Abstract
Liu et al. recently described a group of related temperate bacteriophages that infect Bordetella subspecies and undergo a unique template-dependent, reverse transcriptase-mediated tropism switching phenomenon (Liu et al., Science 295: 2091-2094, 2002). Tropism switching results from the introduction of single nucleotide substitutions at defined locations in the VR1 (variable region 1) segment of the mtd (major tropism determinant) gene, which determines specificity for receptors on host bacteria. In this report, we describe the complete nucleotide sequences of the 42.5- to 42.7-kb double-stranded DNA genomes of three related phage isolates and characterize two additional regions of variability. Forty-nine coding sequences were identified. Of these coding sequences, bbp36 contained VR2 (variable region 2), which is highly dynamic and consists of a variable number of identical 19-bp repeats separated by one of three 5-bp spacers, and bpm encodes a DNA adenine methylase with unusual site specificity and a homopolymer tract that functions as a hotspot for frameshift mutations. Morphological and sequence analysis suggests that these Bordetella phage are genetic hybrids of P22 and T7 family genomes, lending further support to the idea that regions encoding protein domains, single genes, or blocks of genes are readily exchanged between bacterial and phage genomes. Bordetella bacteriophages are capable of transducing genetic markers in vitro, and by using animal models, we demonstrated that lysogenic conversion can take place in the mouse respiratory tract during infection.
Parasite adaptation to dynamic host characteristics is a common theme in biology. We recently identified a unique mechanism of adaptation that governs the interactions between a group of bacterial pathogens belonging to the Bordetella genus and a family of bacteriophages that infect them (21). As pathogens of numerous mammalian species, Bordetella spp. undergo major changes in gene expression as they transition through their infectious cycles (9). As part of their adaptive strategy, Bordetella phages use a novel mechanism to evolve new ligands that allow the use of alternative surface receptors for host cell entry.
Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica are highly related, gram-negative coccobacilli that infect respiratory epithelial surfaces in humans and other mammals (25). In response to a variety of environmental signals, these subspecies modulate virulence gene expression through the BvgAS signal transduction system, which controls a spectrum of gene expression states. BvgAS signaling occurs through a multistep phosphorelay involving the BvgS transmembrane sensor kinase and the BvgA response regulator (41, 42). When the system is active (Bvg+ phase), expression of virulence factors such as adhesins, toxins, and a type III secretion system is induced. When BvgAS is inactive (Bvg− phase), an alternative set of genes are expressed, including motility and urease genes in B. bronchiseptica and virulence-repressed genes in B. pertussis (8).
BPP-1 is a temperate bacteriophage initially found in a clinical isolate of B. bronchiseptica that displays a marked tropism for Bvg+ phase B. pertussis, B. parapertussis, and B. bronchiseptica (21). The primary receptor for BPP-1 is pertactin, an outer membrane autotransporter protein that is only expressed in Bvg+ phase Bordetella spp. At a frequency of approximately 10−6, BPP-1 gives rise to two classes of tropic variants. One class, designated BMP (Bvg minus-tropic phage), has an acquired tropism for Bvg− phase bacteria. The second class, designated BIP (Bvg indiscriminate phage), can infect both Bvg+ and Bvg− phase B. bronchiseptica with equal efficiency. We showed that the tropism determinant mapped to a 134-bp sequence, VR1, located at the 3′ end of the mtd locus (21). Further examination demonstrated that VR1 undergoes site-specific sequence alterations at positions corresponding to adenine residues in a closely related repeat, the template repeat (TR), which is located downstream of VR1 in a noncoding region.
On the basis of our initial genetic analysis, we hypothesize that tropism switching involves the production of a TR-containing RNA intermediate followed by reverse transcription by the product of the phage-encoded reverse transcriptase (Brt) and subsequent integration of a mutagenized cDNA copy of TR at VR1. The mtd, VR1, TR, and brt loci comprise a novel “evolution cassette” that functions to generate diversity in ligand-receptor interactions. The extent of diversity appears to be vast, as the variability system is theoretically capable of generating nearly 1012 polypeptide sequences at the C terminus of Mtd (21).
To better understand the biology of Bordetella phages, we obtained the complete nucleotide sequences of BPP-1, BMP-1, and BIP-1 as part of the Bordetella genome sequencing project. We also carried out genetic and molecular analyses on a second region of variability within the bpp36 locus and on a unique phage methylase encoded by bpm. We demonstrated that all phage types could be used to transduce genetic markers between different strains of Bordetella and, using animal models of B. bronchiseptica colonization, we determined that in vivo lysogenic conversion could take place in the respiratory tract during infection.
MATERIALS AND METHODS
Bacterial strains, phage, plasmids, and media.
Table 1 lists the bacterial strains, phages, and plasmids used in this study. B. bronchiseptica and Escherichia coli were maintained on standard Luria-Bertani (LB) broth and LB agar as described previously (2). For allelic exchange, sucrose-sensitive cointegrants were grown in modified LB broth containing 10% sucrose with no NaCl (LBS). Antibiotics were routinely used at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; streptomycin, 60 μg/ml; rifampin, 20 μg/ml; and chloramphenicol, 20 μg/ml. Bordet-Gengou (BG) agar supplemented with 7.5% defibrinated sheep blood was used for routine growth of B. bronchiseptica. When appropriate, media were supplemented with 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml.
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| B. bronchiseptica | ||
| RB50 | Isolated in 1992 at UCLA from a rabbit, Smr | 2, 9 |
| RB30 | Isolated in 1992 at UCLA from a rabbit, Smr | J.F.M. lab collection |
| RB50Gm | RB50 with Gmr cassette | This work |
| BB3464 | Isolated from feline respiratory tract in 1988 at UC Davis | J.F.M. lab collection |
| ML6401 | BPP-1 lysogen of RB50 | This work |
| ML6403 | BIP-1 lysogen of RB50 | This work |
| ML6405 | BMP-1 lysogen of RB50 | This work |
| RB53 | RB50 bvgS-C3 | 2, 9 |
| RB54 | RB50 ΔbvgAS | 2, 9 |
| ML83DMIN | ML6401 BPP-1Δbpm | This work |
| ML83DM3AN | ML6403 BIP-1Δbpm | This work |
| ML83DM5A | ML6405 BMP-1Δbpm | This work |
| ML89-D36-1A | ML6401 BPP-1Δbbp36 | This work |
| ML89-D36-3A | ML6403 BIP-1Δbbp36 | This work |
| ML89-D36-5A | ML6405 BMP-1Δbbp36 | This work |
| MH406 | RB50Rf wbmD671::mTn5-lacZ1 | This work |
| G2-15 | RB50Rf fhaB3125::mTn5-lacZ1 | This work |
| E. coli | ||
| DH5α | F−hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(argF-lac)U169 φ80dlacZΔM15 | Gibco-BRL |
| XL1 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 lac [F′ proAB+ lacIqZΔM15::Tn10] | Stratagene |
| DH5αλpir | DH5α λpir | J.F.M. lab collection |
| Plasmids | ||
| pMLG13 | oriT Ampr Gmr ColE1 MCS from pBluescript II KS+ | This work |
| pML68-G3 | 1.4-kb EcoRI-BamHI insert containing the complete cI repressor CDS cloned into pMLG13 | This work |
| pRK2013 | Kmrori ColE1 RK2-Mob+ RK2-Tra+ | 11 |
| pML89-D36-6 | pRE112-based Δorf36 allelic exchange vector | This work |
| pML83-112M3A | pRE112-based Δbpm allelic exchange vector | This work |
| pBBR1MCS | Cmr broad-host-range plasmid | ATCC |
| pML93-bpm106 | pBBR1MCS-based plasmid expressing bpm | This work |
| pRE112 | Cmr allelic exchange vector | ATCC |
UCLA, University of California, Los Angeles; UC, University of California; MCS, multiple cloning site; CDS, coding sequence; ATCC, American Type Culture Collection.
Nucleic acid manipulation.
Standard methods were used for preparation of genomic and plasmid DNA, restriction enzyme digestions, agarose gel electrophoresis, DNA ligations, and other DNA manipulations (5, 35). To determine whether noncovalent circularization through base-pairing occurs in the phage genome, PstI-digested phage DNA was heated to 65°C for 30 min and immediately cooled on ice before being loaded onto a 0.5% agarose gel. Restriction enzymes and Taq polymerase were purchased from New England Biolabs, Promega, Roche, or Stratagene and used according to the manufacturer's instructions. Bordetella phage DNA was prepared according to the Qiagen lambda DNA minipreparations from plate lysates (Qiagen), except the column purification step was omitted.
Phage lysates.
For B. bronchiseptica, LB broth and agar were routinely used for phage propagation. Plate lysates were prepared by using the soft-agar overlay method (1). Briefly, 50 μl of an overnight B. bronchiseptica culture was added to 2.5 ml of 0.7% top agar kept molten at 42 to 46°C. Phage lysate was then added in sufficient quantity to cause confluent lysis within 24 h. Phage particles were eluted from the lawn by adding 4 ml of SM buffer (35) to the plate and incubating the plate at 4°C for 3 to 5 h. Resuspended lysates were passed through sterile syringe filters (Corning) (pore size, 0.2 μm) afterward. The phage titer was then determined by serial dilution with RB53 as the tester strain.
Transduction.
Phage lysates from donor strains were added to 300 to 500 μl of recipient strains at a multiplicity of infection of 0.1 to 0.01 and incubated at 37°C for 1 to 2 h. Cells were spun down at 12,000 × g for 5 min, washed twice in 0.85% saline, resuspended in 100 μl of 0.85% saline, and plated on BG (no blood) plates containing kanamycin. For each transduction, a control was performed by plating 100 μl of the corresponding lysate onto BG plates containing the corresponding antibiotics.
Electron microscopy.
Electron microscopy was performed by one of two methods. In the first, concentrated phage samples were diluted 30-fold into a volatile buffer (20 mM ammonium acetate, pH 7.4), then applied to single-carbon support films mounted on freshly cleaved mica as follows. Carbon films were floated onto 200 μl of sample, followed by washes in 20 mM ammonium acetate, pH 7.4, then in distilled H2O, for 2 min each. Samples were stained by floating the carbon onto 0.2% uranyl acetate for 30 s, then picking the film up onto a copper grid, and blotting off excess stain. In the second, carbon-coated Parlodion support films mounted on grids were made hydrophilic immediately before use by high-voltage alternating current glow discharge. Samples were applied directly onto grids and allowed to adhere for 2 min. The grids were rinsed with 3 drops of distilled water, negatively stained with 1% uranyl acetate, and blotted dry with filter paper. Specimens were examined in a Hitachi H-7000 electron microscope at an accelerating voltage of 75 kV.
Sequence determination.
Phage DNA was sonicated and size-fractionated on agarose gels. Plasmid libraries were generated in pUC18 with insert sizes of 1.4 to 2.0 kb. Each clone was sequenced once from each end with ABI Big-Dye terminator chemistry on an ABI 3700 capillary sequencing machines. The final sequences were generated from 705 sequencing reads, giving 7.4-fold total coverage (BMP-1); 875 sequencing reads, giving 8.8-fold total coverage (BIP-1); and 722 sequencing reads, giving 7.0-fold total coverage (BPP-1). All repeats were bridged by clone end read-pairs or end-sequenced PCR products to confirm the assembly.
Plasmid rescue.
To identify the phage integration site, a ColE1-based plasmid with a gentamicin resistance cassette, oriT, and a 1.4-kb insert containing the complete cI repressor coding sequence was transferred into lysogenized B. bronchiseptica strains ML6401 and ML6403 to create two cointegrants. Since the phage genomes did not contain any BamHI site while the vector backbone contained one BamHI site, genomic DNA preparations from the two cointegrants were digested with BamHI. The digested fragments were then self-ligated and transformed into competent E. coli XL1 cells. Gentamicin-resistant transformants were recovered. Two gentamicin-resistant plasmids, pML83-B101 from the ML6401 cointegrant and pML83-B301 from the ML6403 cointegrant, were chosen for further analysis.
Bacterial conjugation.
All B. bronchiseptica conjugations were carried out by triparental matings with the mobilizing strain DH5α(pRK2013) (11). Growth of Escherichia coli donors was inhibited with plates supplemented with streptomycin. B. bronchiseptica transformants or cointegrants were selected on the basis of their resistance to chloramphenicol.
PstI site protection by Bpm.
For expressing bpm from the broad-host-range plasmid pBBR1MCS, a PCR fragment was amplified from the BPP-1 genome with two primers, BpmHF (5′-AGCAAGCTTGCGCAAGCGTGGTCATCG-3′) and BpmBR (5′-AGCGGATCCCCGGTCAGATCAAATCGG-3′). The PCR fragment was amplified with Pfu Turbo (Stratagene) with pML68-16 as the template and cloned into the BamHI and HindIII sites of pBBR1MCS, downstream of the lac promoter Plac, to make pML93-bpm106. To test PstI sensitivity, complementary oligonucleotides were ordered that contained the sequence shown in Table 3 (Invitrogen). For each complementing pair, one oligonucleotide would contain a GATC overhang and the other would contain an AGCT overhang. The annealing was done by mixing 20 μl of each oligonucleotide (100 μM in water) and heating to 100°C for 10 min, followed by cooling in room temperature. The annealed fragments were then cloned into the BamHI and HindIII sites of pBluescript II KS+. The finished constructs were then transformed by themselves or cotransformed with pML93-bpm106 into E. coli DH5α. The plasmids were then purified by miniprep and analyzed by restriction digestions.
TABLE 3.
Summary of Bordetella phage mutantsa
| Phage | Relative plaquing efficiency
|
|
|---|---|---|
| Bvg+ | Bvg− | |
| BPP-1 | 1 | 10−6 |
| BMP-1 | 10−3 | 1 |
| BPP-1Δbpp36 | 1 | 10−4 |
| BMP-1Δbpp36 | 10−3 | 1 |
| BPP-1Δbpm | 1 | 10−6 |
| BMP-1Δbpm | 10−3 | 1 |
Phage titers were determined on Bvg+ and Bvg− bacteria after induction with mitomycin C (∼1011 PFU/ml), and for each strain the higher of the two titers was set arbitrarily to 1.
Construction of Δbpm and Δbbp36 mutants by allelic exchange.
The constructs for introducing in-frame deletions into bpm and bbp36 were made by overlap PCR with three primers each (20). For Δbpm, the primers were ΔMETA5 (5′-AGCGGATCCCAGCCCACCATCTG-3′), DMETB3 (5′-AGCGGATCCGCGTCGGTCGTGTCC-3′), and ΔMET-L was the linker primer (5′-CGCTCATCGCCATGCCGAGCGGCATCGAGCGCGAACC-3′). The final PCR fragment was first cloned into the EcoRI site of pBluescript II KS+, then excised with KpnI and XbaI for cloning into pRE112 to make pML83-112M3A.
For Δbbp36, the three primers were Δ36K-F (5′-AGCGGTACCATGTCCCTCGAAGCAGC-3′), Δ36X-R (5′-AGCTCTAGAATCGCCGCCCACGTGTC-3′), and the linker primer, Δ36-L (5′-CGAACTTGTGGTTGTCGTCGAGGCTCATGGCGTCAGCCCTCCATCGCC-3′). The PCR fragment was cloned into the KpnI and XbaI sites of pRE112 to make pML89-D36-6. Genomic DNA from ML6401 was used as the template in all reactions. The allelic exchange vectors were introduced into B. bronchiseptica strains ML6401, ML6403, and ML6405 via triparental mating. Sucrose-sensitive, chloramphenicol-resistant cointegrants were inoculated into LB overnight and then plated on LB agar plates supplemented with 10% sucrose. The resulting colonies were then screened for sensitivity to chloramphenicol and the in-frame deletion by PCR with primers flanking the target region. Multiple lysogens carrying the expected mutation in phage genes were then selected for further analysis.
High-pressure liquid chromatography and mass spectrometry analyses of phage DNA nucleoside composition.
We used the method described by Magrini et al. with slight modifications to analyze phage DNA composition (24). After hydrolysis and dephosphorylation, the free nucleosides were analyzed by reverse-phase high-pressure liquid chromatography with an analytical RP Microsorb-MV 300 Å C18 column (Varian, Walnut Creek, Calif.) on a Rainin Dynamax SD-200 solvent delivery system with a Rainin Dynamax PDA-2 diode array detector. The elution profiles were monitored at both 215 nm and 254 nm; 50 μl of each sample was loaded onto the column in a 50 mM KH2PO4 (pH 5.8) solvent containing 5% (vol/vol) methanol. The nucleosides were eluded at flow rate of 1 ml/min in an initial 5 to 10% methanol gradient over 20 min, followed by a 10 to 65% methanol gradient over 20 min. The collected fraction containing the peak at Rt = 29 min was subjected to electrospray mass spectrometry performed by the Yale Cancer Center Mass Spectroscopy Resource and the Howard Hughes Medical Institute Biopolymer Laboratory/W. M. Keck Foundation Biotechnology Resource Laboratory. Nucleoside standards (adenosine, N6-methyladenosine, and 5-methylcytosine dissolved in 1 mM deferoxamine mesylate-20 mM sodium acetate at pH 5) were run at the beginning and end of each set of analysis runs.
PCR conditions.
Taq polymerase was used unless specified otherwise. The reaction mixture contained 1× Taq assay buffer B (Promega), 2 mM MgCl2, 5% dimethyl sulfoxide, 1 μM each primer, and 200 μM deoxynucleoside triphosphate mix. PCR cycling conditions were as follows: initial denaturing at 95°C for 5 min, denaturing at 94°C for 1 min, annealing at 55°C unless specified otherwise, and extension at 72°C for 1 min per kb of expected PCR product. The cycle was repeated 29 more times and concluded with a 5-min final extension step.
Animal experiments.
Animals experiments were carried out as previously described (3, 12, 26). Briefly, C57BL/6 mice and Wistar rats were obtained from Charles River Laboratories (Wilmington, Mass.). Inocula were grown at 37°C in Stainer-Scholte broth and normalized by optical density at 600 nm. Rats and mice lightly sedated with halothane were given a dose consisting of 0.5 × 106 to 1.0 × 106 bacteria in 50 μl of phosphate-buffered saline. Colonization of the nasal cavity and a portion of the trachea (∼0.5 cm for mice and ∼1 cm for rats) was quantified by homogenizing each tissue in 200 μl of phosphate-buffered saline, plating aliquots onto BG blood agar, and counting the colonies after 2 days of incubation at 37°C.
Bioinformatics.
Artemis software was used to collate data and facilitate annotation (http://www.sanger.ac.uk/Software/Artemis/) (33). Phage DNA sequences were compared with the EMBL/GenBank entries by BlastN and BlastX (4). Potential coding sequences were identified with codon usage and positional base preference methods, and the predicted protein sequences were searched against a nonredundant protein database with WUBlastP and FastA. Inverted repeats were identified with the Emboss applications (http://www.uk.embnet.org/Software/EMBOSS/Apps). Sequences from the Bordetella sequencing projects are available from the Sanger Center web site (http://www.sanger.ac.uk/Projects/Microbes/). Bacterial signal peptides were predicted with the SignalP program (29). The sequences and annotations have been submitted to the EMBL and GenBank databases. Motifs are described by accession numbers from the Pfam and InterPro motif databases (prefixes PF and IPR, respectively). Rho-independent transcriptional terminators were identified with the TransTerm algorithm (10).
Nucleotide sequence accession number. The genome sequences determined here have been deposited in EMBL/GenBank under accession number AY029185.
RESULTS
Phage derivation and morphological analysis.
BPP-1 was originally isolated from B. bronchiseptica RB30, a rabbit strain, following UV induction. BMP-1 was derived from BPP-1 after repeated rounds of passage on RB54 and selection for tropism switching. BIP-1 was isolated from B. bronchiseptica BB3464, a cat strain, also following UV induction. BPP-1 and BIP-1 readily infect B. pertussis, B. parapertussis, and B. bronchiseptica strains in the Bvg+ phase, while BMP-1 only forms plaques on Bvg− phase B. bronchiseptica.
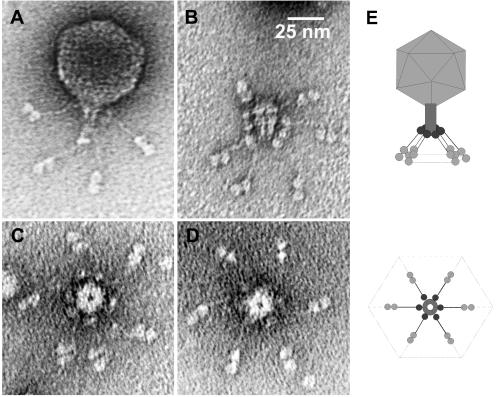
Figure 1 shows the morphology of BPP-1. On the basis of structural characteristics, it belongs to the Podoviridae family of phages with isometric heads and short noncontractile tails, generally similar in appearance to phages T7 and P22. Phage particles have an icosahedral capsid 60 nm in diameter, a short tubular tail with a decorating collar, and six tail fibers with unusual, bilobed globular ends. A remarkable overall hexagonal symmetry, akin to that of a snowflake, is prevalent in the structure of the capsid, the base plate, and the tail fibers. BPP-1 particles are considerably more stable in solution and show greater infectivity than either BMP-1 or BIP-1, presumably due to greater stability of pertactin-tropic Mtd. For this reason, BMP-1 and BIP-1 could not be sufficiently concentrated to obtain high-resolution electron microscopy images. At lower resolution, the morphologies of these phage were indistinguishable from that of BPP-1 (data not shown).
FIG. 1.
BPP-1 virion morphology. Negative-stain transmission electron micrographs of (A) an intact phage particle, (B) an isolated tail (side view), with partially dissociated tail fibers, and (C, D) isolated tails with tail fibers (top view). (E) Schematic diagram (not to scale) showing general particle morphology and hexagonal symmetry.
Determination of phage DNA sequences and attachment sites (attB and attP).
The complete nucleotide sequences of BPP-1, BIP-1, and BMP-1 were independently determined to be 42,493 bp, 42,638 bp, and 42,663 bp, respectively. The length differences were due to a variable number of tandem repeats within the VR2 region of bbp36, and the three phage sequences are identical except for changes at VR1, VR2, two single nucleotide polymorphisms within mtd, and single nucleotide insertions and deletions within a homopolymer tract “G-string” located within bpm (see below). The overall base composition is 65.4% GC for all three genomes and is similar to that of the host bacterium (66% GC for B. bronchiseptica).
In assembling the sequences, the lack of abrupt stops or discontinuities in the template suggested that the genomes of BPP-1, BIP-1, and BMP-1 are circular. Analysis of DNA from purified phage yielded restriction fragments corresponding in size to those that would be expected from a circular genetic map, and partial denaturation failed to reveal evidence for cohesive ends (data not shown). Since BPP-1, BIP-1, and BMP-1 are tailed phages, the packaged genomes are likely to be linear with overlapping permutations.
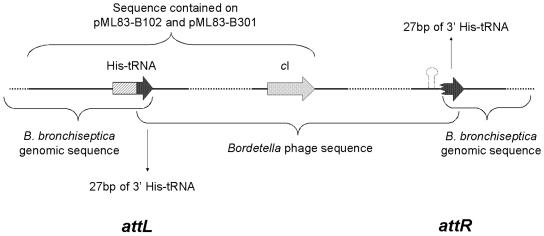
Plasmid rescue was used to clone the phage integration sites. Figure 2 shows the organization of two resulting plasmids, pML83-B102 and pML83-B301, which were derived from RB50 lysogens containing BPP-1 and BIP-1, respectively. Sequence analysis indicated that both plasmids include one of the two junctions, attL, where the Bordetella phage had integrated into the B. bronchiseptica genome. The B. bronchiseptica sequence matched the region containing the single his tRNA locus, and examination of the phage sequence revealed that it contains a 27-bp sequence that is identical to the 3′ end of the his tRNA gene. The last 27 bp of the gene are duplicated when the Bordetella phage integrates into the genome, and this sequence therefore comprises the attP core. As a result, the his tRNA gene is not disrupted. Since the phage genetic map is circular, we numbered the Bordetella phage genome sequence starting with position 1 of the 27-bp attP core.
FIG. 2.
Phage integration site. Plasmids pML83-B102 and pML83-301 contained attL. attR was identified with PCR primers derived from the phage genome and the B. bronchiseptica genome. The Bordetella phage genome contains a 27-bp sequence exactly identical to the 3′ end of the Bordetella his tRNA gene. This allows phage integration into the his tRNA locus without disrupting gene function.
Identification and analysis of phage coding sequences.
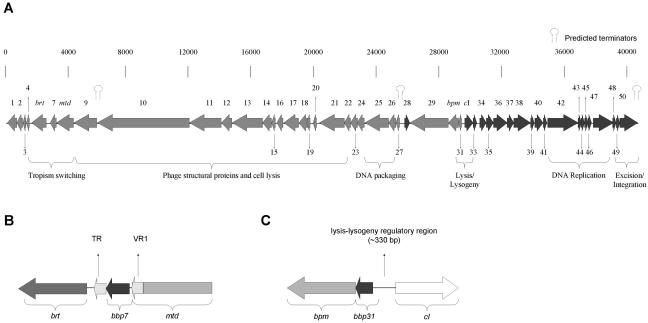
We identified 49 putative coding sequences by analyzing both strands of the phage genome for open reading frames encoding proteins 50 amino acids or longer that also contained plausible translational control signals. All of the predicted coding sequences encode proteins larger than 7 kDa. Most of the predicted coding sequences are named with the prefix Bbp (for Bordetella bronchiseptica phage). Ribosome-binding sites were identified when possible. ATG is the start codon in all except five of the predicted coding sequences. Of the five exceptions, four use GTG and one is predicted to use TTG as its start codon. Similar to lambdoid phages, the genome is organized in a major leftward unit of expression, which includes bbp1 through bbp31, and a rightward unit of expression, containing cI through bbp50. A stem-loop structure at the 3′ end of bpp50 is predicted to form a rho-independent transcription terminator which may prevent the extension of transcription from prophage sequences into adjacent host loci (Fig. 2 and 3a). Specific information regarding each coding sequence is listed in Table 2, and a schematic representation of the phage genome is shown in Fig. 3. Particularly noteworthy features of the genome are briefly described below.
FIG. 3.
Predicted Bordetella phage coding sequences. (A) Arrows represent predicted coding sequences encoding proteins of more than 7 kDa. Functional assignments for several gene clusters are indicated. See the text and Table 2 for details. (B) Schematic representation of the tropism switch region. brt encodes a reverse transcriptase. VR1 is located at the 3′ end of mtd. TR, also required for tropism switching, is not predicted to be part of a coding region and is located downstream from bbp7. (C) Schematic representation of the lysis/lysogeny region. cI encodes the phage repressor, and bbp31 encodes a putative Cro-like protein. bpm encodes an adenine DNA methylase, and its coding sequence is located immediately downstream of bpb31. Bpm does not appear to play a role in the lysis versus lysogeny decision.
TABLE 2.
Bordetella phage BPP-1, BIP-1, and BMP-1 coding sequencesa
| Gene | Position | No. of codons | Mass (kDa) | Related GenBank or Pfam entries (GenBank accession no.) | BlastP E-value | Predicted function | Comments, additional homology or motifs |
|---|---|---|---|---|---|---|---|
| bbp1 | 772-140 | 210 | 21.5 | Unknown | Contains a putative signal sequence and two transmembrane domains (aa 13-35 and aa 114-136) | ||
| bbp2 | 1264-773 | 163 | 17.6 | Neisseria meningitidis MC58 hypothetical protein (AAF41413) | 1e-41 | Unknown | |
| bbp3 | 1517-1266 | 83 | 8.9 | Unknown | Contains a putative transmembrane domain (aa 55-77) | ||
| bbp4 | 1775-1527 | 82 | 9.1 | Agapetes chortechinii and Orobanche fasciculata MatK | Unknown | May be related to type II intron RNA maturases (Pfam01348) | |
| brt | 2742-1756 | 328 | 38.0 | Streptococcus pneumoniae maturase-related protein (AAC38715); Pfam00078 | 3e-11 | RT | Component of phage evolution cassette (see text) |
| bbp7 | 3407-3021 | 128 | 14.5 | Unknown | |||
| mtd | 4583-3438 | 381 | 39.5 | Tail protein | No significant similarity to other phage tail fiber proteins; contains VR1 (see text) | ||
| bbp9 | 6125-4599 | 508 | 50.7 | E. coli K5 eliminase ElmA (CAA65353) | 2e-7 | Lysin | C-terminal region (aa 216-454) has A, V, and N-rich 17-aa degenerate repeats |
| bbp10 | 12345-6187 | 2,052 | 223.0 | Sinorhizobium meliloti phage PBC5 Orf33 | 5e-28 | Lysin | Contains a β/γ crystallin motif signature (Pfam PS00225; aa 866-881) |
| bbp11 | 14504-12342 | 720 | 78.4 | Sinorhizobium meliloti transglycosylase (CAC45637) | 3e-8 | Lysin | Contains a soluble lytic transglycosylase motif (Pfam01464; aa 295-413) also found in phages T7 (protein D) and PRD1 (protein p7) (27, 34) |
| bbp12 | 15205-14510 | 231 | 23.1 | Roseophage SI01 gp7 | 0.025 | Tail protein | Similar to roseophage SI01 gp7 (540 aa; E = 0.025) and lambda orf-401 (401 aa; E = 0.78); A- and N-rich internal repeats between residues 31 and 185 |
| bbp13 | 17262-15217 | 681 | 73.7 | L. pneumophila hypothetical protein (CAC33469) | 1e-5 | Unknown | Contains an ATP/GTP binding site motif (aa 183-190); encoded in the structural region of the prophage (23) |
| bbp14 | 17933-17262 | 223 | 30.0 | L. pneumophila hypothetical protein (CAC33470) | 0.006 | Unknown | Encoded in the structural region of the prophage (23) |
| bbp15 | 18215-18009 | 68 | 7.1 | Unknown | |||
| bbp16 | 18650-18228 | 140 | 14.5 | Unknown | |||
| bbp17 | 19701-18706 | 331 | 36.4 | Unknown | |||
| bbp18 | 20404-19715 | 229 | 24.3 | Putative Salmonella LT2 endoprotease (AAL20911) | 4e-13 | Protease | |
| bbp19 | 20708-20361 | 115 | 13.4 | Unknown | |||
| bbp20 | 21080-20751 | 109 | 11.7 | Unknown | |||
| bbp21 | 22760-21093 | 555 | 61.9 | L. pneumophila hypothetical protein (CAC33475) | 7e-6 | Tail-head connector | Also similar to the head-tail connectors of Podoviridae phage, including cyanophage C60 (E = 0.016), T3, T7, and phiYe-03 (both E = 0.019) (23) |
| bbp22 | 23169-22762 | 135 | 14.3 | RNA polymerase β subunit | 1e-8 | Unknown | Contains a putative transmembrane domain (aa 4-23) and a Glu-rich region (aa 26-95) including three tandem repeats of the sequence AQQQ (aa 33-44) |
| bbp23 | 23173-23631 | 152 | 16.1 | Unknown | |||
| bbp24 | 24092-23628 | 154 | 16.2 | Acetyltransferase (GNAT) family, Pfam00583 | 0.001 | Acetyltransferase | Contains GCN5-related N-acetyltransferase motif (aa 71-155); also similar to a number of bacterial acetyltransferases from Streptomyces spp., e.g., streptothricin acetyltransferase (191 aa; E = 0.099) and Brucella melitensis BMEI0158 (152 aa; E = 0.032)/PICK> |
| bbp25 | 25737-24136 | 533 | 58.9 | Methanobacterium phage psiM2 large terminase subunit (AAC27048) | 1e-6 | Terminase | Similar over the central region (aa 109-400) to terminase-like proteins and over a shorter region (aa 75-155) to large phage terminase subunits; contains an ATP/GTP binding site motif near the N terminus (aa 54-61) |
| bbp26 | 26318-25734 | 194 | 21.5 | Bacteriophage HK620 small terminase subunit (AAK28890) | 1e-7 | Terminase | |
| bbp27 | 26573-26337 | 78 | 8.8 | Transcriptional regulator | Weak similarity (36% identity over 50 aa) to a 96-aa hypothetical protein in E. coli O157:H7 (gene ECs4956) that is similar to phage Mu gp9; contains a (HTH) motif (aa 40-60) and is also downstream of a 104-bp sequence that contains two closely spaced inverted repeats | ||
| bbp28 | 27165-26809 | 118 | 13.0 | Unknown | Only putative gene encoded on the opposite strand in the left arm of the genome; it has several attributes indicative of a gene inserted into a progenitor genome, e.g., upstream transcription and translation signals; contains a putative transmembrane helix or possible N-terminal signal sequence (aa 7-29) | ||
| bbp29 | 29741-27177 | 854 | 93.3 | Bacteriophage PSA primase (CAC85608) | 5e-62 | Primase | N-terminal half (aa 21-316) has similarity to repA (43) from Synechococcus sp. PCC7942 plasmid pUH24 (E = 4e-09); C-terminal half is highly similar to primase or helicase proteins from Bacillus subtilis phage φ 105, Listeria monocytogenes phage PSA, Lactobacillus phages A2 and adh, and Streptomyces phage φ C31; contains an ATP/GTP binding motif (PS00017; aa 587-594) |
| bpm | 30513-29758 | 251 | 27.4 | Xylella fastidiosa site-specific DNA-methyltransferase (AAF85112) | 3e-51 | Adenine DNA methyltransferase | Highly similar to a number of methylases from bacteria, archaea, and viruses (see text); contains several motifs, including a DNA methylase N-4/N-6 family signature (Pfam PF01555; aa 85-237) and a S-adenosyl-l-methionine binding domain motif (Pfam PS50193; aa 192-251) |
| bbp31 | 30758-30510 | 82 | 9.3 | Bacteriophage APSE-1 protein P2 (AAF03947) | 3e-6 | cI repressor (Cro) | Contains HTH DNA-binding motif (Pfam01381; aa 22-77); transcribed in the opposite orientation to cI, as is cro in lambdoid phages |
| cI | 30843-31514 | 223 | 24.5 | Phage 434 repressor protein cI (AAA72530) | 1e-20 | Repressor of lysis | Homolog of lambdoid cI-like repressors from a number of phages and bacteria, including Providencia rettgeri conjugative genomic island R391 (E = 3e-30), phage 434 cI repressor, and phage APSE-1 protein P1 (E ≤ 1e-12 for both); contains HTH DNA-binding motif (Pfam01381; aa 11-63) and a peptidase family S24 motif (PF00717; aa 91-208) |
| bbp33 | 31511-31726 | 71 | 8.3 | Unknown | Weakly similar to gene XF2112 in Xylella fastidiosa 9a5c (E = 0.39); contains two predicted transmembrane regions (aa 5-22, aa 32-51) | ||
| bbp34 | 31858-32289 | 143 | 15.5 | Unknown | |||
| bbp35 | 32295-32771 | 158 | 15.6 | Unknown | |||
| bbp36 | 32776-33705 | 309 | 32.2 | Xanthomonas campestris ice nucleation protein (CAA37140) | 8e-8 | Unknown | Homologous to ice nucleation proteins (36, 46, 48); contains VR2 (see text) |
| bbp37 | 33702-34133 | 143 | 14.7 | Unknown | |||
| bbp38 | 34130-35386 | 418 | 45.1 | Bacteriophage APSE-1 protein P51 (AAF03994) | 4e-87 | Unknown | Also similar over entire length to two proteins in X. fastidiosa (genes XF0686 and XF2522; E = 1e-125), Staphylococcus aureus phages φ12 (an APSE P51-like protein; E = 7e-34) and S. aureus subsp. aureus MW2 prophage φ-Sa-2mw (gene MW1427; E = 3e-30) |
| bbp39 | 35383-35553 | 56 | 6.5 | Unknown | Not found in the corresponding syntenic region of APSE-1 or X. fastidiosa, suggesting it was laterally transferred to Bordetella phage lineage | ||
| bbp40 | 35559-36107 | 185 | 19.5 | Bacteriophage APSE-1 protein P50 (AAF03993) | 4e-44 | Unknown | Also similar to two proteins in X. fastidiosa (XF0685 and XF2523, E ≤ 6e-37); S. aureus phage φ12 (185aa), and prophage φ-Sa-2mw (MW1426; E = 4e-07) |
| bbp41 | 36225-36413 | 62 | 7.4 | Unknown | |||
| bbp42 | 36413-38479 | 688 | 76.0 | Bacteriophage APSE-1 P45 (AAF03988) | 1e-50 | DNA polymerase | Also similar to DNA polymerases from X. fastidiosa (XF0683, XF2290/XF2291, and XF2525, E = 0.0 to 1e-50), phage SPO2 (E = 6e-76) and S. aureus phage φ12 (E = 2e-50); see text for amino acid motifs encoded |
| bbp43 | 38568-38831 | 87 | 10.2 | Bacteriophage APSE-1 P44 (AAF03987) | 2e-14 | Unknown | Also similar over its entire length to proteins from X. fastidiosa (genes XF0682, XF2292, and XF2526; E = 6e-20) and S. aureus phage φ12 (96 aa; E = 5e-05) |
| bbp44 | 38828-39046 | 72 | 8.4 | Unknown | |||
| bbp45 | 39043-39249 | 68 | 7.5 | Unknown | |||
| bbp46 | 39246-39470 | 74 | 8.8 | Unknown | In the syntenic region of X. fastidiosa and APSE-1 two unrelated proteins are present in this position (genes XF2293/XF2294 and P42/43, respectively) | ||
| bbp47 | 39467-40861 | 464 | 52.4 | Bacteriophage APSE-1 P41 (AAF03984) | 1e-173 | DEAD box helicase | Contains a DEAD/DEAH box helicase motif (IPR001410; aa 4-203) and an ATP/GTP binding site motif (IPR001687; aa 29-36); also similar to three proteins in X. fastidiosa (genes XF0680, XF2295, and XF2528, ≤65% identity to all), S. aureus phage φ12 phage helicase (40% identity) |
| bbp48 | 40858-41085 | 75 | 8.6 | Bacteriophage PSA gp52 (CAC85610) | 1e-5 | Unknown | Also similar to the C-terminal regions of Listeria innocua protein lin2585 (214 aa), Streptococcus pneumoniae phage MM1 Orf26, a protein in Streptococcus pyogenes phage φNIH1.1 (89 aa) and spyM18_1792 and spyM18_0737 in S. pyrogenes strain MGA8232 (E ≤ 1e-03 for all) |
| bbp49 | 41078-41287 | 69 | 7.7 | Bacteriophage 933W putative excisionase (AAD25407) | 0.079 | Excisionase | Similar to excisionase from phages or prophages specific for E. coli 0157:H7 (phages 933W, Stx2-converting bacteriophage I, VT2-Sa, CP-933V; E = 0.1 for all); contains a putative transmembrane domain (aa 4-26) |
| bbp50 | 41263-42468 | 401 | 45.1 | Bacteriophage φCTX integrase (AAD14164) | 3e-27 | Integrase | The closest homolog is Pseudomonas aeruginosa phage φCTX integrase (389 aa; E = e-27); similar integrases are found in S. typhimurium LT2 prophages Gifsy-2 (430 aa) and Fels-1 (441 aa), S. typhi strain CT18 prophage 10 (446 aa), and several phages or prophages of E. coli (e.g., CP-933V, Sakai-VT1, P27, 933W, Sakai-VT2; E ≤ e-20 for all); in addition, phage APSE-1 protein P38 (390 aa) is significantly similar (E = 0.005); contains a phage integrase motif of the tyrosine site-specific recombinase family (IPR002104; aa 195-375) |
RT, reverse transcriptase; aa, amino acid; HTH, helix-turn-helix.
(i) Tropism switching cassette region: bbp4, bbp7, brt, TR, and mtd.
Bbp4 is a small predicted protein containing a region (amino acids 53 to 72) with 45 to 60% identity to sequences in ribosomal maturases (matK) from a number of flowering plants (e.g., Agapetes schortechinii). This segment is part of a longer sequence pattern for type II intron maturases (Pfam PF01348) which are involved in group II intron splicing. The proximity of bbp4 gene to brt, mtd, and the TR element raises the possibility that it is part of the phage tropism-switching module.
Brt (Bordetella reverse transcriptase) contains a region (amino acids 72 to 265) that matches the Pfam entry PF00078 rvt (reverse transcriptase) with an E-value of 3e-14. Using a His-6-tagged derivative of Brt, we previously demonstrated that the protein does indeed have reverse transcriptase activity (21). Deletion and site-directed mutagenesis experiments also showed that the reverse transcriptase domain is required for the Bordetella phage to undergo tropism switching.
As shown in Fig. 3B, located within the 278-bp intergenic region between brt and bbp7 is the 134-bp TR sequence. Depending on the particular phage isolate, TR sequences are 81 to 99% identical to the closely linked VR1 sequence. Differences between TR and VR1 occur at positions in VR1 corresponding to adenine residues in TR, of which there are 23. The sequence of TR is invariant, it is required for tropism switching, and synonymous substitution experiments indicate that TR acts as a template in the DNA diversity-generating process (21). Although the predicted product of bbp7 has no significant matches in the database, its location is intriguing. The ATG start codon is 30 bp downstream from the stop codon of mtd, and the bbp7 stop codon lies immediately upstream of the beginning of TR. It is therefore possible that bbp7 plays an as yet undiscovered role in tropism switching.
The mtd gene contains the 134-bp VR1 segment that has been shown to be the receptor tropism determinant (21). Two single nucleotide polymorphisms located outside of VR1 in mtd were identified when the three phage genomes were compared. These two nucleotides, located at bp 605 and 652, have not been observed to undergo variation associated with tropism switching. Instead, they differed based on which lysogenic B. bronchiseptica isolate the particular phage was derived from. Although the polymorphisms result in amino acid substitutions, they appear to have no effect on host tropism. Preliminary results indicate that Mtd binds to phage receptors on the Bordetella cell surface (Doulatov et al., unpublished data), and experiments to determine the precise location of Mtd in mature phage particles are currently under way.
(ii) bbp9 through bbp21: phage structural genes.
The region encompassing bbp9 to bbp21 is predicted to encode phage structural and assembly-related proteins. Three coding sequences in this region are likely to encode products with enzymatic activity. Bbp9 contains a region that is similar to the E. coli eliminase, ElmA, which depolymerizes capsular polysaccharide (18) and Bbp11 contains a soluble lytic transglycosylase motif, which is commonly found in phage structural proteins with murine hydrolytic activity, and they appear to facilitate penetration of the peptidoglycan layer during cell entry (19, 34). Bbp18 is highly similar over its central region (amino acids 67 to 172) to the corresponding segment of Salmonella enterica serovar Typhimurium phage LT2 endoprotease. Phage proteases are typically involved in cleavage of structural proteins during assembly, suggesting that Bbp18 may be a phage assembly-related protease. Although most expected structural components of BPP-1 and its family members are difficult to predict on the basis of sequence similarity alone, Bbp12 displays weak similarity to tail fiber proteins from other tailed phages, and Bbp21 is predicted to encode the head-tail connector.
(iii) bbp25 and bbp26: DNA packaging.
Bbp25 is similar over the central region to terminase-like proteins in Mesorhizobium loti and the archaeon Methanosarcina acetivorans strain C2A. It is also similar over a shorter region to large terminase subunits from two archaeophages, psiM2 (Methanobacterium, E = 1e-06) and psiM100 (Methanothermobacter, E = 1e-06). There is also a predicted ATP/GTP binding site motif near the N terminus, characteristic of large phage terminases. Bbp26 is most similar over its central region (amino acids 26 to 161) to the central region (amino acids 11 to 132) of enterobacteriophage HK620 small terminase subunit (140 amino acids; E = 3e-05). bbp25 and bbp26 are therefore likely to encode the large and small terminase subunits, respectively, which form part of the DNA packaging machinery.
(iv) bbp29 and bpm.
Bbp29 appears to be a two-domain protein involved in DNA replication. The N-terminal half (amino acids 21 to 316) has greatest similarity to RepA from the cyanobacterial Synechococcus sp. strain PCC7942 plasmid pUH24, which encodes an essential replication protein (43). The C-terminal half is highly similar to primase and helicase proteins from a number of phages and contains an ATP/GTP binding motif.
Bpm (for Bordetella phage methylase) is highly similar to a number of methylases from bacteria, archaea, and viruses. The most similar proteins in the database are site-specific DNA methyltransferases from two Xanthomonas species. In addition, highly similar proteins are found in the Streptomyces coelicolor genome, the F plasmid of E. coli K-12, Yersinia pestis plasmid pMT1, E. coli virulence plasmid pO157 (E < 7e-20 for all), and many other bacteria and plasmids. Related proteins are found in numerous bacteriophages and archaeophages, the most similar of which is an adenine methyltransferase from an archaeal halophilic virus, φCh1 (E = 3e-14). Bpm contains several motifs, including a DNA methylase N-4/N-6 family signature, an S-adenosyl-l-methionine binding domain motif, and two motifs shared by all adenine methylases, an N-terminal Asp-Pro-Pro-Tyr motif and a C-terminal Phe-X-Gly-X-Gly (FXGXG) sequence (15, 40). One common feature of this family of methylases is that they are usually not part of restriction-modification systems. In some species, such as Sinorhizobium meliloti and Caulobacter crescentus, they are essential for viability (47). A functional analysis of Bpm is described below.
(v) bbp31 and the cI lysis repressor.
bbp31 is located directly adjacent to the cI repressor homolog and is transcribed in the opposite direction (Fig. 3C). Bbp31 is highly similar to phage APSE-1 protein P2, a Cro repressor homolog, and it contains a predicted helix-turn-helix DNA-binding motif. cI is a homolog of lambda cI-like repressors from a variety of phages, including 434, P22, HK97, and lambda, and it also contains a predicted helix-turn-helix DNA-binding motif. To test the predicted repressor function of the protein, we constructed in-frame deletions in the cI loci of BPP-1 and BIP-1. Plaques produced by the ΔcI mutants were less turbid than wild-type plaques, and ΔcI mutants were unable to form lysogens. Expression of the cI gene alone on a broad-host-range vector, pMMB207, in sensitive B. bronchiseptica strains was sufficient to confer resistance to phage lysis (data not shown). These observations are consistent with the hypothesis that cI is the lysis repressor and together point to cI and Bbp31 as controlling the lysis-lysogeny switch.
(vi) bbp36.
Bbp36 has sequence similarity to ice nucleation proteins of several bacterial plant pathogens, such as Xanthomonas campestris, Pseudomonas syringae, and Erwinia uredovora (36, 46, 48). All members of this class of proteins, including Bbp36, contain imperfect repeats of a consensus octapeptide. The nonrepetitive N-terminal regions of the Bbp36 protein and the ice nucleation proteins show the highest similarity, but they are also similar at the nonrepetitive portions of their C termini. SignalP analysis suggests that Bbp36 carries a signal peptide. This raises the possibility that, like ice nucleation proteins, Bbp36 may be exported to the cell surface during the lysogenic phase. The bbp36 gene contains the second major region of variability, VR2, which is described in detail below.
(vii) bbp42 and bbp47.
bbp42 is predicted to encode a multidomain DNA polymerase with high sequence similarities to DNA polymerases from numerous bacteria and phages. Amino acid motifs found in Bbp42 include a 3′-5′ exonuclease motif, a class II aminotransferase motif, a DNA-directed DNA polymerase domain, and an N-6 adenine-specific DNA methylase segment. Bbp47 is highly similar to a number of helicase-like proteins found in phages, bacteria, archaea, and eukaryotes. It contains a DEAD/DEAH box helicase motif and a predicted ATP/GTP binding site. The Bbp42 and Bbp47 proteins are likely to constitute the phage DNA replication machinery.
(viii) bbp49 and bbp50: lysogeny genes.
The last functional module in the right arm is predicted to encode two proteins involved in excision and integration. Bbp49 displays weak sequence similarity to several phage excisionases, and Bbp50 is highly similar to numerous integrase proteins. Bbp50 contains a phage integrase motif of the tyrosine site-specific recombinase family.
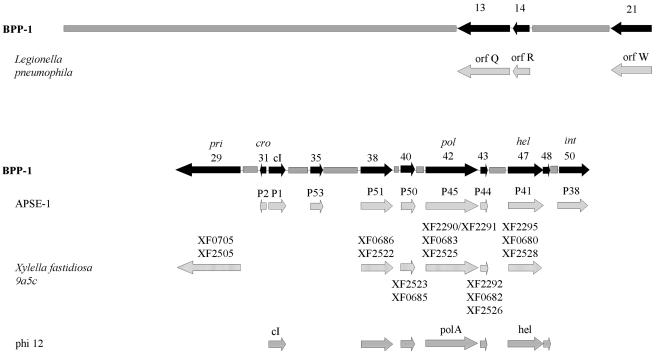
Syntenic regions.
As shown in Fig. 4, several phage and prophage genomes that contain regions with similar coding sequences in the same order as in the Bordetella phage genome were identified, implying evolutionary relatedness. On the left arm, a short region of partial synteny was found with a 30-kb unstable genetic element in Legionella pneumophila which is apparently of phage origin and is responsible for phase-variable expression of a virulence-associated lipopolysaccharide (23). The right arm of the phage genome displays partial synteny with nine loci encoded by APSE-1, a Podoviridae phage that infects a secondary endosymbiont of the pea aphid Acyrthosiphon pisum (44). This genomic similarity includes the divergently expressed bbp31 and cI loci. Partially overlapping syntenic regions were also found with three cryptic prophages in Xylella fastidiosa 9a5c, a bacterial citrus pathogen (37), and Staphylococcus aureus phage φ12.
FIG. 4.
Syntenic regions conserved between Bordetella phages and other phages and phage-like elements. The genome of BPP-1 is represented by a solid grey bar or black arrows for BPP-1 genes that have homologs present in a similar order in other genomes. Loci from similarly ordered genomic regions in other phages or phage-related elements are represented below the relevant BPP-1 gene. Similarly ordered genomic regions are found in a Legionella pneumophila defective prophage element, insect endosymbiont phage APSE-1, three Xylella fastidiosa 9a5c cryptic prophages, and Staphylococcus aureus phage φ12.
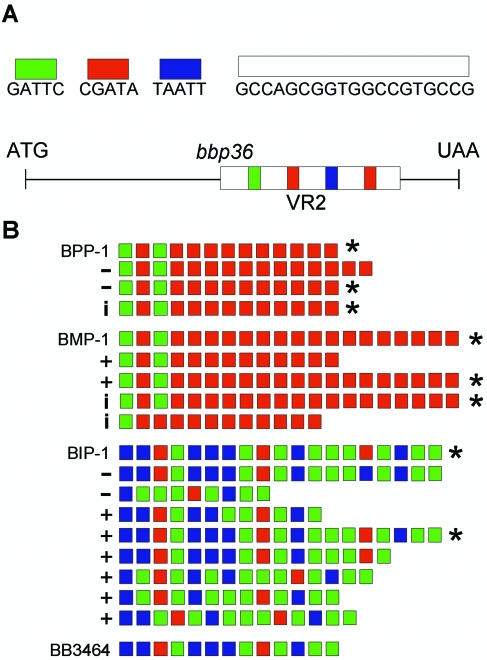
Variable region 2.
Sequence comparisons between BPP-1, BIP-1, and BMP-1 revealed a striking heterogeneity near the 3′ end of bbp36. This region, which was designated VR2, consists of a series of identical 19-bp repeats separated by one of three 5-bp spacers (Fig. 5A). To investigate a possible relationship between VR2 and tropism switching, we determined the VR2 sequences of multiple phage isolates which were derived from different parents and represent all three host tropism types (Fig. 5B). Each VR2 contained a variable number (n) of the 19-bp cassettes separated by n − 1 spacers. Although a diversity of patterns were observed, in no instance was the bbp36 reading frame altered by variation in VR2.
FIG. 5.
Structure and variability of the VR2 segment of bbp36. (A) Graphic representation of VR2 and bbp36. The 19-bp cassette and the three 5-bp spacers are represented by color-coded bars. (B) VR2 sequence variations. The VR2 sequences are represented by color-coded 5-bp spacers. Derivatives of BPP-1, BMP-1, and BIP-1 are listed below the parental phage. Isolates with different tropisms but identical VR2 sequence are marked with *. +, BPP tropism; −, BMP tropism; i, BIP tropism.
As shown in Fig. 5B, in several cases we observed identical VR2 sequences in phages with different host tropisms. This was in contrast to previous results with VR1 (21). Given the lack of correspondence between VR2 variability and tropism switching, we introduced an in-frame deletion into bbp36 to directly measure the effects on phage infectivity, specificity, or tropism switching. BPP-1Δbbp36 and BMP-1Δbbp36 produced viable phages that retained their parental specificity and were fully capable of tropism switching (Table 3). Although VR2 varies at high frequency, its function is not related to host specificity. The pattern of VR2 variability can be accounted for by a slipped-strand mispairing mechanism. Slipped-strand mispairing occurs as a result of slippage of DNA polymerase during replication of highly repetitive templates, resulting in occasional insertion or deletion of repeat units (6). Sampling of randomly selected BPP-1 progeny suggested that at least 10% carried a different VR2 sequence. This high frequency is remarkable, considering the apparent lack of selective pressure for bbp36 function or variability.
Bpm encodes a DNA adenine methylase with novel site specificity.
Examination of the bpm gene from BPP-1, BIP-1, and BMP-1 revealed a variable stretch of G residues located 13 bp upstream of the highly conserved FXGXG motif. The BPP-1 sequence contained eight G's, BIP-1 contained nine, and BMP-1 contained ten. In both BIP-1 and BMP-1, the additional guanosine residues result in frameshift mutations. Homopolymer tracts such as the G-string sequence in bpm are associated with an increased frequency of frameshift mutations (20, 39) and are sometimes used as mechanisms to promote phase variability (14). The fact that both BMP-1 and BIP-1 contained frameshift mutations suggested that they occur frequently during routine passage and/or are associated with tropism switching.
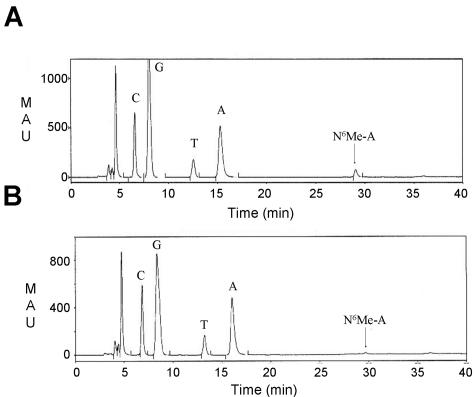
The first hint that bpm may encode a functional methylase came from analyzing the Bordetella phage genome by restriction endonuclease digestion. We found that BPP-1 DNA was resistant to PstI, while BMP-1 and BIP-1 DNA was not. To determine if protection from PstI digestion correlated with expression of bpm, an in-frame deletion was introduced into the bpm locus in BPP-1. Phage DNA purified from the Δbpm mutant was no longer resistant to PstI digestion (data not shown). DNA purified from BPP-1 and BIP-1 phage particles was subjected to hydrolysis and dephosphorylation to produce nucleosides for analysis by reversed-phase high-pressure liquid chromatography. As shown in Fig. 6, a peak at Rt = 29 min, which corresponds to the expected peak for N6-methyladenine, was present in BPP-1 DNA and greatly diminished in DNA prepared from BIP-1. No peak corresponding to 5-methylcytosine was detected in either sample. The peak corresponding to N6-methyladenine was collected and subjected to electrospray mass spectroscopy analysis. The result confirmed the identity of the peak as N6-methyladenine (expected mass, 265.12; actual mass, 266.14). Taken together, the results support the conclusion that Bpm is a DNA adenine methylase.
FIG. 6.
High-pressure liquid chromatography analysis of BPP-1 and BIP-1 DNA. (A) Elution profile of BPP-1 nucleosides. (B) Elution profile of BIP-1 nucleosides. The N6-methyladenine (Me-A) peak (downward arrow at Rt = 29 min) is significantly smaller in BIP-1 than in BPP-1, after taking into account the different scales. MAU, milli-absorbance units.
An EcoRI fragment from BPP-1 containing the bpm coding sequence plus 8 kb of surrounding sequence was cloned into pUC19 to obtain pML68-16. Of the four PstI sites (three in the insert and one in the multiple cloning site of the plasmid), only the site on the vector backbone was susceptible to PstI digestion. This raised the possibility that Bpm does not methylate all PstI sites. Cotransformation of pML68-16 with several other plasmids bearing PstI sites from a variety of sources further corroborated the observation that only a subset of available PstI sites were protected by Bpm. Since all six PstI sites on the Bordetella phage genome were apparently methylated, sequences surrounding these sites were compared (Table 4). The alignment, centered around each PstI site, revealed additional sequence features that were shared by all of the protected sites.
TABLE 4.
Protection of PstI sites by Bpma
| Phage | Sequence | PstI protection |
|---|---|---|
| Bordetella phages | ggcgAGaaaccgCTGCAGagattcatcctc | + |
| cgccAGcctgcgCTGCAGcgttgcttcccg | + | |
| ttgaAGcccggcCTGCAGttgcgcctccat | + | |
| gaccAGaaaggaCTGCAGcgacccctgttg | + | |
| caccAGtaactgCTGCAGcaccgcctgacc | + | |
| gccgAGcgtgcgCTGCAGttcatcggcaag | + | |
| Variants | gaccAGaaaggaCTGCAGcgacccctctag | + |
| agctcGaaaggaCTGCAGcgaccccgatcc | − | |
| gatcAGgggtcgCTGCAGtcctttcgagct | + | |
| gatcAcgggtcgCTGCAGagcttatcgata | − | |
| gatcAGaaaggaCTGCAGcgacagcttcga | + | |
| gatcAGgggtcgCTGCAGagcttatcgata | + |
The core CTGCAG site (underlined) is shown with 12 bp of flanking sequence on each side. +, sequences protected from digestion; −, susceptible sequences. Note that protected sequences all have a conserved AG (boldfaced) dinucleotide 6 bp upstream of the core site.
A series of constructs containing modified sequences were made with synthetic oligonucleotides, and plasmids containing them were cotransformed with a broad-host-range plasmid expressing bpm. The constructs were purified and assayed for resistance to PstI digestion. The results are shown in Table 4 and are consistent with the conclusion that in addition to the core CTGCAG sequence, the AG dinucleotide located 6 bp upstream of the PstI site is also required for adenine methylation. Most DNA methylase enzymes identified to date have recognition sequences that are contiguous and range from 4 to 8 bp (http://rebase.neb.com). Bpm, on the other hand, is not only a relatively small DNA methylase (predicted size, 27.4 kDa) but it also appears to have a nonpalindromic recognition site that stretches over 14 bp. Three of the six CTGCAG sites on the Bordetella phage genome contain the required AG dinucleotide on only one strand, raising the possibility that these PstI-resistant sites are hemimethylated.
The BPP-1Δbpm mutant did not reveal any qualitative or quantitative defect in plaquing or tropism switching compared to wild-type BPP-1 (Table 3). Analysis of the available B. pertussis, B. parapertussis, and B. bronchiseptica genomic sequences suggests the presence of several restriction-modification systems (http://www.sanger.ac.uk/Projects/Microbes/). One hypothesis is that bpm protects phage DNA from host restriction. The increased specificity conferred by the AG dinucleotide would result in modification of only a subset of host PstI sites.
Generalized transduction.
To facilitate genetic analysis of Bordetella subspecies, we tested the ability of BPP-1ΔcI to serve as a generalized transducing phage. Two B. bronchiseptica mTn5-lacZ1 (Kmr) transposon mutants in Bvg-regulated genes were used as donor strains for these experiments. One strain carried a transposon insertion in fhaB, the structural gene for filamentous hemagglutinin, which is expressed in the Bvg+ phase. The other strain carried a transposon insertion in wbmD, which is part of the lipopolysaccharide biosynthetic locus and is expressed in the Bvg− phase. Wild-type B. bronchiseptica RB50 was used as the recipient strain. The overall transduction frequency was approximately 10−7 transductions per PFU. Furthermore, it was confirmed that the β-galactosidase activity of RB50-derived transductants grown under Bvg+ or Bvg− conditions matched well with those measured in the donor strains (data not shown). Similar results were obtained with TnphoA B. pertussis donor strains and B. pertussis 18323 as the recipient.
Since the fhaB locus is proximal to bvgAS, we tested for cotransduction of Kmr and bvgAS markers from RB54 (ΔbvgS) to RB50 with BIP-1ΔcI and vice versa. Approximately 88% of the Kmr transductants were also Bvg+ when RB54 was the recipient and 80% of the transductants were Bvg− when RB50 was the recipient. These experiments demonstrate that BPP-1 and BIP-1 can be used as tools for generalized transduction.
In vivo lysogenic conversion.
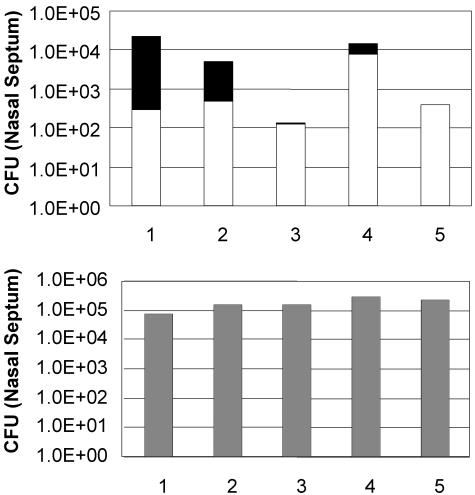
Since BPP-1 uses the Bvg+ phase protein pertactin as a receptor, we tested whether in vivo lysogenic conversion could occur in the mouse respiratory tract. Equal numbers of RB30 (lysogenic for BPP-1) and RB50 marked with gentamicin resistance (RB50Gm) were coinoculated (≈5.5 × 104 CFU/animal). Bordetella organisms were recovered from the nasal septum and the trachea of each animal 25 days postinoculation. Gentamicin-resistant and -sensitive colonies were counted, and the recovered RB50Gm colonies were characterized to determine if they had acquired resistance to BPP-1 (Fig. 7). To confirm that BPP-1 resistance was due to lysogeny, the presence of phage sequences was verified either by PCR detection of the cI gene or by production of infectious phage particles. In four of five mice, there were detectable levels of RB50Gm lysogenized with BPP-1, indicating that in vivo transmission of BPP-1 could indeed take place during respiratory tract infection, as has been demonstrated for other phages, such as CTXφ (45). In several animals, recovery of RB50 was lower than that of RB30, possibly due to phage killing.
FIG. 7.
Colonization of murine respiratory tract by RB30 and RB50Gm. Colonization of the nasal cavity 25 days postinoculation with equal numbers of RB30 (a BPP-1 lysogen) and RB50Gm. The proportion of RB50Gm derivatives (white bars) lysogenized by BPP-1 is indicated by the black portion of the histogram bars. The bottom graph shows the colonization efficiency of the RB30 lysogen strain. The overall quantity of RB50 recovered is lower than that of RB30, presumably due to phage killing.
DISCUSSION
BPP-1, BIP-1, and BMP-1 are among the first phages that infect bacteria from the beta subdivision of the proteobacteria to be completely sequenced. Our analysis indicates that BPP-1 is a novel genomic hybrid, combining characteristics of lambda-like genome organization with the presence of T7-like structural genes (Table 2, see below). The overall gene organization is modular and shows a high degree of mosaicism, as demonstrated by multiple segments with similarity to genes from diverse bacteria and phages. The most notable feature of these phages is the presence of a unique, template-dependent, reverse transcriptase-mediated tropism-switching mechanism encoded in an “evolution cassette” on the left arm of the phage genome. Preliminary results suggest that similar modules exist in other phage and bacterial genomes (S. Doulatov et al., unpublished data).
The tail and capsid morphology of BPP-1 groups it with Podoviridae according to the classification used by the International Committee on the Taxonomy of Viruses. The International Committee on the Taxonomy of Viruses phage classifications, based primarily on tail morphology, have recently been questioned due to the lack of correlation with genome characteristics and evolutionary relatedness (31). Based on genome and proteome features, it was suggested that the Podoviridae are more accurately segregated into several groups, in which short-tailed P22 clusters with long-tailed lambdoid phages due to their genetic similarities, while short-tailed T7-like phages form a separate group. There is no known genetic relationship between P22 and T7, which have entirely different lifestyles (temperate versus lytic) and transcriptional control mechanisms.
The organization of the BPP-1 genome is distinctly lambdoid, with two major clusters (left and right arms) that differ according to the direction of transcription. Structural and assembly proteins appear to be encoded on the left arm and DNA metabolism functions on the right, demarcated by a lambda-like divergent expression region encoding cI and Cro homologs. This contrasts with the unidirectional organization of genes in T7-like phages. Similarities in genome organization are found with phage APSE-1. Like BPP-1, APSE-1 is a short-tailed phage with a lambda-like genome (44). Synteny between regions of the BPP-1 and APSE-1 genomes, most strikingly in the segment containing bbp38 through bbp47, is detectable even at the DNA level and indicates their close relationship. This is remarkable considering that their bacterial hosts occupy very different niches, the mammalian respiratory tract for B. bronchiseptica versus intercellular and intracellular locations within the pea aphid A. pisum for the endosymbiotic host of APSE-1 (44). Furthermore, the APSE-1 host is a member of the Enterobacteriaceae, which is phylogenetically distant from the bordetellae. The same syntenic region shows similarities to several other phage genomes (Fig. 4). This implies a common ancestry for these phages and suggests that they have lambda-like mechanisms of DNA metabolism.
Structural features of BPP-1 indicate commonalities with T7-like phages. The capsid diameter is identical to that of T7 (60 nm [13]), but larger than APSE-1 (45 to 55 nm [44]). The BPP-1 tail is also similar in shape to the tail of T7 (Fig. 1). BPP-1 has some structural features that are absent from T7, most notably the bilobed, globular structures at the tips of the tail fibers. However, similar tail fiber ends have been described for some capsule-specific T7-family members (e.g., E. coli strain K-235 φ1.2 [16] and Klebsiella phage K11 [32]). These tail fibers have capsule-lytic hydrolase activity, most likely localized at the tips. A similar function may also reside in the globular ends of BPP-1 tail fibers, perhaps involving the bbp9 gene product.
Several polypeptides with similarity to T7-like phage proteins are predicted to be encoded in the BPP-1 structural gene region. The position of bbp11 in the genome and its murein transglycosylase sequence motif suggest significant homology with transglycosylases in T7-like phages, which participate in creating a passage through the peptidoglycan layer to allow DNA entry during infection (19). Sequence similarities between Bbp21 and head-tail connector proteins from several T7-like phages provide further evidence for conservation of structural features. Sequence similarity could indicate regions of Bbp21 that interact with other conserved structural proteins and/or with DNA, since the connector is the portal for DNA. Interestingly, the most highly conserved sequences in BPP-1 are those predicted to encode proteins that interact with DNA (i.e., helicase, DNA polymerase, methylase, cI and Cro repressors, integrase, large and small terminases, head-tail connector).
The majority of Bordetella phage proteins predicted by our analysis lack strong similarities to proteins in the GenBank database. One possible explanation is that relatively few phages that infect bacteria that are phylogenetically related to Bordetella have been analyzed in detail. The hybrid architecture of the BPP-1 genome supports emerging views of bacteriophage phylogeny and evolution (30). Phages such as BPP-1, P22, and APSE-1, with a lambda-like genome and a short-tail structural gene cassette, suggest a “braided” rather than a vertical lineage for tailed phages. These hybrids support the idea that regions encoding protein domains, single genes, or blocks of genes are readily exchanged between bacterial and phage genomes. The likelihood that more hybrid phage genomes exist suggests that segregation of characteristics is not as limited as previously thought, and a combinatorial continuum of variety may exist among phages.
Perhaps the most remarkable characteristic of the Bordetella phages analyzed here is their propensity to undergo targeted DNA sequence variation. VR1, as part of a larger “diversity generating cassette,” allows the phages to undergo host tropism switching (21). This ability has an obvious evolutionary advantage, as it confers an expanded host range. VR2 appears to undergo a significantly different type of variation, likely mediated by slipped-strand mispairing. Although the advantage conferred by VR2 variability remains to be determined, it is intriguing that the product is a predicted secreted protein with similarities to surface proteins on other gram-negative bacteria. Finally, the homopolymeric tract in bpm causes inactivation of the Bpm DNA adenine methylase upon acquisition of frameshift mutations, which also appear to occur at high frequency. Neither the functional role of the Bpm methylase nor the significance of phase variation is currently known.
The sequence analysis reported here, along with previous studies (21), suggests numerous applications for BPP-1 derivatives, gene products, and genetic elements. Phage-encoded proteins, including holins and lysins, have recently been used as effective antimicrobial agents (7, 22, 28), and several products (Bbp9, Bbp11, and Bbp18) encoded in the Bordetella phage genome are predicted to have antimicrobial activities. The completed sequences allowed the construction of ΔcI mutants, which can be used as generalized transducing phages for transferring markers between B. pertussis, B. parapertussis, and B. bronchiseptica. The identification of attachment sites and integration genes could facilitate the development of single-copy genomic integration vectors for the Bordetella genus and possibly other related bacteria, and the unusual site specificity of Bpm may be useful for introducing strand-specific methylation of adenine residues at defined locations.
Perhaps the most interesting potential applications of these Bordetella phages derive from their ability to switch tropism. This could, for example, provide a significant advantage for their use in phage therapy (38). Bordetella infections are confined to respiratory epithelial surfaces, which should be accessible to therapeutically administered phages. Phage variants arising via the tropism-switching mechanism encoded on the left arm of the genome could potentially overcome mutations in receptor proteins that would otherwise confer resistance to infection. Finally, further characterization of Brt, TR, and other cis- and trans-acting elements that promote variability in VR1 could lead to the development of novel genetic systems for evolving desired functional attributes in heterologous proteins of interest.
Acknowledgments
We thank members of the J. F. Miller laboratory for constructive input throughout the course of this project.
M.L. was supported by a research fellowship from the American Lung Association and training grant GM-08042 to the UCLA-CalTech Medical Scientist Training Program from the NIH. A.H. is a predoctoral trainee recipient of Microbial Pathogenesis Training Grant 2-T32-AI-07323. This work was supported by NIH grant AI38417 (J.F.M.). The sequencing of the BPP-1, BMP-1, and BIP-1 genomes was supported by the Wellcome Trust.
REFERENCES
- 1.Adams, M. H. 1959. Bacteriophages. Interscience Publishers, Inc., New York, N.Y.
- 2.Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611-620. [DOI] [PubMed] [Google Scholar]
- 3.Akerley, B. J., D. M. Monack, S. Falkow, and J. F. Miller. 1992. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J. Bacteriol. 174:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, and D. D. Moore. 1995. Short protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 6.Belkum, A., S. Scherer, L. Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernhardt, T. G., I. N. Wang, D. K. Struck, and R. Young. 2001. A protein antibiotic in the phage Qbeta virion: diversity in lysis targets. Science 292:2326-2329. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. A., and V. J. DiRita. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519-565. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, P. A., and J. F. Miller. 2000. Bordetella, p. 619-674. In E. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 10.Ermolaeva, M. D., H. G. Khalak, O. White, H. O. Smith, and S. L. Salzberg. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27-33. [DOI] [PubMed] [Google Scholar]
- 11.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harvill, E. T., P. A. Cotter, M. H. Yuk, and J. F. Miller. 1999. Probing the function of Bordetella bronchiseptica adenylate cyclase toxin by manipulating host immunity. Infect Immun. 67:1493-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hausmann, R., and M. Messerschmid. 1988. Inhibition of gene expression of T7-related phages by prophage P1. Mol. Gen. Genet. 212:543-547. [DOI] [PubMed] [Google Scholar]
- 14.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 15.Kaszubska, W., C. Aiken, C. D. O'Connor, and R. I. Gumport. 1989. Purification, cloning and sequence analysis of RsrI DNA methyltransferase: lack of homology between two enzymes, RsrI and EcoRI, that methylate the same nucleotide in identical recognition sequences. Nucleic Acids Res. 17:10403-10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwiatkowski, B., B. Boschek, H. Thiele, and S. Stirm. 1982. Endo-N-acetylneuraminidase associated with bacteriophage particles. J. Virol. 43:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landt, O., H. P. Grunert, and U. Hahn. 1990. A general method for rapid site-directed mutagenesis with the polymerase chain reaction. Gene 96:125-128. [DOI] [PubMed] [Google Scholar]
- 18.Legoux, R., P. Lelong, C. Jourde, C. Feuillerat, J. Capdevielle, V. Sure, et al. 1996. N-Acetyl-heparosan lyase of Escherichia coli K5: gene cloning and expression. J. Bacteriol. 178:7260-7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehnherr, H., A. M. Hansen, and T. Ilyina. 1998. Penetration of the bacterial cell wall: a family of lytic transglycosylases in bacteriophages and conjugative plasmids. Mol. Microbiol. 30:454-457. [DOI] [PubMed] [Google Scholar]
- 20.Levinson, G., and G. A. Gutman. 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4:203-221. [DOI] [PubMed] [Google Scholar]
- 21.Liu, M., R. Deora, S. R. Doulatov, M. Gingery, F. A. Eiserling, A. Preston, J. Duncan, R. W. Simons, P. A. Cotter, J. Parkhill, and J. F. Miller. 2002. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295:2091-2094. [DOI] [PubMed] [Google Scholar]
- 22.Loeffler, J. M., D. Nelson, and V. A. Fischetti. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170-2172. [DOI] [PubMed] [Google Scholar]
- 23.Luneberg, E., B. Mayer, N. Daryab, O. Kooistra, U. Zahringer, M. Rohde, et al. 2001. Chromosomal insertion and excision of a 30 kb unstable genetic element is responsible for phase variation of lipopolysaccharide and other virulence determinants in Legionella pneumophila. Mol. Microbiol. 39:1259-1271. [DOI] [PubMed] [Google Scholar]
- 24.Magrini, V., D. Salmi, D. Thomas, S. K. Herbert, P. L. Hartzell, and P. Youderian. 1997. Temperate Myxococcus xanthus phage Mx8 encodes a DNA adenine methylase, Mox. J. Bacteriol. 179:4254-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattoo, S., A. K. Foreman-Wykert, P. A. Cotter, and J. F. Miller. 2001. Mechanisms of Bordetella pathogenesis. Front. Biosci. 6:E168-E186. [DOI] [PubMed] [Google Scholar]
- 26.Mattoo, S., J. F. Miller, and P. A. Cotter. 2000. Role of Bordetella bronchiseptica fimbriae in tracheal colonization and development of a humoral immune response. Infect. Immun. 68:2024-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moak, M., and I. J. Molineux. 2000. Role of the Gp16 lytic transglycosylase motif in bacteriophage T7 virions at the initiation of infection. Mol. Microbiol. 37:345-355. [DOI] [PubMed] [Google Scholar]
- 28.Nelson, D., L. Loomis, and V. A. Fischetti. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by with a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 98:4107-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 30.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumer, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. J. Jacobs, R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 31.Rohwer, F., and R. Edwards. 2002. The Phage Proteomic Tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudolph, C., E. Freund-Molbert, and S. Stirm. 1975. Fragments of Klebsiella bacteriophage no. 11. Virology 64:236-246. [DOI] [PubMed] [Google Scholar]
- 33.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 34.Rydman, P. S., and D. H. Bamford. 2000. Bacteriophage PRD1 DNA entry uses a viral membrane-associated transglycosylase activity. Mol. Microbiol. 37:356-363. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Schmid, D., D. Pridmore, G. Capitani, R. Battistutta, J. R. Neeser, and A. Jann. 1997. Molecular organisation of the ice nucleation protein InaV from Pseudomonas syringae. FEBS Lett. 414:590-594. [DOI] [PubMed] [Google Scholar]
- 37.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. The Xylella fastidiosa Consortium of the Organization for Nucleotide Sequencing and Analysis. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 38.Summers, W. C. 2001. Bacteriophage therapy. Annu. Rev. Microbiol. 55:437-451. [DOI] [PubMed] [Google Scholar]
- 39.Tautz, D., M. Trick, and G. A. Dover. 1986. Cryptic simplicity in DNA is a major source of genetic variation. Nature 322:652-656. [DOI] [PubMed] [Google Scholar]
- 40.Trautner, T. A., B. Pawlek, U. Gunthert, U. Canosi, S. Jentsch, and M. Freund. 1980. Restriction and modification in Bacillus subtilis: identification of a gene in the temperate phage SP beta coding for a BsuR specific modification methyltransferase. Mol. Gen. Genet. 180:361-367. [DOI] [PubMed] [Google Scholar]
- 41.Uhl, M. A., and J. F. Miller. 1996a. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J. Biol. Chem. 271:33176-33180. [DOI] [PubMed] [Google Scholar]
- 42.Uhl, M. A., and J. F. Miller. 1996b. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 15:1028-1036. [PMC free article] [PubMed] [Google Scholar]
- 43.Van der Plas, J., R. Oosterhoff-Teertstra, M. Borrias, and P. Weisbeek. 1992. Identification of replication and stability functions in the complete nucleotide sequence of plasmid pUH24 from the cyanobacterium Synechococcus sp. PCC 7942. Mol. Microbiol. 6:653-664. [DOI] [PubMed] [Google Scholar]
- 44.van der Wilk, F., A. M. Dullemans, M. Verbeek, and J. F. van den Heuvel. 1999. Isolation and characterization of APSE-1, a bacteriophage infecting the secondary endosymbiont of Acyrthosiphon pisum. Virology 262:104-113. [DOI] [PubMed] [Google Scholar]
- 45.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 46.Warren, G., and L. Corotto. 1989. The consensus sequence of ice nucleation proteins from Erwinia herbicola, Pseudomonas fluorescens and Pseudomonas syringae. Gene 85:239-242. [DOI] [PubMed] [Google Scholar]
- 47.Wright, R., C. Stephens, and L. Shapiro. 1997. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J. Bacteriol. 179:5869-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao, J. L., and C. S. Orser. 1990. Conserved repetition in the ice nucleation gene inaX from Xanthomonas campestris pv. translucens. Mol. Gen. Genet. 223:163-166. [DOI] [PubMed] [Google Scholar]