Abstract
Several mycoplasma species are known to glide in the direction of the membrane protrusion (head-like structure), but the mechanism underlying this movement is entirely unknown. To identify proteins involved in the gliding mechanism, protein fractions of Mycoplasma mobile were analyzed for 10 gliding mutants isolated previously. One large protein (Gli349) was observed to be missing in a mutant m13 deficient in hemadsorption and glass binding. The predicted amino acid sequence indicated a 348,758-Da protein that was truncated at amino acid residue 1257 in the mutant. Immunofluorescence microscopy with a monoclonal antibody showed that Gli349 is localized at the head-like protrusion's base, which we designated the cell neck, and immunoelectron microscopy established that the Gli349 molecules are distributed all around this neck. The number of Gli349 molecules on a cell was estimated by immunoblot analysis to be 450 ± 200. The antibody inhibited both the hemadsorption and glass binding of M. mobile. When the antibody was used to treat gliding mycoplasmas, the gliding speed and the extent of glass binding were inhibited to similar extents depending on the concentration of the antibody. This suggested that the Gli349 molecule is involved not only in glass binding for gliding but also in movement. To explain the present results, a model for the mechanical cycle of gliding is discussed.
Mycoplasmas are parasitic, small-genome bacteria that lack a peptidoglycan layer (28). Several mycoplasma species, including Mycoplasma pneumoniae, M. genitalium, M. pulmonis, M. gallisepticum, and M. mobile, have distinct cell polarity and exhibit gliding motility, a smooth translocation of cells across solid surfaces in the direction of the tapered end (17, 22, 36). The gliding motility of mycoplasmas is believed to be involved in pathogenicity, but the mechanisms underlying gliding motility have not been investigated well (19). Mycoplasmas have no surface structures, such as flagella or pili, or any homologs of genes that encode such structures. Neither do they have genes related to other bacterial motility systems or to eukaryotic motor proteins (5, 11, 13, 21). These facts suggest that mycoplasmas glide by an entirely unknown mechanism.
M. mobile, isolated in the early 1980s from the gills of a freshwater fish, is the fastest-gliding mycoplasma (18). M. mobile glides on glass in the direction of its tapered end, where its so-called head-like structure is. Its average speed is 2.0 to 4.5 μm/s, about 3 to 7 times its cell length per second (32), and its maximum force can reach as high as 27 pN (23). It binds easily to glass and glides smoothly without pausing regardless of its growth stage. These distinct characteristics have allowed for detailed analyses of its gliding (9, 23-25, 32, 33) as well as for the isolation of gliding mutants, which are characterized by reduced or deficient gliding or by enhanced speed (26). However, no proteins related to gliding have been identified. In this study, we identified a huge protein that is truncated in a nonadhesive mutant and that is responsible for hemadsorption and glass-binding during gliding.
MATERIALS AND METHODS
Strains and culture conditions.
M. mobile strain 163K (ATCC 43663) and its mutants (26) were grown at 25°C in Aluotto medium, consisting of 2.1% heart infusion broth, 0.56% yeast extract, 10% horse serum, 0.025% thallium acetate, and 0.005% ampicillin (1), to an optical density at 600 nm of around 0.07, which corresponds to 7 × 108 CFU/ml.
Triton X-100 extraction.
The cultured cells were centrifuged at 12,000 × g for 10 min at 4°C and washed three times with phosphate-buffered saline (PBS). The cells were suspended in 20 mM Tris-HCl (pH 7.5), 0.15 M NaCl, and 0.1 mM phenylmethylsulfonyl fluoride and extracted with 1% (vol/vol) Triton X-100. After incubation at 37°C for 20 min, the suspension was centrifuged at 25,000 × g for 15 min at 4°C, and the Triton-insoluble fraction was recovered as a pellet. The insoluble fraction was analyzed by sodium dodecyl sulfate (SDS)-5, 10, and 15% polyacrylamide gel electrophoresis (PAGE) and stained with silver. The molecular mass was estimated by a broad-range protein marker (New England BioLabs, Inc., Beverly, Mass.).
Cloning and sequencing.
The Triton-insoluble fraction obtained from a 150-ml culture was subjected to SDS-5% PAGE and stained with Coomassie brilliant blue. The Gli349 protein bands were excised and equilibrated with Tris-SDS buffer, consisting of 125 mM Tris-HCl (pH 6.8) and 10% SDS. The bands were put into three wells, each 7 mm wide, 1 mm thick, and 15 mm deep. The bands in each well were overlaid with 5 μl of a solution containing 2 μg of V8 protease. Gli349 was partially digested and separated on a gel according to the Cleveland method (6), transferred to an Immobilon-PSQ membrane (Millipore, Inc., Billerica, Mass.), and stained with amido black. Edman degradation of 18- and 20-kDa protein bands revealed N-terminal amino acid sequences of eight residues, EITNLVQG and EVSDQNII, respectively. Four degenerate DNA sequences were designed from the amino acid sequences as overlapping nested primers: M1-18-5F (5′-GANATHACNAAYYTNGTNC-3′), M1-18-5S (5′-HACNAAYYTNGTNCARGG-3′), M1-20-3F (5′-DATDATRTTYTGRTCNSWNA-3′), and M1-20-3S (5′-RTTYTGRTCNSWNACNTC-3′). The primary PCR was performed with the primers M1-18-5F and M1-20-3F with chromosomal DNA obtained by the Genomic-tip system (Qiagen, Hilden, Germany) as a template. A 5-kb DNA fragment was amplified by the secondary PCR with the primers M1-18-5S and M1-20-3S and sequenced by using ABI PRISM 310 (Applied Biosystems, Foster City, Calif.). The regions outside the 5-kb fragment were sequenced directly, starting with primers inside the 5-kb fragment, by using the direct genomic DNA sequencing protocol (Qiagen). A region of about 30 kb was sequenced. The gli349 gene of mutant m13 was amplified by PCR as described above and sequenced.
Immunoblotting analysis and estimation of the number of Gli349 molecules.
Whole-cell lysate was fractionated by SDS-10% PAGE and subjected to immunoblotting analysis. This analysis used a hybridoma medium containing a mouse monoclonal antibody obtained by immunizing a BALB/c mouse with intact M. mobile cells (unpublished data). The concentration of monoclonal antibody in the ascitic fluid was titrated by enzyme-linked immunosorbent assay (30) with a mouse immunoglobulin G1 (IgG1) antibody (Ancell, Bayport, Minn.) as the standard. The antibody bound to the target protein on the membrane was detected by using anti-mouse IgG horseradish peroxidase conjugate (Promega, Madison, Wis.) and a chemiluminescence reaction (Western lightning; PerkinElmer Life Sciences Inc., Wellesley, Mass.).
The number of Gli349 molecules on a cell surface was estimated based on the amount of antibody needed to saturate its binding sites on a cell. Various concentrations (0, 0.0008, 0.002, 0.008, 0.02, 0.08, 0.2, 0.8, 2, 8, and 20 μg/ml) of the anti-Gli349 antibody and of a mock mouse IgG1 antibody (Ancell) were incubated with mycoplasma cells at 5 × 108 CFU/ml in 1 ml of Aluotto medium at 4°C for 3 h. The cells bound with antibodies were collected by centrifugation at 12,000 × g for 4 min at 4°C, washed three times with PBS, and subjected to SDS-PAGE with the bound antibodies. The antibody bound to cells was detected by immunoblotting on an X-ray film with the anti-mouse IgG antibody conjugated with horseradish peroxidase. The band intensity was measured by NIH Image (version 1.61) from band images scanned by a transmission scanner (GT9800F; Epson, Nagano, Japan). The amount of antibody was calculated by using various amounts of the anti-Gli349 and the mock antibodies applied to immunoblots as standards. The linear correlation was confirmed between the amount of standard antibody and its band intensity. Four measurements were taken from each independent culture, and the average was obtained.
Immunofluorescence microscopy.
Cultured cells were collected by centrifugation at 12,000 × g for 4 min at 20°C and suspended in fresh Aluotto medium at a 10-fold-higher concentration. Then, 200-μl suspension samples were put on coverslips and incubated at 25°C for 1 h. After the medium was removed, the attached cells were fixed with 3.0% (wt/vol) paraformaldehyde and 0.1% (vol/vol) glutaraldehyde in PBS. The following procedure was done as described previously (34, 35), except that the cells were not treated with Triton X-100 or dried. The fixed cells were stained with 0.2 μg of monoclonal antibody/ml. The secondary antibody was a goat antibody labeled with Cy3. The cells were observed with Olympus BX50 and IX71 microscopes and photographed with an ORCA-ER charge-coupled device (CCD) camera (Hamamatsu Photonics, Hamamatsu, Japan) attached to the IX71 microscope.
Immunoelectron microscopy.
The cultured cells were collected by centrifugation and suspended in fresh Aluotto medium at a 50-fold-higher concentration. The cell suspension was placed on a collodion-coated nickel grid for 10 min at room temperature to let mycoplasmas bind to the grid. After removal of the medium, the cells were fixed with 3.0% (wt/vol) paraformaldehyde and 0.1% (vol/vol) glutaraldehyde in PBS for 10 min at room temperature and subsequently fixed for 30 min at 4°C. The cells were washed three times with PBS and incubated for 1 h at room temperature with hybridoma medium diluted 1:10 with PBS and containing 0.2 μg of anti-Gli349 antibody/ml. After three washes with PBS, labeling for 1 h at room temperature was performed with an electron microscopy goat anti-mouse IgG (heavy plus light chains) conjugated to 10-nm-diameter gold beads (BB International, Cardiff, United Kingdom) diluted 1:20 with PBS. The cells were washed three times with PBS, negatively stained with 2% ammonium molybdate (7), and observed with a transmission electron microscope at a tilt angle of 10°.
Hemadsorption assay.
Colonies grown on 0.7% agar medium for 6 days were overlaid with sheep red blood cells (RBCs) suspended in PBS, as previously described (26, 37), and observed under an inverted microscope. The antibody's effect on hemadsorption was tested by adding it to the RBC suspension before the suspension was overlaid on colonies. A monoclonal antibody against a 57.5-kDa cell surface protein of M. mobile was used as a mock antibody. The colonies were photographed with an Olympus CK2 inverted microscope equipped with a Photometrics CoolSNAP CCD camera (Roper, Atlanta, Ga.).
Inhibition of gliding.
Cultured cells were collected by centrifugation and suspended in Aluotto medium at a 30-fold-higher concentration. The cell suspension was poured into a tunnel (8-mm interior width, 26-mm length, and 60-μm wall thickness) constructed by taping a coverslip to a glass side. The glasses were cleaned with saturated ethanolic KOH (3). The suspension was then incubated for 5 min at room temperature. The nonbinding cells were removed by a flow of 200 μl of Aluotto medium. The effects of the antibody on the gliding M. mobile cells were observed after the Aluotto medium was replaced with medium containing various concentrations of anti-Gli349 monoclonal antibody. The antibody against a 57.5-kDa cell surface protein was used as a mock antibody. The cell movements were observed under an Olympus BX50 microscope. The images were recorded by a WV-BP510 CCD camera (Panasonic, Osaka, Japan) onto mini-DV tapes and were transferred to a personal computer via DV Raptor (Canopus Co., Kobe, Japan). To make still images, the video footage was converted into sets of 20 frames for 2 s by using Video Editor, version 2.0, editing software (Ulead Systems, Inc., Taipei, Taiwan). An average image was produced from each set of frames inverted and the last inverted frame subtracted with Scion Image PC, beta version 4.0.2, software (Scion Corp., Frederick, Md.). To measure the number of bound cells and their gliding speeds, the video was converted into sets of 6 frames per 2 s by using Video Editor, version 6.0 (Ulead Systems). The number of cells bound to a glass surface 65.3 μm wide by 48.9 μm long was counted. The initial number of cells was more than 150. To measure gliding speeds, the mass center of cells gliding in the square was determined by Scion Image, and their speeds were calculated from their displacements, as described by Miyata et al. (23). Speed as reported here is the average speed of 10 gliding cells (or lower at the points where the log10 bound cell ratio was less than −1.5 because there were fewer than 10 gliding cells in the field).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper were deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB074422 (wild-type) and AB074421 (m13).
RESULTS
Identification of protein missing in nongliding mutant.
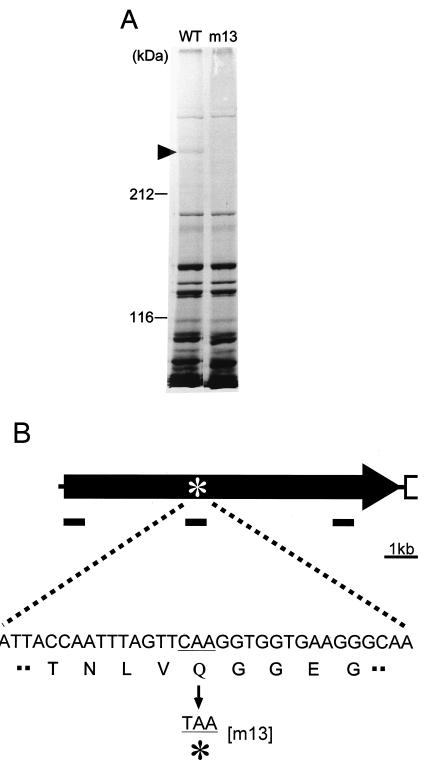
Ten gliding mutants were obtained previously by UV irradiation (26). To identify the mutated genes, we examined the protein profiles of Triton X-100-insoluble fractions for the wild-type strain and these gliding mutants. As the adhesion protein, P1, of M. pneumoniae is found in Triton-insoluble fractions (29), we focused on the Triton-insoluble fraction of M. mobile. M. mobile cells were treated with 1% Triton X-100 at 37°C for 20 min followed by washing with PBS and were then divided into Triton-insoluble and Triton-soluble fractions. The former were analyzed by SDS-PAGE at three different concentrations of polyacrylamide for all strains, resulting in the detection of about 40 major protein bands. Only one band was determined to be missing in a mutant; the position of this band was extrapolated to an apparent mass of 240 kDa in the wild-type strain (Fig. 1A). The mutant m13 is characterized by an inability to bind to glass and consequently by an inability to glide.
FIG. 1.
Mutation of Gli349 protein in nonbinding mutant m13. (A) The Triton-insoluble fractions were subjected to SDS-5% PAGE and stained by a silver staining method. The arrowhead shows a protein missing in the mutant m13. Molecular masses are indicated on the left. WT, wild type. (B) The gli349 gene is represented by the arrow. The site of the point mutation in mutant m13 is indicated by an asterisk. Short solid bars indicate the approximate positions of amino acid sequences determined by Edman degradation (not drawn to scale).
Cloning and sequencing of gli349 gene.
Focused protein bands were collected from the Triton X-100-insoluble fraction following separation by 5% gel electrophoresis. The bands were then partially digested with V8 protease and fractionated electrophoretically according to a method previously reported (6). The 8 N-terminal amino acid residues of two peptide digests were sequenced by Edman degradation. DNA primers were designed from the amino acid sequences, and the DNA sequence of the gene encoding the missing protein was determined (Fig. 1B). A long open reading frame (ORF) was found to encode a polypeptide of 3,183 amino acids. The predicted molecular mass and isoelectric point of the protein, which we refer to as Gli349, were 349 kDa and 4.9, respectively. Edman degradation analysis of the N terminus of the whole protein resulted in a sequence corresponding to the predicted N-terminal sequence of the ORF. The amino acid sequences deduced by the partial digests corresponded to the sequences of amino acids 1251 to 1258 and 2847 to 2854, respectively. These results shows that the ORF is at least translated from amino acids 1 to 2854 and not processed in this region. Sequence analysis of the gene in mutant m13 revealed a nonsense mutation at amino acid residue 1257 (Fig. 1B). A CAA codon in the wild-type strain is mutated to a TAA nonsense codon in mutant m13. The primary structure of the gene was significantly similar to that of the ORF MYPU_2110 from the gliding mycoplasma M. pulmonis, which is a mouse pathogen (5). The amino acid sequences were similar in all regions; 22% identity and 40% similarity were found especially in the region expanding from amino acid residues 623 to 2929 of the gli349 gene of M. mobile. Neither protein sequence has any cysteine residue. Using SOSUI, an algorithm to predict a membrane-spanning segment, a transmembrane segment preceded by a positively charged region can be predicted near the N terminus for both sequences (14). No significant similarity to other protein sequences was detected, and no motifs, such as a P-loop involved in the motor proteins, were found.
Truncated gli349 gene product in mutant m13.
The anti-Gli349 monoclonal antibody was obtained by screening a hybridoma established after immunizing a BALB/c mouse with intact M. mobile cells. Cell lysates from the wild-type strain and mutant m13 were examined by immunoblot analysis with a hybridoma medium containing the anti-Gli349 monoclonal antibody (Fig. 2). The antibody recognized only the Gli349 protein band in the whole-cell lysate of the wild-type strain. In mutant m13, a band was detected with an approximate mass of 137 kDa, which corresponds to the size of the truncated polypeptide predicted by the DNA sequence.
FIG. 2.
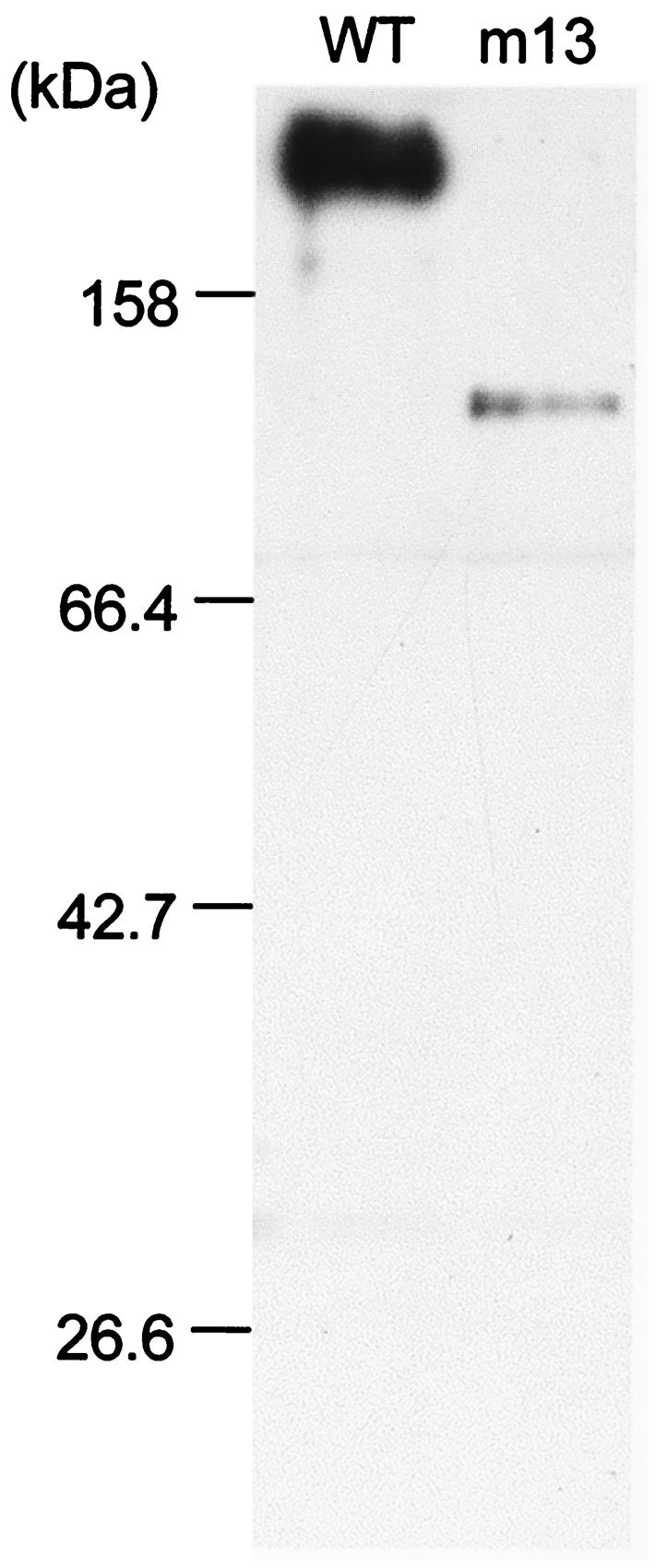
Immunoblot analysis of Gli349 in the wild-type (WT) strain and mutant m13. Cell lysates of the strains were analyzed by immunoblotting following SDS-10% PAGE with an anti-Gli349 monoclonal antibody. Molecular masses are indicated on the left.
Subcellular localization and number of Gli349 molecules.
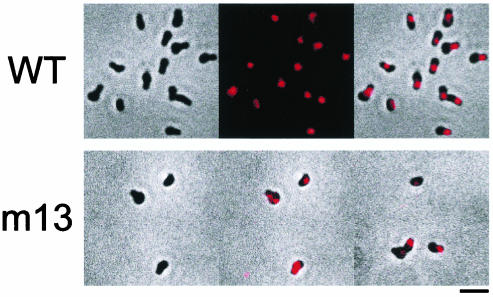
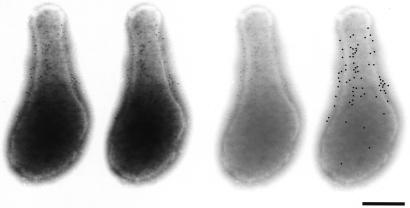
The localization of Gli349 in the wild-type cells was examined by immunofluorescence microscopy (Fig. 3, upper panel). The results clearly showed that Gli349 is located at the head-like structure's basal position, which we call the neck. The localization of the truncated product in mutant m13 was examined by immunofluorescence microscopy (Fig. 3, lower panel), but no signal was found with the same exposure time as that for the wild-type cells. A 10-times-longer exposure revealed the signal on the cell surface, but its position was not uniform, unlike the case for the wild-type cells. Neck localization of wild-type Gli349 was confirmed by using electron microscopy to examine subcellular localization at a higher resolution (Fig. 4). Three-dimensional immunoelectron microscopic images revealed gold particles on all sides of a cell.
FIG. 3.
Subcellular localization of Gli349 examined by immunofluorescence microscopy. The upper panels show wild-type (WT) cells. (Left) Phase-contrast image; (middle) antibody-stained image; (right) merge of the phase-contrast and antibody-stained images. The lower panels show m13 mutant cells. (Left) Merge obtained with the same exposure time for immunofluorescence as that for the wild-type cells; (middle) same field as shown in the left panel but with a 10-times-longer exposure; (right) another merge field obtained with the longer exposure time. Bar, 2 μm.
FIG. 4.
Subcellular localization of Gli349 examined by immunoelectron microscopy. Gli349 was labeled with 10-nm-diameter gold particles. The left two images are a stereo pair. The right two images are pale presentations of the one on the left to show the distribution of gold particles in the body region. The positions of particles in the center-right image are shown on the far-right identical image marked as artificial dots. Most cells found on an electron microscopy grid showed a similar distribution of gold particles. A stereoscopic view is achieved by tilting the specimen 10 degrees. Bar, 0.2 μm.
The number of Gli349 molecules on the cell surface was examined based on the amount of antibody needed to saturate its binding sites on a cell. Various concentrations (0 to 20 μg/ml) of the antibody were incubated with mycoplasma cells at 5 × 108 CFU/ml, and the amount of antibody bound to the cell surface was titrated by centrifugation and immunoblotting. The amount increased linearly with the antibody concentration used, and saturation occurred at the concentration of 0.8 μg/ml. The number of Gli349 molecules on the cell surface was estimated to be 445 ± 197 per cell. The mock antibody used as a control did not bind to the cells.
To learn Gli349's association with the structures included in the Triton-insoluble fraction, we examined the existence of this protein in the extract and in the insoluble fraction. Sixty percent of the Gli349 was found in the soluble fraction, and the remainder was found in the insoluble fraction, which was slightly solubilized after two additional extractions.
Inhibition of hemadsorption by antibody.
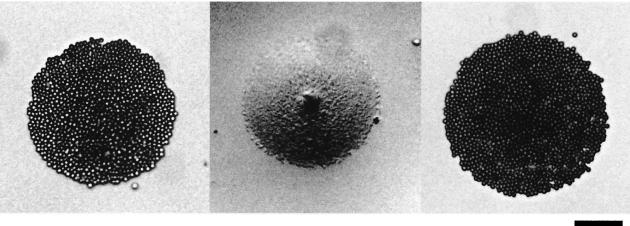
The mutant m13 is deficient in both the glass binding and static binding activities of animal cells, as estimated by the adsorption of RBCs to colonies formed on an agar plate (hemadsorption activity) (26). Therefore, Gli349 protein may be involved in both activities. To examine the possible involvement of this protein in static binding, the effect of the antibody on hemadsorption activity was examined (Fig. 5). RBC suspension with the addition of 0, 0.1, or 1 μg of anti-Gli349 antibody/ml was overlaid onto mycoplasma colonies. When no antibody was added, the colonies bound to RBCs across the entire area. The addition of 1 μg of anti-Gli349 antibody/ml inhibited the binding, whereas the same concentration of antibody against another surface protein showed no effect. The addition of 0.1 μg of anti-Gli349 antibody/ml resulted in no significant reduction in hemadsorption activity.
FIG. 5.
Inhibitory effects of anti-Gli349 antibody on hemadsorption activity. Colonies on an agar plate were overlaid with an RBC suspension containing antibodies. No antibody, 1 μg of anti-Gli349 antibody/ml, and 1 μg of mock antibody/ml were added to the colonies shown in the left, middle, and right panels, respectively. Bar, 50 μm.
Inhibition of gliding by antibody.
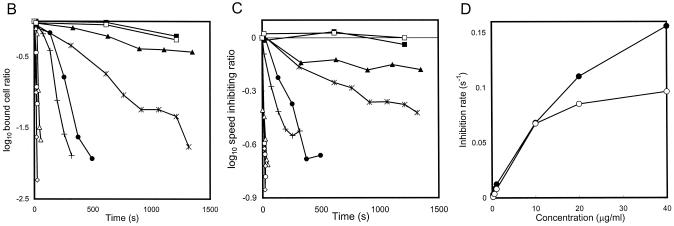
The effects of the antibody on gliding were examined (Fig. 6). Mycoplasma cells suspended in a fresh medium without antibody were inserted into a tunnel slide, and the medium was replaced by that containing various concentrations of the antibody, ranging from 0 to 40 μg/ml. Gliding cells were then observed continuously. When the medium had a 40-μg/ml concentration of antibody, all mycoplasmas lost their binding to glass, resulting in Brownian motion, whereas the same concentration of an antibody against another surface protein showed no effects (Fig. 6A). Detailed analyses showed that, with time, the anti-Gli349 antibody reduced both glass binding and gliding speed, although both were affected slightly by the addition of a mock antibody (Fig. 6B and C). The inhibition rates were estimated from the time required to reach 0.1 and 0.5 of the start values for glass binding and gliding speed, respectively. The rates were similar between glass binding and gliding speed at a wide range of antibody concentrations (Fig. 6D).
FIG. 6.
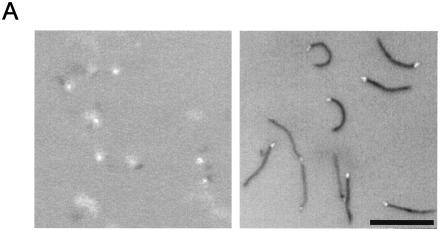
Inhibitory effects of anti-Gli349 antibody on glass binding and gliding speed. (A) Still cell images were obtained at 20 s after addition of 40 μg/ml of anti-Gli349 (left) or of mock antibody (right). Videos of 2-s duration observed by phase-contrast microscopy were averaged together and subtracted from the last frame. The focal plane was at the glass surface where the cells were gliding. Bar, 10 μm. (B) The number of cells bound to a glass surface with an area of 3,200 μm2 was counted after the addition of 0 (▪), 0.1 (▴), 0.2 (*), 0.5 (•), 1 (+), 10 (Δ), 20 (○), and 40 (⋄) μg of anti-Gli349 antibody/ml or of 40 μg of mock antibody (□)/ml. The ratio of the number of cells on the glass to the number of cells at time zero is presented for each time point. (C) Gliding speed after the addition of the antibody is represented as the ratio to the initial speed. The speeds were averaged for the cells on glass. The same symbols as those in panel B are used. (D) Inhibition rate (decreasing ratio per second) was calculated from the time required to reach 0.1 and 0.5 of the start values for glass binding (•) and gliding speed (○), respectively.
DISCUSSION
We fractionated M. mobile proteins by Triton X-100 extraction and looked for differences in protein contents between the wild-type strain and the mutants. Only in the Triton-insoluble fraction was Gli349 obviously missing from the mutant m13, which lacks both hemadsorption and glass binding activities (Fig. 1) (26). This suggested that Gli349 is responsible for hemadsorption and glass binding in gliding. This inference was proven by the specific inhibitory effects of an anti-Gli349 antibody (Fig. 5 and 6A).
The ORF for the protein truncated in the mutant m13 encoded a 349-kDa protein. The ORF contained some TGA triplets generally used as stop codons, suggesting that the TGA codon of M. mobile encodes tryptophan, as observed with other mycoplasmas (39). A homologous gene was found in M. pulmonis but not in the recently determined genome of Mycoplasma hyopneumoniae, (C. Minion, personal communication), although this species is believed to be more closely related to M. mobile (38). This might be consistent with the fact that neither gliding motility nor a tapered cell pole has been reported for M. hyopneumoniae thus far. Several cytadherence proteins have been identified from M. pneumoniae (2, 8, 16) and some other mycoplasma species (4, 10, 12, 15, 40), but none have shown any similarities to Gli349 in size or in primary structure. This suggests that adhesion proteins are diversified among mycoplasma species.
In immunoblotting, the monoclonal antibody recognized the Gli349 protein band for the wild-type strain and a 137-kDa band for mutant m13 (Fig. 2). The 137-kDa band is presumably a peptide of the N-terminal region and is truncated at the mutated site because the molecular mass estimated from the electrophoresis corresponded to the predicted molecular mass obtained from the sequence. Evidently, the epitope for the monoclonal antibody resides in this fragment. The existence of Gli349 in the other nine gliding mutants, including six strains with deficient or reduced glass binding activity (26), was examined by immunoblotting, but none showed significant differences from the wild-type strain. This suggests that additional proteins are involved in glass binding (data not shown).
Immunofluorescence and immunoelectron microscopy revealed that Gli349 molecules are localized to the neck of a cell (Fig. 3 and 4). Based on the results of previous studies, we expected that the gliding proteins would be located at the distal end of the head-like structure because gliding cells that each carried a bead attached to its tail were reoriented to glide upstream by the flow of fluid to the tail (23) and because an elongated head-like structure found in a very old culture moved forward, sometimes leaving the cell body in one position, resulting in a stretched head-like structure (25). The localization of Gli349 at the cell neck is consistent with these previous observations. The three-dimensional immunoelectron microscopic images revealed that Gli349 molecules are distributed around the cell neck, indicating that Gli349 molecules on the cell surface exist also on sides other than that facing the glass surface.
The truncated protein in m13 mutant cells appeared to be present in reduced amounts and was randomly localized on the cell surface (Fig. 3, lower panel). As permeabilization of cells in the staining procedure by a previously described method (34, 35) did not increase the signal intensity (data not shown), it is unlikely that the major part of the truncated protein exists inside the cell. We failed to detect the truncated molecule by immunoblot analysis in the supernatant of m13 culture (data not shown). These facts suggest that the truncated molecules were exported from inside the cell and anchored to the cell membrane but were not localized normally. Possibly, the physical interaction with other structures involving the truncated region is essential for the normal localization of the Gli349 molecule on a cell.
The maximum force generated in gliding was previously measured as 27 pN (23). Considering this rather large force and the possible participation of this protein in the gliding mechanism as explained below, the Gli349 molecule should be supported by a robust structure. Such a structure may be included in the Triton-insoluble fraction, in which 40% of Gli349 molecules were detected. One transmembrane stretch preceded by a cluster of positively charged residues was predicted at the N terminus of Gli349 (14). A monoclonal antibody raised against the cell surface affected hemadsorption and glass binding from the outside (Fig. 5 and 6). These observations suggest that Gli349 is a membrane-anchored protein, a large part of which is outside the cell, and also that the molecule is supported by the robust structure. Gli349 shares this feature with the cytadherence-related proteins of M. pneumoniae, linked to a Triton-insoluble structure, called the Triton shell (27, 29), which is thought to involve cytoskeletal structures, i.e., a filamentous network and a rod structure (20).
The anti-Gli349 antibody inhibited hemadsorption (Fig. 5), and it removed the gliding mycoplasmas from the glass in a concentration-dependent manner (Fig. 6B and D). The monoclonal antibody against a 57.5-kDa protein had no effect on either activity, although it also recognized the protein from the outside. Two other antibodies that recognized the surface proteins showed effects on neither gliding nor glass binding (data not shown). These observations show that Gli349 is responsible for both static binding of animal cells and glass binding essential for gliding. This is consistent with the previous observations that the gliding mutants showed similar extents of reduction in both activities, suggesting that these two activities share the same machinery, in M. mobile (26).
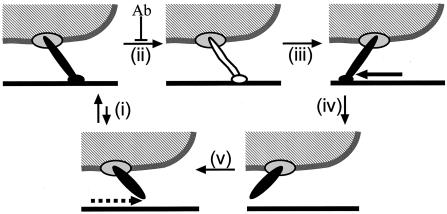
The addition of antibody against Gli349 reduced gliding speed with time in a concentration-dependent manner, in a way similar to that for glass binding (Fig. 6B, C, and D). As the antibody ultimately removed the gliding mycoplasmas from the glass surface, the binding of the antibody should release Gli349 molecules from the glass surface. Then how did the antibody reduce the gliding speed? This can be explained by an assumption that the gliding mechanism involves the movement of the Gli349 molecule, which can be inhibited by the antibody. One possible model is presented in Fig. 7, where the cell propels itself by repeating a cycle of binding the Gli349 protein as follows: (i) binding of Gli349 to glass, (ii) transfer of generated force to Gli349, (iii) stroke of Gli349, (iv) release of Gli349 from glass, and (v) return of Gli349 to its original form. Here, we know neither the actual molecular shape nor the manner of movement. If we assume that the step i is inhibited by the antibody, the reduction of speed should be caused by the lack of force needed to thrust a cell, which resulted from a decrease in the number of working molecules. However, the actual maximal force of gliding, 27 pN, is 1,800 times larger than 15 fN, the force required to thrust an unbound M. mobile cell in a medium (23, 24, 31). Therefore, it is unlikely that step i is inhibited by the antibody. If we assume that step ii is inhibited by the antibody, the Gli349 molecule would generate drag force, thereby reducing speed. In this case, the ultimate release of cells from glass can occur through a backward transition in step i.
FIG. 7.
One possible schematic explanation for the effect of the antibody. A mycoplasma cell is represented by a hatched area and gray line. The Gli349 molecule is presented as a rod sticking out of the cell and supported by a scaffold represented by an ellipse on the cell. The glass surface is represented by a solid line. The cyclic movement of Gli349, including binding, stroke, and release, thrusts the cell body. The Gli349 molecule is represented differently for the bound, free, and force-transmitting forms. The binding of antibody inhibits step ii and removes Gli349 from the glass surface through the backward transition of step i. See Discussion for details.
Acknowledgments
We thank Howard C. Berg of Harvard University for comments on the manuscript and Shintaro Seto of Osaka City University for helpful discussions.
This work was supported in part by grants-in-aid for scientific research (C) and for science research on priority areas (motor proteins, genome science, and infection and host response) from the Ministry of Education, Science, Sports, Culture, and Technology of Japan to M.M.
REFERENCES
- 1.Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58. [Google Scholar]
- 2.Baseman, J. B., R. M. Cole, D. C. Krause, and D. K. Leith. 1982. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J. Bacteriol. 151:1514-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, H. C., and L. Turner. 1993. Torque generated by the flagellar motor of Escherichia coli. Biophys. J. 65:2201-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boguslavsky, S., D. Menaker, I. Lysnyansky, T. Liu, S. Levisohn, R. Rosengarten, M. Garcia, and D. Yogev. 2000. Molecular characterization of the Mycoplasma gallisepticum pvpA gene which encodes a putative variable cytadhesin protein. Infect. Immun. 68:3956-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleveland, D. W., S. G. Fischer, M. W. Kirschner, and U. K. Laemmli. 1977. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J. Biol. Chem. 252:1102-1106. [PubMed] [Google Scholar]
- 7.Cole, R. M. 1983. Transmission electron microscopy: basic techniques, p. 43-50. In S. Razin and J. S. Tully (ed.), Methods in mycoplasmology. Academic Press, New York, N.Y.
- 8.Feldner, J., U. Gobel, and W. Bredt. 1982. Mycoplasma pneumoniae adhesin localized to tip structure by monoclonal antibody. Nature 298:765-767. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, M., H. Kirchhoff, R. Rosengarten, G. Kerlen, and K.-H. Seack. 1987. Gliding movement of Mycoplasma sp. nov. strain 163K on erythrocytes. FEMS Microbiol. Lett. 40:321-324. [Google Scholar]
- 10.Fleury, B., D. Bergonier, X. Berthelot, E. Peterhans, J. Frey, and E. M. Vilei. 2002. Characterization of P40, a cytadhesin of Mycoplasma agalactiae. Infect. Immun. 70:5612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J.-F. Tomb, B. A. Dougherty, K. F. Bott, P.-C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 12.Goh, M. S., T. S. Gorton, M. H. Forsyth, K. E. Troy, and S. J. Geary. 1998. Molecular and biochemical analysis of a 105 kDa Mycoplasma gallisepticum cytadhesin (GapA). Microbiology 144:2971-2978. [DOI] [PubMed] [Google Scholar]
- 13.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B.-C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirokawa, T., B. C. Seah, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, T., S. Artiushin, and F. C. Minion. 1997. Cloning and functional analysis of the P97 swine cilium adhesin gene of Mycoplasma hyopneumoniae. J. Bacteriol. 179:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, P. C., R. M. Cole, Y. S. Huang, J. A. Graham, D. E. Gardner, A. M. Collier, and W. A. Clyde, Jr. 1982. Mycoplasma pneumoniae infection: role of a surface protein in the attachment organelle. Science 216:313-315. [DOI] [PubMed] [Google Scholar]
- 17.Kirchhoff, H. 1992. Motility, p. 289-306. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas—molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 18.Kirchhoff, H., and R. Rosengarten. 1984. Isolation of a motile mycoplasma from fish. J. Gen. Microbiol. 130:2439-2445. [DOI] [PubMed] [Google Scholar]
- 19.Kirchhoff, H., R. Rosengarten, W. Lotz, M. Fischer, and D. Lopatta. 1984. Flask-shaped mycoplasmas: properties and pathogenicity for man and animals. Isr. J. Med. Sci. 20:848-853. [PubMed] [Google Scholar]
- 20.Krause, D. C. 1996. Mycoplasma pneumoniae cytadherence: unravelling the tie that binds. Mol. Microbiol. 20:247-253. [DOI] [PubMed] [Google Scholar]
- 21.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 22.Miyata, M. 2002. Cell division, p. 117-130. In R. Herrmann and S. Razin (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluver Academic/Plenum Publishers, London, England.
- 23.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyata, M., W. S. Ryu, and H. C. Berg. 2001. Force-velocity relationship of mycoplasma gliding. Biophys. J. 80:70. [Google Scholar]
- 25.Miyata, M., and A. Uenoyama. 2002. Movement on the cell surface of gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215:285-289. [DOI] [PubMed] [Google Scholar]
- 26.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311-1320. [DOI] [PubMed] [Google Scholar]
- 27.Razin, S., and E. Jacobs. 1992. Mycoplasma adhesion. J. Gen. Microbiol. 138:407-422. [DOI] [PubMed] [Google Scholar]
- 28.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regula, J. T., G. Boguth, A. Gorg, J. Hegermann, F. Mayer, R. Frank, and R. Herrmann. 2001. Defining the mycoplasma 'cytoskeleton': the protein composition of the Triton X-100 insoluble fraction of the bacterium Mycoplasma pneumoniae determined by 2-D gel electrophoresis and mass spectrometry. Microbiology 147:1045-1057. [DOI] [PubMed] [Google Scholar]
- 30.Roggendorf, M., R. Wigand, F. Deinhardt, and G. G. Frosner. 1982. Enzyme-linked immunosorbent assay for acute adenovirus infection. J. Virol. Methods 4:27-35. [DOI] [PubMed] [Google Scholar]
- 31.Rosengarten, R., M. Fisher, H. Kirchhoff, G. Kerlen, and K.-H. Seack. 1988. Transport of erythrocytes by gliding cells of Mycoplasma mobile 163K. Curr. Microbiol. 16:253-257. [Google Scholar]
- 32.Rosengarten, R., and H. Kirchhoff. 1987. Gliding motility of Mycoplasma sp. nov. strain 163K. J. Bacteriol. 169:1891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosengarten, R., A. Klein-Struckmeier, and H. Kirchhoff. 1988. Rheotactic behavior of a gliding mycoplasma. J. Bacteriol. 170:989-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seto, S., and M. Miyata. 2003. Attachment organelle formation represented by localization of cytadherence protein and formation of the electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu, T., and M. Miyata. 2002. Electron microscopic studies of three gliding mycoplasmas, Mycoplasma mobile, M. pneumoniae, and M. gallisepticum, by using the freeze-substitution technique. Curr. Microbiol. 44:431-434. [DOI] [PubMed] [Google Scholar]
- 37.Sobeslavsky, O., B. Prescott, and R. M. Chanock. 1968. Adsorption of Mycoplasma pneumoniae to neuraminic acid receptors of various cells and possible role in virulence. J. Bacteriol. 96:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weisburg, W. G., J. G. Tully, D. L. Rose, J. P. Petzel, H. Oyaizu, D. Yang, L. Mandelco, J. Sechrest, T. G. Lawrence, J. Van Etten, J. Maniloff, and C. R. Woese. 1989. A phylogenetic analysis of the mycoplasmas: basis for their classification. J. Bacteriol. 171:6455-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamao, F., A. Muto, Y. Kawauchi, M. Iwami, S. Iwagami, Y. Azumi, and S. Osawa. 1985. UGA is read as tryptophan in Mycoplasma capricolum. Proc. Natl. Acad. Sci. USA 82:2306-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Q., and K. S. Wise. 1996. Molecular basis of size and antigenic variation of a Mycoplasma hominis adhesin encoded by divergent vaa genes. Infect. Immun. 64:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]