the transglycosylase active site. In a previous study, a similar alteration of the active site of VirB1 abolished its ability to complement tumor formation on Kalanchoë diagremontiana leaves (39). In contrast to that, we found that the alteration of the active-site Glu residue by Ala reduced the activity of the modified gene product but did not lead to a loss of complementation. The difference to previous results may be explained by the higher expression from the trc promoter used in this work and by the choice of assay system. Mushegian et al. did not monitor the level of VirB1 and used the K. diagremontiana leaf puncture assay (39), which is inherently difficult to quantify, whereas we chose a variety of different assay systems. In contrast to the results obtained with VirB1E→A, VirB1sE→A and TraLE→A did not complement the virB1 defect in PC1001. Thus, the orthologs can exclusively substitute for the activity of VirB1 from the putative transglycosylase active site. The complementation by the wild-type genes may be efficient due to the strong expression from the trc promoter, but since specific antisera were not available, we could not directly monitor protein levels. In contrast, VirB1 may interact with other VirB proteins (35, 53), and due to the limited sequence similarity, VirB1s and TraL may not be able to fulfill these transglycosylase-independent function(s). The substantial residual activity of VirB1E→A is in accord with the hypothesis that VirB1 may have additional functions, and previous work from other groups lends further support to this notion (37, 53). Further studies are required to dissect the different contributions of VirB1 transglycosylase activity and VirB protein interactions to T4SS function(s).
Acknowledgments
We are indebted to David O'Callaghan (Nîmes, France) and August Böck (Munich, Germany) for continued support and discussions. We thank Günter Koraimann (Graz, Austria) for discussions and for the communication of results prior to publication.
This work was supported by grants from the DAAD/Procope, the Deutsche Forschungsgemeinschaft DFG (Ba 1416-2/2), and the European Union Frame Programme 5 (contract QLK2-CT-2001-01200) to C.B. and by an NSF grant (MCB9817149) to A.N.B.
REFERENCES
- 1.Anderson, L. B., A. Vogel Hertzel, and A. Das. 1996. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. USA 93:8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, C., M. Llosa, S. Zhou, and P. C. Zambryski. 1997. C-terminal processing and cellular localization of VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens. J. Bacteriol. 179:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1366. [DOI] [PubMed] [Google Scholar]
- 4.Baron, C., Y. R. Thorstenson, and P. C. Zambryski. 1997. Biochemical analysis of the complex between the lipoprotein VirB7 and VirB9 in the membranes of Agrobacterium tumefaciens. J. Bacteriol. 179:1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer, M., R. Eferl, G. Zellnig, K. Terferle, A. Dijkstra, G. Koraimann, and G. Högenauer. 1995. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J. Bacteriol. 177:4279-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayer, M., R. Iberer, K. Bischof, E. Rassi, E. Stabentheiner, G. Zellnig, and G. Koraimann. 2001. Functional and mutational analysis of P19, a DNA transfer protein with muramidase activity. J. Bacteriol. 183:3176-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaupré, C. E., J. Bohne, E. M. Dale, and A. N. Binns. 1997. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 179:78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohne, J., A. Yim, and A. N. Binns. 1998. The Ti plasmid increases the efficiency of Agrobacterium tumefaciens as a recipient in virB-mediated conjugal transfer of an IncQ plasmid. Proc. Natl. Acad. Sci. USA 95:7057-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cellini, C., V. S. Kalogeraki, and S. C. Winans. 1997. The hydrophobic TraM protein of pKM101 is required for conjugal transfer and sensitivity to donor-specific bacteriophage. Plasmid 37:181-188. [DOI] [PubMed] [Google Scholar]
- 11.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 13.Das, A., L. B. Anderson, and Y.-H. Xie. 1997. Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J. Bacteriol. 179:3404-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das, A., and Y.-H. Xie. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dijkstra, A. J., and W. Keck. 1996. Peptidoglycan as a barrier to transenvelope transport. J. Bacteriol. 178:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dijkstra, B. W., and A.-M. W. H. Thunnissen. 1994. Holy proteins II: the soluble lytic transglycosylase. Curr. Opin. Struct. Biol. 8:810-813. [DOI] [PubMed] [Google Scholar]
- 17.Eisenbrandt, R., M. Kalkum, R. Lurz, and E. Lanka. 2000. Maturation of IncP pilin precursors resembles the catalytic dyad-like mechanism of leader peptidases. J. Bacteriol. 182:6751-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez, D., T. A. T. Dang, G. M. Spudich, X.-R. Zhou, B. R. Berger, and P. J. Christie. 1996. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 178:3156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez, D., G. M. Spudich, X.-R. Zhou, and P. J. Christie. 1996. The Agrobacterium tumefaciens virB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J. Bacteriol. 178:3168-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer, W., R. Haas, and S. Odenbreit. 2002. Type IV secretion systems in pathogenic bacteria. Int. J. Med. Microbiol. 292:159-168. [DOI] [PubMed] [Google Scholar]
- 21.Fischer, W., J. Püls, R. Buhrdorf, B. Gebert, S. Odenbreit, and R. Haas. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337-1348. [DOI] [PubMed] [Google Scholar]
- 22.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garfinkel, D. J., R. B. Simpson, L. W. Ream, F. F. White, M. P. Gordon, and E. W. Nester. 1981. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell 27:143-153. [DOI] [PubMed] [Google Scholar]
- 24.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hapfelmeier, S., N. Domke, P. C. Zambryski, and C. Baron. 2000. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J. Bacteriol. 182:4505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Höltje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakubowski, S. J., V. Krishnamoorthy, and P. J. Christie. 2003. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 185:2867-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koraimann, G. 2003. Cell wall degrading enzymes in macromolecular transport systems of Gram-negative bacteria. Cell. Mol. Life Sci. 60:2371-2378. [DOI] [PMC free article] [PubMed]
- 29.Krall, L., U. Wiedemann, G. Unsin, S. Weiss, N. Domke, and C. Baron. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, R. B., and A. Das. 2002. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 43:1523-1532. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Lai, E.-M., and C. I. Kado. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180:2711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai, E.-M., and C. I. Kado. 2000. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 8:361-369. [DOI] [PubMed] [Google Scholar]
- 34.Lessl, M., and E. Lanka. 1994. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77:321-324. [DOI] [PubMed] [Google Scholar]
- 35.Liu, Z., and A. N. Binns. 2003. Functional subsets of the VirB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010. J. Bacteriol. 185:3259-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 37.Llosa, M., J. Zupan, C. Baron, and P. C. Zambryski. 2000. The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis. J. Bacteriol. 182:3437-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maniatis, T. A., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Mushegian, A. R., K. J. Fullner, E. V. Koonin, and E. W. Nester. 1996. A family of lysozyme-like virulence factors in bacterial pathogens. Proc. Natl. Acad. Sci. USA 93:7321-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutus, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 41.Pohlman, R. F., H. D. Genetti, and S. C. Winans. 1994. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol. Microbiol. 14:655-668. [DOI] [PubMed] [Google Scholar]
- 42.Rohde, M., J. Püls, R. Buhrdorf, W. Fischer, and R. Haas. 2003. A novel sheathed surface organelle of the Helicobacter pylori type IV secretion system. Mol. Microbiol. 49:219-234. [DOI] [PubMed] [Google Scholar]
- 43.Sagulenko, E., V. Sagulenko, J. Chen, and P. J. Christie. 2001. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 183:5813-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range of 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt-Eisenlohr, H., N. Domke, C. Angerer, G. Wanner, P. C. Zambryski, and C. Baron. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181:7485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt-Eisenlohr, H., N. Domke, and C. Baron. 1999. TraC of IncN plasmid pKM101 associates with membranes and extracellular high molecular weight structures in Escherichia coli. J. Bacteriol. 181:5563-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stahl, L. E., A. Jacobs, and A. N. Binns. 1998. The conjugal intermediate of plasmid RSF1010 inhibits Agrobacterium tumefaciens virulence and VirB-dependent export of VirE2. J. Bacteriol. 180:3933-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tabor, S., and C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 50.van Asselt, E. J., K. H. Kalk, and B. W. Dijkstra. 2000. Crystallographic studies of the interactions of Escherichia coli lytic transglycosylase Slt35 with peptidoglycan. Biochemistry 39:1924-1934. [DOI] [PubMed] [Google Scholar]
- 51.van Asselt, E. J., A. M. Thunissen, and B. W. Dijkstra. 1999. High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment. J. Mol. Biol. 291:877-898. [DOI] [PubMed] [Google Scholar]
- 52.Vergunst, A. C., B. Schrammeijer, A. den Dulk-Ras, C. M. de Vlaam, T. J. Regensburg-Tuink, and P. J. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 53.Ward, D., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the A. tumefaciens vir-encoded type IV secretion system reveals novel protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winans, S. C., and G. C. Walker. 1985. Conjugal transfer system of the N incompatibility plasmid pKM101. J. Bacteriol. 161:402-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yanisch-Perron, C., J. Viera, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC18 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 56.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative DNA transfer processes. Harwood Academic Publishers, Amsterdam, The Netherlands.