Abstract
Pathogens of the bacterial genus Bordetella cause respiratory disease in humans and animals. Although virulence and host specificity vary across the genus, the genetic determinants of this diversity remain unidentified. To identify genes that may underlie key phenotypic differences between these species and clarify their evolutionary relationships, we performed a comparative analysis of genome content in 42 Bordetella strains by hybridization of genomic DNA to a microarray representing the genomes of three Bordetella species and by subtractive hybridization. Here we show that B. pertussis and B. parapertussis are predominantly differentiated from B. bronchiseptica by large, species-specific regions of difference, many of which encode or direct synthesis of surface structures, including lipopolysaccharide O antigen, which may be important determinants of host specificity. The species also exhibit sequence diversity at a number of surface protein-encoding loci, including the fimbrial major subunit gene, fim2. Gene loss, rather than gene acquisition, accompanied by the proliferation of transposons, has played a fundamental role in the evolution of the pathogenic bordetellae and may represent a conserved evolutionary mechanism among other groups of microbial pathogens.
Bacteria of the genus Bordetella are important pathogens that cause respiratory disease in humans and animals. B. pertussis is the causative agent of whooping cough (8), which is responsible for up to 500,000 annual deaths worldwide in unvaccinated populations (52) and is increasing in incidence in some countries with established vaccination programs, including the United States (e.g., see references 15, 30, and 38). Although most Bordetella species can cause similar disease in the upper respiratory tract, their host specificities vary dramatically. B. pertussis infects only humans; B. parapertussis strains can be classified in two groups, one of which infects only humans and one of which infects only sheep (16, 17, 25); B. bronchiseptica causes respiratory infections in a wide variety of mammals and birds but only rarely in humans (22, 41, 51).
These three Bordetella species are closely related at the nucleotide sequence level (4, 24), and their relatedness has been further established using a variety of common molecular strain typing techniques (reviewed in reference 29). Phylogenetic analyses of the genus Bordetella, using pulsed-field gel electrophoresis, multilocus enzyme electrophoresis (MLEE), and IS typing data, suggested that B. pertussis, human-derived B. parapertussis, and sheep-derived B. parapertussis arose independently from B. bronchiseptica-like ancestors (46, 47). Comparative genome sequencing of a representative strain of each species (33) confirmed that nucleotide sequence similarity is very high in conserved regions of the genome but demonstrated that B. pertussis and B. parapertussis evolved by genome decay from a B. bronchiseptica-like ancestor.
Although a number of critical conserved virulence mechanisms have been identified in the bordetellae, the genetic basis of these species' phenotypic variability remains unclear. We reasoned that a comparative analysis based on genome content of numerous isolates of each species might clarify evolutionary relationships between the species and identify genes that determine key differences in phenotypes, such as virulence and host specificity. Recent advances in comparative genomic methods have illuminated the remarkable extent of variation between microbial genomes, even among strains of the same bacterial species (reviewed in references 9, 20, 21, and 42). We describe here the use of microarray-based comparative genome hybridization (CGH) and suppressive subtractive hybridization (SSH) to examine the relationships between genome content, host species, and pathogenicity.
MATERIALS AND METHODS
Bordetella DNA microarray design and construction.
Three thousand six hundred ninety-one probes representing 93% of the B. pertussis Tohama I predicted coding sequences were amplified by PCR. More than 95% of the B. pertussis-derived probes were at least 95% identical to a sequence in the other two genomes. An additional 589 probes were amplified for B. bronchiseptica RB50 and B. parapertussis 12822 predicted coding sequences that were not at least 95% similar to an existing probe. Microarrays were constructed using standard techniques, printing each probe at least twice on every array. Complete details are in supplementary methods. (All supplementary materials referred to within this article are available at http://relman.stanford.edu/supplements/CGH2003.)
Strains and genomic DNA preparation.
Bordetella strains used in this study are described in supplementary Table 1 (see the above URL). Single colonies of purity-confirmed cultures were inoculated into modified Stainer-Scholte media and grown to saturation prior to harvesting. Genomic DNA preparation is described in supplementary Methods (see the above URL).
Fluorescence labeling of genomic DNA.
Genomic DNA was labeled by incorporation of dye-coupled nucleotides based on protocols found at the Brown Lab Mguide website (http://cmgm.stanford.edu/pbrown/protocols/index.html). Test DNA samples were labeled with Cy5. The mixed reference sample, comprising a 1:1:1 mixture of genomic DNA from B. pertussis Tohama I, B. parapertussis 12822, and B. bronchiseptica RB50, was labeled with Cy3. Two micrograms of genomic DNA (in 10 mM Tris) was combined with 20 μl of 2.5× random octamers (BioPrime labeling kit; Invitrogen, Carlsbad, Calif.) in a total volume of 41 μl. The DNA was boiled for 5 min, placed on ice for 2 min, and centrifuged at 16,000 × g. The DNA was mixed with 5 μl of deoxynucleoside triphosphate (dNTP) mix (120 nM [each] dATP, dCTP, and dGTP; 60 nM dTTP in Tris-EDTA), 2.5 μl of 1 mM Cy5-dUTP or Cy3-dUTP (Amersham Biosciences, Little Chalfont, Buckinghamshire, England), and 1 μl of Klenow (40 U/μl; BioPrime labeling kit; Invitrogen). The reaction was incubated at 37°C for 2 h and then stopped by addition of 5 μl of 0.5 M EDTA (pH 8.0).
Microarray hybridization and scanning.
Hybridization was performed as described in protocols found at the Brown Lab Mguide website (http://cmgm.stanford.edu/pbrown/protocols/index.html). Test and reference labeled DNA samples were combined and then washed two times using Microcon YM-30 columns (Millipore, Billerica, Mass.), according to the manufacturer's instructions. One hundred fifty micrograms of yeast tRNA (ATCC, Manassas, Va.) was added to block nonspecific binding. The probe volume was adjusted to 12 μl with water, and then 2.55 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.45 μl of 10% sodium dodecyl sulfate were added. Fourteen microliters of the probe was placed on the array and covered with a 25- by 25-mm no. 1 glass coverslip. The arrays were sealed in hybridization chambers (Die-Tech, San Jose, Calif.) with 20 μl of 3× SSC to maintain humidity and incubated at 65°C for 2 h to overnight.
Arrays were washed in 0.5× SSC, 0.005% sodium dodecyl sulfate for approximately 30 s, 0.1× SSC for 30 s, 0.05× SSC for 1 min, and 0.025× SSC for 1 min. The first wash was performed at 65°C, and the remaining washes were performed at room temperature. Slides were dried by centrifugation for 5 min at 50 × g and scanned using a GenePix 4000B scanner and GenePix 3.0 software (Axon Instruments, Union City, Calif.).
Data filtering.
Spots were screened visually to identify those of low quality. Signals from the remaining spots were converted into Cy5/Cy3 ratios after subtracting local background intensity from each spot in both channels. The ratios for each experiment were normalized by adding a constant to the Cy5 signal so that the mean ratio across the array was 1.0. The data set was filtered to include only spot measurements with a net Cy3 (reference DNA) fluorescence intensity above 50 U, greater than 60% of pixels with intensity greater than one standard deviation above the mean Cy3 background intensity, and with usable data from at least 26 of the 42 arrays. Background-subtracted hybridization signals from replicate probes were averaged. After image analysis and data filtering, 4,140 independent probes remained in the data set.
Normalization against mixed reference.
The mixed reference DNA sample contained equal amounts of genomic DNA from each of the three sequenced strains, so each probe sequence was represented in proportion to its presence in one or more of the sequenced strains. The reference signal was normalized to account for differences in predicted gene abundance in the reference mixture. Based on hybridization experiments, a probe sequence was considered absent from a sequenced strain if its top BLASTN hit had less than 80% sequence identity over at least 100 bp. For each BLAST hit exceeding 80% identity the species sequence dose, d, was calculated as d = (% identity − 80)/20. The total sequence dose, D, in the reference DNA sample was calculated as the sum of d from all three genomes, divided by 3. For each probe, the background-subtracted Cy5/Cy3 intensity ratio was multiplied by its D value to obtain measurements normalized for sequence copy number in the reference.
Determination of threshold log intensity ratios.
Because data distributions varied from experiment to experiment, the threshold was calculated independently for each individual array based on its data distribution. Let x = 0.0025n where n is the number of data points from a single experiment. In this data set, x ranged from 8.94 to 10.33. For each array, a histogram of the log ratio values with a bin size of 0.1 was computed. The bin value of the first bin to the left of the “main” peak (centered near zero) that had fewer than x measurements was chosen as the threshold. In other words, the threshold was selected by determining the point at which the left tail of the distribution representing the “detected” sequences contained very few data points. The decision to use 0.0025 as the multiplier for the calculation of x was empirically determined to maximize the sensitivity and specificity for arrays of the three sequenced strains (see Results).
For determination of sequences that vary between or within species, a conservative approach was used to further refine “not detected” calls. Reasoning that rare false-positive calls were unlikely to cluster by species or genome order, “undetected” data points were counted only if the same probe was not detected in a second strain of the same species or an adjacent probe sequence was not detected in the same strain.
Phylogenetic analysis.
The data set was recoded as a binary representation of detected (1) and undetected (0) genes based on whether the log ratio hybridization signal was greater than or less than the threshold ratio (see above), respectively. Maximum-parsimony analysis of the data set was performed using PAUP (version 3.0; Sinauer Associates) with a Camin-Sokal model (13). This algorithm was chosen because it requires no assumption that evolutionary rates are equivalent among lineages. All character states were assumed to evolve independently, although a more exacting model would treat the absence of adjacent genes as a single deletion event. The complete gene complement was designated the ancestral state (state = 1), and the undetected genes resulting from deletion events were a derived state (state = 0), which produced a rooted phylogenetic tree. Bootstrap values (18) were calculated from 100 bootstrapped datasets using the BOOT, MIX, and CONSENSE programs of the PHYLIP software package (version 3.5c [http://evolution.genetics.washington.edu/phylip/]).
Phylogenetic analysis treated each identified prophage as a single character state. This was done because inclusion of multiple sequences from each prophage would give unwarranted weight to prophage loss events and potentially bias the phylogenetic analysis. Furthermore, bacteriophage are inherently mobile and possibly exchanged among Bordetella strains by lateral transfer and therefore do not accurately represent the evolutionary history of the core genome.
Confirmation of microarray-identified regions of difference.
Ten regions of difference (RDs) were chosen randomly from regions where at least two adjacent sequences were variable between the sequenced strain and another strain of the same species and thus were not detected using genome sequence data. Four of these regions were variable among B. pertussis strains, three were variable among B. parapertussis strains, and three were variable among B. bronchiseptica strains. Primers were designed flanking one junction of 10 putative RDs. Primer sequences are in supplementary Table 2 (see the above URL). For each RD, PCR was performed with one strain for which the region was expected to be present and one strain for which it was expected to be absent. In addition, a conserved fragment of approximately 3 kb was amplified from each DNA preparation to confirm that DNA was amplifiable. Each PCR contained 1× GC genomic PCR buffer, 1 M GC Melt, 1.1 mM Mg(OAc)2, 0.2 mM dNTPs, and 1× Advantage-GC genomic polymerase mix (all from BD Biosciences Clontech, Palo Alto, Calif.), plus 200 nM (each) primer and 1 ng of genomic DNA, in a reaction volume of 25 μl. Touchdown PCR cycling conditions were as follows: initial denaturation at 95°C for 1 min; 10 cycles of 94°C for 15 s, 65°C (decreased by 1°C per cycle) for 30 s, 68° for 3 min; 25 cycles of 94°C for 15 s, 55°C for 30 s, 68° for 3 min; and a final extension at 68°C for 3 min. PCR products were run on agarose gels and visualized with ethidium bromide to confirm the presence of a band of the expected size.
In every case tested, no PCR product of the expected size was detected in a strain predicted to be missing an RD. In one instance a larger PCR product was detected, suggesting that the strain missing the RD might possess an insertion that occurred close to the junction and disrupted or deleted the sequence found at that position in the sequenced strain.
Subtractive hybridization.
Subtractive hybridization was carried out with Bpe159 as the tester and a mixed driver pool of genomic DNA from the three sequenced strains of Bordetella. The PCR-Select bacterial genome subtraction kit (Clontech) was used, with the following modifications to optimize the protocol for the high-GC genome of Bordetella: the incubation temperatures of the two rounds of hybridization were increased to 66°C, 1.3% dimethyl sulfoxide and 1.3 M betaine were added to the primary PCR, the primary PCR was diluted 1:15 instead of 1:40 for use as the secondary PCR template, and 12 to 15 cycles of amplification were performed for the secondary PCR. After the generation of a clone library containing the products of the subtractive hybridization, inserts were screened for presence in Bpe159. This was accomplished by amplifying approximately 300 inserts and printing them on microarrays. These screening microarrays were hybridized with Bpe159 and mixed reference genomic DNA. No spots had ratios significantly higher than the average. The two spots with the highest ratio of Bpe159/mixed reference signal were sequenced to confirm that they were present in both DNA pools.
In addition, B. pertussis strain 18323 was subjected to SSH analysis against B. pertussis Tohama I using the same methodology. Approximately 40% of the 300 fragments printed on the screening microarrays had a high ratio of 18323/Tohama I signal and were chosen for sequencing.
Amplification of fim2.
Oligonucleotides were designed to amplify by PCR either the Tohama I or RB50 fim2 gene (supplementary Table 2). The forward primer was in a conserved region at the 5′ end, and two reverse primers at the 3′ end were specific for the two genes (supplementary Table 2). The PCR mix contained 1× PCR buffer II (Applied Biosystems, Foster City Calif.), 1.5 mM MgCl2, 0.6 mM dNTPs, 20 pmol of each primer, 1 ng of genomic DNA, and 1.25 U of AmpliTaq DNA polymerase (other than buffer, PCR reagents were from Perkin-Elmer, Boston, Mass.). The reaction conditions were 96°C for 3 min; 35 cycles of 95°C for 1 min, 57°C for 1 min, 72°C for 1 min; and a final extension at 72°C for 3 min. PCR products were run on 2.5% agarose gels and visualized with ethidium bromide staining.
Nucleotide sequence accession numbers.
The sequences of two fim2 amplification products have been submitted to GenBank under the accession numbers AY289620 (Bpp4) and AY289621 (Bbr77).
RESULTS AND DISCUSSION
Interspecific comparative genome hybridization.
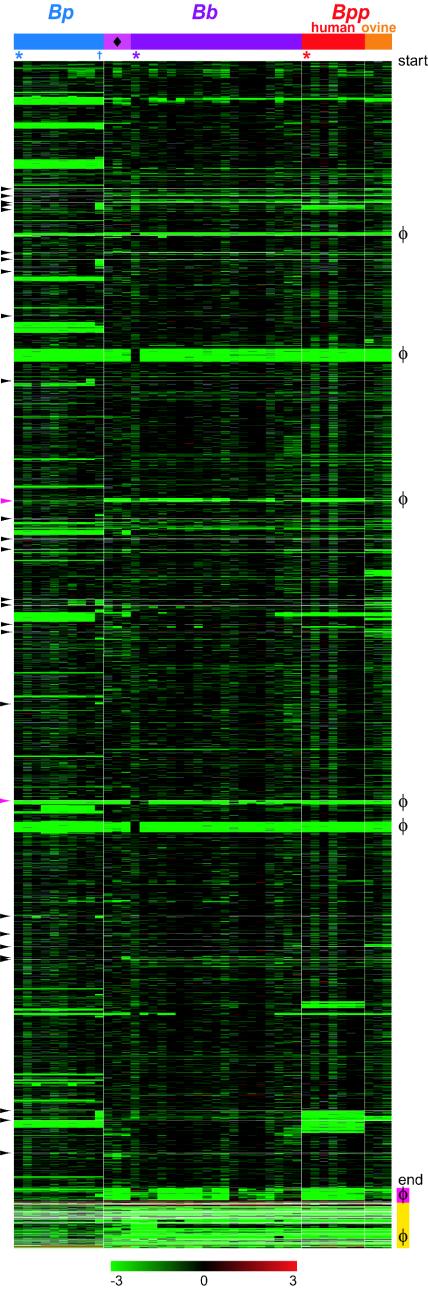
A PCR product-based DNA microarray representing 93% of B. pertussis Tohama I, 75% of B. parapertussis 12822, and 73% of B. bronchiseptica RB50 predicted coding sequences was designed using draft genome sequence data (see the above URL for supplementary methods). Using this array, a collection of 42 B. pertussis, B. parapertussis, and B. bronchiseptica strains (supplementary Table 1) was surveyed by CGH. The 10 B. pertussis strains included 7 low-passage-number clinical isolates collected between 1989 and 1997 in the United States and The Netherlands and the laboratory strains Tohama I, Wellcome 28, and 18323. The 10 B. parapertussis strains comprised 3 strains isolated from sheep and 7 strains isolated from humans. The 22 B. bronchiseptica strains were isolated from a variety of host species, including one human. The labeled genomic DNA of each strain was hybridized to the microarray together with a reference composed of a 1:1:1 mixture of genomic DNA from each of the three sequenced species. Comparison of the Bordetella CGH profiles revealed extensive variation between and within species (Fig. 1 and supplementary data). Bordetella strains exhibited multiple RDs, defined as blocks of two or more adjacent probes that hybridize poorly to at least one strain (Fig. 1).
FIG.1.
CGH of 42 Bordetella strains. Each column represents a strain, ordered and color-coded according to the tree topology shown in Fig. 2. The three sequenced strains are marked by asterisks. B. pertussis strain 18323 is indicated by the dagger. Each row represents a microarray probe, arranged in RB50 gene order from “start” to “end,” with RB50 genome coordinates listed on the left. φ indicates clusters of phage genes. Arrowheads indicate regions of >5 kb not represented by a probe. Magenta bar, duplicated sequences in RB50. The chromosomal locations of the largest of these duplications, a degenerate prophage, are marked by magenta arrowheads. Yellow bar, genes not present in the RB50 sequence, arranged in Tohama I gene order. The values below the color bar at the bottom refer to log2(test/reference intensities). Missing data are gray. Bp, B. pertussis; Bb, B. bronchiseptica; Bpp, B. parapertussis.
For phylogenetic analysis and enumeration of variable genes, a threshold log ratio was calculated for each hybridization experiment and used to differentiate detected sequences from undetected sequences, such that probes with values below the threshold were called undetected. Accuracy of the method was estimated by comparing, for the three sequenced genomes, calls based on hybridization data to the “gold standard” of sequence data (33). A sequence was classified as “known absent” based on sequence data if it had less than 80% similarity to the microarray probe sequence. For all three sequenced genomes, sensitivity (sequences correctly called “not detected” from array data/total known absent sequences) and specificity (sequences correctly called “detected”/total known present sequences) were calculated. Detection sensitivity for the three sequenced genomes ranged from 94.5 to 100%, and specificity ranged from 98.8 to 99.1% (supplementary Table 3 [see the above URL]). Additionally, two dupilcate hybridizations of a B. pertussis genome were compared to determine reproducibility of the method. Overall, 99% of the gene calls were concordant between the replicates.
Phylogenetic analysis of Bordetella.
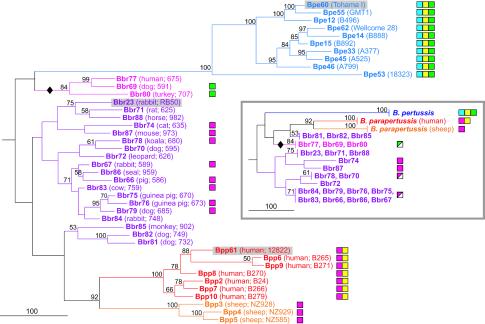
Variation in patterns of gene content often provides important insights into microbial evolution and population structure (21). Maximum parsimony, previously used to analyze CGH datasets (36, 40), was employed for phylogenetic analysis of our CGH data. Because gene acquisition appears to be infrequent in Bordetella (33, 49), polarity was assumed such that the complete gene complement comprised the ancestral state and deletion events were an irreversible derived state. This analysis produced two equally likely phylogenies (Fig. 2, large tree and inset). Both support the proposition that B. pertussis and B. parapertussis are more closely related to separate lineages of B. bronchiseptica than to each other (47) and that human and sheep isolates of B. parapertussis derive from a common ancestor but form separate clades. Although B. pertussis is monophyletic, the 18323 strain is very distantly related to the others, as previously suggested by other typing methods (10, 43, 47); 18323 may represent a B. pertussis subspecies. IS element distribution is also useful for determining evolutionary relationships. IS1663 was detected in all B. pertussis strains and in two B. bronchiseptica isolates (Fig. 2). IS1001 was previously detected in B. parapertussis and a subset of B. bronchiseptica strains. The presence of IS1002 in B. pertussis and human-derived B. parapertussis has been proposed to result from lateral transfer of IS1002 to B. parapertussis from a coinfecting B. pertussis (46). The simplest explanation for IS distribution is that after an early split in the B. bronchiseptica lineage, one group acquired IS1663 and then eventually gave rise to B. pertussis; the other group acquired IS1001 (which was sporadically lost from B. bronchiseptica) and then eventually gave rise to B. parapertussis. This is consistent with the topology of the large tree in Fig. 2, which we assert to be the more likely. In this phylogeny, B. pertussis is most closely associated with a distinct clade of B. bronchiseptica that conspicuously includes the only human isolate in the study. The CGH-based trees are also consistent with 16S ribosomal DNA-based phylogenies.
FIG. 2.
Phylogeny of Bordetella strains based on CGH data. Phylogenetic associations were inferred using a maximum-parsimony algorithm, and confidence intervals were determined from 100 bootstrapped datasets. The scale bar represents 100 evolutionary events (steps), and bootstrap values of ≥50% are indicated (19). The presence of Bordetella IS elements, as detected by CGH, is indicated for each strain: blue boxes, IS481; yellow, IS1002; and green, IS1663. The presence of IS1001 (magenta), which was not represented on the array, is taken from reference 46. In that study, the presence of IS1001 was not determined for B. pertussis, excepting strains Tohama I and 18323. In parentheses following each strain number are the strain's host and alias. Sequenced strains are shaded. The diamond indicates the B. bronchiseptica clade that may be associated with B. pertussis. The inset shows an overview tree of an equally likely phylogeny, created by collapsing all branches supported by bootstrap values of ≥50.
Our CGH-based phylogeny shares similarity with a MLEE-based tree that supported the independent derivation of B. pertussis, human-derived B. parapertussis, and sheep-derived B. parapertussis from the B. bronchiseptica lineage (47); however, the CGH-based tree strongly supports the conclusion that the two subspecies of B. parapertussis are more closely related to each other than to any B. bronchiseptica strain. The CGH-based phylogeny, supported by IS typing data, also identifies a deep-branching clade of B. bronchiseptica that is associated with B. pertussis, which is not supported by the MLEE-based tree; in fact, the three members of this clade represent two electrophoretic types (ET14 and ET29) that differ at 6 out of 15 loci. These discrepancies may be due to differences in phylogenetic methods but may also reflect the relatively small number of gene content differences between individual B. bronchiseptica strains.
Gene content diversity.
To understand the molecular differences that distinguish the Bordetella species, the subset of genes that was not detected in at least one strain of each species was determined (see supplementary methods). Based on our phylogenetic analysis, human- and sheep-derived B. parapertussis were treated separately. Eight hundred twenty probes were not detected in at least one B. pertussis strain, and 93.9% of these were located in RDs; 477 (88.7% in RDs) and 440 (86.4% in RDs) were not detected in at least one human-derived and sheep-derived B. parapertussis strain, respectively; and 555 (87.4% in RDs) were not detected in at least one B. bronchiseptica strain. Although RDs were sometimes located at similar RB50 coordinates in multiple species, the endpoints of the RDs were often slightly different between species, suggesting that similar deletions arose independently in the different lineages. At least seven RDs represent either intact or cryptic prophage. Ten randomly chosen RDs were independently confirmed by PCR analysis (supplementary methods and supplementary Table 2). Functional categorization of the genes that were not detected in each species indicated that most encode cell envelope, periplasmic and exported proteins, global regulatory factors, transport proteins, laterally acquired elements (prophage), and hypothetical open reading frames (ORFs) (supplementary Fig. 1). A caveat for interpretation of these data is that gene inactivation by frameshift, point mutation, IS insertion, or regulatory mutation is not discernible by hybridization, so CGH may underestimate the number of nonfunctional genes. For example, although the ptx-ptl locus, encoding pertussis toxin, is detected by CGH in all three species (data not shown), it is known to be transcriptionally silent in B. parapertussis and B. bronchiseptica (5), presumably due to a small number of single-nucleotide differences in the ptxA promoter region (33).
Species-specific sequences.
Evolution of pathogenesis often involves the acquisition of a small number of genes that enhance virulence or influence host range (32, 53). Eleven ORFs and two intergenic regions from B. pertussis Tohama I were unique to B. pertussis (supplementary Table 4). Nine ORFs fell into two contiguous gene clusters. With 61 to 63% GC content, both of these clusters deviate from the genome average (67.7%), suggesting that they may have been horizontally acquired. Each region contains a racemase fragment interrupted by an IS481 element in Tohama I, suggesting that recombination may have rearranged an ancestral insertion containing all nine genes. These sequences were not detected in B. pertussis 18323, indicating that the putative acquisition event occurred after divergence of this strain. Because 18323-like strains have been isolated from pertussis patients as recently as 1993 (10), these genes may not be necessary for B. pertussis pathogenesis or human adaptation. Another B. pertussis-specific gene, BP0507, encodes a hypothetical protein that is 46% similar to an internal region of the P.69 fragment of B. pertussis pertactin, an important adhesin and protective antigen.
One hundred forty-three sequences represented on the array were uniquely detected in B. bronchiseptica (supplementary Table 4). Sixty of these genes, mostly prophage components, were detected only in the sequenced RB50 strain. Among the 83 remaining B. bronchiseptica-specific sequences, 58 are prophage components. The remaining 25 probes represent 21 ORFs. These include BB0124 to BB0127 and BB0130, located in a contiguous block just upstream of the proposed lipopolysaccharide (LPS) O-antigen biosynthesis locus (see below), bbuR, which encodes a proposed transcriptional regulator of urease expression (28), and bfrA, which encodes a ferric siderophore receptor previously shown to be B. bronchiseptica specific (6).
None of the probes on the array hybridized uniquely to either human- or sheep-derived B. parapertussis. Because only 75% of the B. parapertussis 12822 genome was represented on the array, B. parapertussis-specific genes cannot be ruled out, but sequence alignment with the RB50 and Tohama I genomes identified a maximum of 33 possible B. parapertussis 12822-specific ORFs in the unrepresented fraction of the 12822 genome, some of which may be present in other B. bronchiseptica or B. pertussis strains. Furthermore, a sheep-derived B. parapertussis strain has not yet been sequenced and may contain genes not present in the three sequenced genomes.
Molecular discrimination of Bordetella species in the clinical setting is imperfect. The ideal diagnostic markers would be present in every strain of the target species and absent from all strains of the other species. In this data set, two B. pertussis sequences and three B. bronchiseptica sequences met these criteria. The B. pertussis-specific sequences were BP3385 and the B. pertussis BP0426-BP0427 intergenic region. The B. bronchiseptica-specific sequences were bfrA, BB4708, and fimN, although only the 3′ end of fimN is unique. These sequences could potentially be used as the basis for a sensitive and specific diagnostic assay.
Intraspecific variation.
B. pertussis is relatively homogeneous by some typing techniques (e.g., MLEE [47]) but exhibits heterogeneity in pulsed-field gel electrophoresis patterns (7, 23), probably due to extensive genome rearrangements (44). Among the B. pertussis strains examined in this study, 414 sequences were variably detected. The most divergent B. pertussis strain in this study was 18323, consistent with results with other typing methods (4, 10, 43, 47). This strain apparently represents an independently evolving branch of the B. pertussis lineage with very low representation in human infections. While this variant is pathogenic in humans (10), it is important to appreciate its genomic uniqueness when using strain 18323 to study B. pertussis pathogenesis. When 18323 was omitted from the analysis, only 181 sequences were variable, and 124 of these clustered within 26 RDs. All of the sequences that were variably detected among B. pertussis strains were also detected on the RB50 chromosome. In the Tohama I genome, many of these RDs are flanked by IS elements, suggesting that these deletions may have resulted from recombination between perfect repeats. The stability of gene content in B. pertussis is notable given the high frequency of genome rearrangement observed in natural populations (44). The majority of B. pertussis-variable genes with assigned functions encode cell envelope components, exported, periplasmic, and lipoproteins, transport proteins, carbon and fatty acid degradation enzymes, and transcriptional regulators. Of potential relevance to pathogenesis, several iron acquisition genes were variably detected. A locus encoding the ferric citrate uptake regulators, FecI and FecR, and three putative iron uptake proteins was absent from six B. pertussis strains. Probes in the alcaligin biosynthesis operon showed consistent but small decreases in hybridization in three B. pertussis strains, as well as other strains throughout the genus, suggesting that the nucleotide sequence might be divergent.
The most striking observation about the B. parapertussis strains was the clear distinction between human- and sheep-derived isolates. Several RDs distinguished these two subspecies, including ORFs encoding transcriptional regulators, cell surface proteins, and transporters. The genomic region from cheR/cheX to tar/cheM was not detected in sheep-derived B. parapertussis. If chromosomal order is conserved in all B. parapertussis strains, this apparent deletion removes several components of the chemotaxis and motility apparatus, which may explain why sheep-derived B. parapertussis strains are nonmotile (2).
Within each of the two B. parapertussis subspecies, gene content was highly conserved, with only 105 variably detected sequences in the human-derived strains and 71 variably detected sequences in the sheep-derived strains. The variable genes with assigned functions encode cell envelope, exported proteins, transporters, and transcriptional regulators. In one sheep-derived B. parapertussis isolate, genes encoding components of the type III secretion apparatus and an adjacent sigma factor were not detected, suggesting that type III secretion may not be required for pathogenesis of B. parapertussis in sheep. Genes encoding two putative adhesins, fhaS and BP3204/BPP0104, were variably detected among the B. parapertussis strains, implying a diversity of adherence mechanisms within this species.
B. bronchiseptica, with 453 variable sequences, exhibited the most heterogeneity of gene content among the species examined. The genome of B. bronchiseptica RB50, which has been isolated from the environmental gene pool as a result of laboratory passage, harbors three putative prophage that are not found in any other strain in this survey and a fourth prophage that is fully intact only in this strain (Fig. 1). Aside from its unusual prophage content, RB50 appeared to be a typical B. bronchiseptica strain. The presence of two other prophage was also variable within the species. The other variable genes with assigned functions primarily encode cell envelope, exported proteins, transporters, and transcriptional regulators. The putative adhesin BP3204/BPP0104 was not detected in three B. bronchiseptica isolates or sheep-derived B. parapertussis. Seven B. bronchiseptica strains exhibited slightly reduced hybridization across the alcaligin biosynthesis locus, as observed for B. pertussis.
Variation in surface molecules.
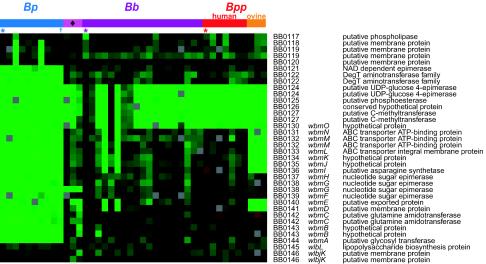
LPS is the predominant cell-surface glycolipid of gram-negative bacteria and an important virulence factor for many pathogens. Gene content of the Bordetella LPS biosynthetic locus was highly variable among the strains (Fig. 3). As previously observed (37), the O-antigen biosynthesis locus was not detected in any B. pertussis strains. BB0124 to BB0127 and B0130 were not detected in 12 B. bronchiseptica isolates and all B. parapertussis isolates. The LPS O-antigen locus previously reported for B. bronchiseptica strain C7635E (37) differs in this region from the RB50 locus (33). Because probes for the wbmPQRS genes identified in C7635E were not included on the array, the presence of these genes could not be determined, so alternative ORFs may be present in this locus in some B. bronchiseptica strains. The ORFs in this region may encode proteins, formyltransferases in particular, that modify the terminal sugar of O antigen (48). Genomes of two B. bronchiseptica strains and all sheep-derived B. parapertussis strains did not hybridize to BB0131 to BB0136 (wbmIJKLMN) and BB0140 to BB0141 (wbmDE). These strains may lack key O-antigen biosynthetic enzymes and transport proteins, which could result in variant LPS structures. Three other B. bronchiseptica strains had unique LPS locus gene content patterns. These results provide a molecular explanation for LPS structural diversity between and within Bordetella species (45) and could have important ramifications for virulence and host association.
FIG. 3.
Gene content variation and structural differences in the LPS O-antigen biosynthesis region. CGH data for the O-antigen biosynthesis region in 42 Bordetella strains are shown, displayed as in Fig. 1, with strain groups color coded according to the tree topology shown in Fig. 2. Some ORFs are represented by multiple probes. Coding sequence designations, gene names, and annotations are included. The color scale is as in Fig. 1.
Bordetella autotransporters (e.g., BrkA, Vag-8, and SphB1) also have been implicated in host cell interactions and virulence (reviewed in reference 26). This family of proteins is distinguished by the presence of a shared domain that inserts into the gram-negative bacterial outer membrane and translocates the attached domain to the cell surface. Several of the Bordetella autotransporters were variably detected (supplementary Fig. 2), including bapA, bapC, and three uncharacterized proteins.
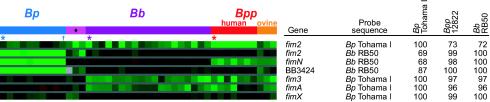
Fimbriae are Bordetella adhesins, major antigens, and determinants of serotype (50). Absence of both fimN and a novel putative fimbrial subunit, BB3424/BPP1684, was confirmed by CGH for all B. pertussis strains (Fig. 4). Sequence divergence as low as 2% could be discerned visually as blocks of consistent, slightly negative log ratios within the species that diverged from the probe sequence. fim2 was represented by two probes that are only 67% identical. The RB50 fim2 probe hybridized to all B. bronchiseptica and B. parapertussis strains but not to B. pertussis strains. Unexpectedly, the Tohama I-derived fim2 probe hybridized to all three ovine B. parapertussis strains and 16 of the 22 B. bronchiseptica strains, as well as to the B. pertussis strains. This indicated that sheep-derived B. parapertussis strains and some B. bronchiseptica strains might have both a Tohama I-like fim2 gene and an RB50-like fim2 gene. Species-specific fim2 PCR was performed on three sheep-derived B. parapertussis strains and four B. bronchiseptica strains that hybridized to both probes (supplementary methods). Tohama I-specific primers failed to amplify a fragment, but PCR with the RB50-specific primers amplified a product in all seven strains (data not shown). Amplicons from a sheep-derived B. parapertussis (Bpp4) strain and a B. bronchiseptica (Bbr77) strain were cloned and sequenced. The 518-bp Bpp4 gene fragment was 98% identical to the RB50 sequence and was predicted to encode a protein with a Thr→Arg substitution and a three-amino-acid deletion relative to the RB50 protein (supplementary Fig. 3). The 528-bp Bbr77 fragment was 97% identical to the RB50 sequence and encoded a protein with five amino acid substitutions relative to the RB50 protein. Because these sequences are less than 63% identical to that of the Tohama I-derived probe, they do not account for the hybridization signals obtained with that probe in the CGH experiments. Therefore, these strains probably possess a second fim2 gene that is similar to the Tohama I orthologue. Loss of fimbrial structural alleles in bordetellae may be responsible in part for a restriction in host range.
FIG. 4.
Gene content variation and sequence polymorphism in fimbriae genes. Strain groups are color coded according to the tree topology shown in Fig. 2. For each probe, the sequenced strain from which it was amplified is listed, as well as the percent sequence identity of the probe to each of the three sequenced genomes. The color scale and symbols are as in Fig. 1.
Subtractive hybridization of B. pertussis strains.
An important limitation of microarray-based CGH studies is that only sequences on the array can be assayed. To investigate the potential genetic diversity in B. pertussis strains that have not been sequenced, SSH experiments were performed. SSH of B. pertussis 18323, which is genetically distinct from Tohama I (31), against a Tohama I driver pool produced 91 clones that did not hybridize to Tohama I genomic DNA, all of which were highly similar to sequences in the B. bronchiseptica RB50 or B. parapertussis 12822 genome (data not shown). All of the sequences identified by SSH that were also represented as probes on the microarray were detected in 18323 and not detected in Tohama I, further verifying the accuracy of the methods. SSH of a recent clinical isolate, Bpe159, against a mixed driver pool comprising Tohama I, RB50, and 12822 produced 174 clones, all of which upon screening hybridized to the driver pool. These results provide further evidence that gene acquisition by B. pertussis has been rare.
Pathogen evolution by gene loss.
Gene loss events may have driven the speciation of B. pertussis and B. parapertussis by altering regulatory networks, modifying metabolic pathways (27), or eliminating antigenic proteins that made them susceptible to immune surveillance. These genomic modifications could have resulted in novel virulence characteristics that made the strains more effective at infecting a particular host (e.g., humans). Alternatively, genome decay may have occurred subsequent to host restriction due to a loss of selective pressure against gene loss and deleterious transposition events (3, 32). In this scenario, the initial niche restriction may have been attributable to a more subtle genetic change, such as inactivation or modification of a key surface molecule or regulatory factor. In fact, CGH revealed extensive variation in the complement of regulatory proteins and in the presence and sequence of genes responsible for cell surface structures including LPS, fimbriae, and autotransporters. These two hypotheses are not mutually exclusive.
The decayed genomes of B. pertussis and B. parapertussis are also characterized by high IS element loads (46, 47). Of particular note, many inferred genomic rearrangements between B. bronchiseptica and the derived species are closely associated with IS elements (33), suggesting that recombination between identical copies of IS elements is an important mechanism in the genesis of inversions, transpositions, and deletions. Genome rearrangement may be the inevitable consequence of accumulating high copy numbers of one or more repetitive sequences. Adoption of a host-restricted lifestyle may have relaxed selection against some deleterious transposition events and subsequent rearrangements. Furthermore, modelling experiments have suggested that selection against increased transposon copy number could be diminished in small populations, as might have been encountered by the original B. pertussis and B. parapertussis progenitors (11).
Genome decay is frequently observed in the genomes of obligate intracellular pathogenic bacteria (3, 32), but among extracellular pathogens the inferred evolution of B. pertussis and B. parapertussis from a B. bronchiseptica-like ancestor is most similar to the proposed evolution of Yersinia pestis from Yersinia pseudotuberculosis (1, 34). In both cases, relative to the ancestral species, the derived species exhibit extensive genomic deletions and rearrangements, accumulation of pseudogenes, and high copy numbers of IS elements that result in genomic fluidity. Genes encoding uptake and transport functions are often inactivated, while central and intermediate metabolism genes remain largely intact. The derived strains also share phenotypic features, including expression of rough LPS (37, 39, 45) and limited capability to survive outside the host (1, 14, 35). A fundamental difference is that gene acquisition has played a fundamental role in the evolution of Y. pestis (12) but does not appear to have contributed significantly to Bordetella evolution. Instead, the molecular events that initially resulted in modified host adaptation and virulence traits in B. pertussis and B. parapertussis were probably modifications or deletions of existing genes or genome rearrangements with global regulatory consequences.
By elucidating the molecular differences between the Bordetella species, we may achieve an understanding of how host adaptation occurs. Ultimately, our results could help to explain the mechanisms of vaccine-induced immunity, which may have played a role in the reemergence of B. pertussis (30) and potentially to guide the design of improved vaccines.
Acknowledgments
This work was funded by NIH grants R01 AI039587 and R03 AI547970 to D.A.R. Microarray design software development was funded by a Stanford University Office of Technology and Licensing Innovation Research Incentive Award. C.A.C. received Research Training Fellowships from the American Lung Association (ALA) and ALA-California. M.M.B. was supported by a Smith Stanford Graduate Fellowship.
We are grateful to Julian Parkhill and the PSU of the Sanger Institute for making genome sequence assemblies and annotation available prior to publication. We also thank Frits Mooi, Gary Sanden, James Musser, Sylvia Yeh, and Jeff F. Miller for providing bacterial strains, Trevor Hastie, Hester Bootsma, and Dimitri Diavatopoulos for helpful discussions, Ikro Yoon for software development, Brett Petersen for protocol development, and Hassya Kedem, Jennifer Maynard, Christine Dieterich, Elisabeth Bik, and Isabelle da Piedade for assistance with array production.
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akerley, B. J., and J. F. Miller. 1993. Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J. Bacteriol. 175:3468-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, J. O., and S. G. Andersson. 1999. Insights into the evolutionary process of genome degradation. Curr. Opin. Genet. Dev. 9:664-671. [DOI] [PubMed] [Google Scholar]
- 4.Arico, B., R. Gross, J. Smida, and R. Rappuoli. 1987. Evolutionary relationships in the genus Bordetella. Mol. Microbiol. 1:301-308. [DOI] [PubMed] [Google Scholar]
- 5.Arico, B., and R. Rappuoli. 1987. Bordetella parapertussis and Bordetella bronchiseptica contain transcriptionally silent pertussis toxin genes. J. Bacteriol. 169:2847-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beall, B., and T. Hoenes. 1997. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology 143(Part 1):135-145. [DOI] [PubMed] [Google Scholar]
- 7.Bisgard, K. M., C. D. Christie, S. F. Reising, G. N. Sanden, P. K. Cassiday, C. Gomersall, W. A. Wattigney, N. E. Roberts, and P. M. Strebel. 2001. Molecular epidemiology of Bordetella pertussis by pulsed-field gel electrophoresis profile: Cincinnati, 1989-1996. J. Infect. Dis. 183:1360-1367. [DOI] [PubMed] [Google Scholar]
- 8.Bordet, J., and O. Gengou. 1906. Le microbe de la coqueluche. Ann. Inst. Pasteur 20:731-741. [Google Scholar]
- 9.Boucher, Y., C. L. Nesbo, and W. F. Doolittle. 2001. Microbial genomes: dealing with diversity. Curr. Opin. Microbiol. 4:285-289. [DOI] [PubMed] [Google Scholar]
- 10.Boursaux-Eude, C., S. Thiberge, G. Carletti, and N. Guiso. 1999. Intranasal murine model of Bordetella pertussis infection. II. Sequence variation and protection induced by a tricomponent acellular vaccine. Vaccine 17:2651-2660. [DOI] [PubMed] [Google Scholar]
- 11.Brookfield, J. F., and R. M. Badge. 1997. Population genetics models of transposable elements. Genetica 100:281-294. [PubMed] [Google Scholar]
- 12.Brubaker, R. R. 1991. Factors promoting acute and chronic diseases caused by yersiniae. Clin. Microbiol. Rev. 4:309-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camin, J. H., and R. R. Sokal. 1965. A method for deducing branching sequences in phylogeny. Evolution 19:311-326. [Google Scholar]
- 14.Cassiday, P. K., G. N. Sanden, C. T. Kane, S. M'Boup, and J. M. Barbaree. 1994. Viability of Bordetella pertussis in four suspending solutions at three temperatures. J. Clin. Microbiol. 32:1550-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2002. Pertussis—United States, 1997-2000. Morb. Mortal. Wkly. Rep. 51:73-76. [PubMed] [Google Scholar]
- 16.Cullinane, L. C., M. R. Alley, R. B. Marshall, and B. W. Manktelow. 1987. Bordetella parapertussis from lambs. N.Z. Vet. J. 35:175. [DOI] [PubMed] [Google Scholar]
- 17.Eldering, G., and P. L. Kendrick. 1938. Bacillus parapertussis: a species resembling both Bacillus pertussis and Bacillus bronchisepticus but identical with neither. J. Bacteriol. 35:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein, J., and H. Kishino. 1993. Is there something wrong with the bootstrap on phylogenies? A reply to Hillis and Bull. Syst. Biol. 42:193-200. [Google Scholar]
- 20.Field, D., D. Hood, and R. Moxon. 1999. Contribution of genomics to bacterial pathogenesis. Curr. Opin. Genet. Dev. 9:700-703. [DOI] [PubMed] [Google Scholar]
- 21.Fitzgerald, J. R., and J. M. Musser. 2001. Evolutionary genomics of pathogenic bacteria. Trends Microbiol. 9:547-553. [DOI] [PubMed] [Google Scholar]
- 22.Goodnow, R. A. 1980. Biology of Bordetella bronchiseptica. Microbiol. Rev. 44:722-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardwick, T. H., P. Cassiday, R. S. Weyant, K. M. Bisgard, and G. N. Sanden. 2002. Changes in predominance and diversity of genomic subtypes of Bordetella pertussis isolated in the United States, 1935 to 1999. Emerg. Infect. Dis. 8:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khattak, M. N., and R. C. Matthews. 1993. Genetic relatedness of Bordetella species as determined by macrorestriction digests resolved by pulsed-field gel electrophoresis. Int. J. Syst. Bacteriol. 43:659-664. [DOI] [PubMed] [Google Scholar]
- 25.Linnemann, C. C., and E. B. Perry. 1977. Bordetella parapertussis. Recent experience and a review of the literature. Am. J. Dis. Child. 131:560-563. [DOI] [PubMed] [Google Scholar]
- 26.Locht, C., R. Antoine, and F. Jacob-Dubuisson. 2001. Bordetella pertussis, molecular pathogenesis under multiple aspects. Curr. Opin. Microbiol. 4:82-89. [DOI] [PubMed] [Google Scholar]
- 27.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMillan, D. J., M. Mau, and M. J. Walker. 1998. Characterisation of the urease gene cluster in Bordetella bronchiseptica. Gene 208:243-251. [DOI] [PubMed] [Google Scholar]
- 29.Mooi, F. R., H. Hallander, C. H. Wirsing von Konig, B. Hoet, and N. Guiso. 2000. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur. J. Clin. Microbiol. Infect. Dis. 19:174-181. [DOI] [PubMed] [Google Scholar]
- 30.Mooi, F. R., I. H. van Loo, and A. J. King. 2001. Adaptation of Bordetella pertussis to vaccination: a cause for its reemergence? Emerg. Infect. Dis. 7:526-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno Lopez, M. 1990. The genus Bordetella. By Manuel Moreno Lopez, 1952. Enferm. Infecc. Microbiol. Clin. 8:480-485. (In Spanish.) [PubMed] [Google Scholar]
- 32.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1099. [DOI] [PubMed] [Google Scholar]
- 33.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. G. Holden, C. R. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeño-Tárraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, R. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 34.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 35.Porter, J. F., R. Parton, and A. C. Wardlaw. 1991. Growth and survival of Bordetella bronchiseptica in natural waters and in buffered saline without added nutrients. Appl. Environ. Microbiol. 57:1202-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preston, A., A. G. Allen, J. Cadisch, R. Thomas, K. Stevens, C. M. Churcher, K. L. Badcock, J. Parkhill, B. Barrell, and D. J. Maskell. 1999. Genetic basis for lipopolysaccharide O-antigen biosynthesis in Bordetellae. Infect. Immun. 67:3763-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preziosi, M. P., A. Yam, S. G. Wassilak, L. Chabirand, A. Simaga, M. Ndiaye, M. Dia, F. Dabis, and F. Simondon. 2002. Epidemiology of pertussis in a West African community before and after introduction of a widespread vaccination program. Am. J. Epidemiol. 155:891-896. [DOI] [PubMed] [Google Scholar]
- 39.Prior, J. L., J. Parkhill, P. G. Hitchen, K. L. Mungall, K. Stevens, H. R. Morris, A. J. Reason, P. C. Oyston, A. Dell, B. W. Wren, and R. W. Titball. 2001. The failure of different strains of Yersinia pestis to produce lipopolysaccharide O-antigen under different growth conditions is due to mutations in the O-antigen gene cluster. FEMS Microbiol. Lett. 197:229-233. [DOI] [PubMed] [Google Scholar]
- 40.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 41.Register, K. B., A. Boisvert, and M. R. Ackermann. 1997. Use of ribotyping to distinguish Bordetella bronchiseptica isolates. Int. J. Syst. Bacteriol. 47:678-683. [DOI] [PubMed] [Google Scholar]
- 42.Schoolnik, G. K. 2002. Functional and comparative genomics of pathogenic bacteria. Curr. Opin. Microbiol. 5:20-26. [DOI] [PubMed] [Google Scholar]
- 43.Stibitz, S., and M. S. Yang. 1997. Genomic fluidity of Bordetella pertussis assessed by a new method for chromosomal mapping. J. Bacteriol. 179:5820-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stibitz, S., and M. S. Yang. 1999. Genomic plasticity in natural populations of Bordetella pertussis. J. Bacteriol. 181:5512-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Akker, W. M. 1998. Lipopolysaccharide expression within the genus Bordetella: influence of temperature and phase variation. Microbiology 144(Part 6):1527-1535. [DOI] [PubMed] [Google Scholar]
- 46.van der Zee, A., H. Groenendijk, M. Peeters, and F. R. Mooi. 1996. The differentiation of Bordetella parapertussis and Bordetella bronchiseptica from humans and animals as determined by DNA polymorphism mediated by two different insertion sequence elements suggests their phylogenetic relationship. Int. J. Syst. Bacteriol. 46:640-647. [DOI] [PubMed] [Google Scholar]
- 47.van der Zee, A., F. Mooi, J. Van Embden, and J. Musser. 1997. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J. Bacteriol. 179:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vinogradov, E., M. S. Peppler, and M. B. Perry. 2000. The structure of the nonreducing terminal groups in the O-specific polysaccharides from two strains of Bordetella bronchiseptica. Eur. J. Biochem. 267:7230-7237. [DOI] [PubMed] [Google Scholar]
- 49.von Wintzingerode, F., G. Gerlach, B. Schneider, and R. Gross. 2002. Phylogenetic relationships and virulence evolution in the genus Bordetella. Curr. Top. Microbiol. Immunol. 264:177-199. [PubMed] [Google Scholar]
- 50.Willems, R., A. Paul, H. G. van der Heide, A. R. ter Avest, and F. R. Mooi. 1990. Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO J. 9:2803-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woolfrey, B. F., and J. A. Moody. 1991. Human infections associated with Bordetella bronchiseptica. Clin. Microbiol. Rev. 4:243-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.World Health Organization. 1999. Pertussis vaccines: W. H. O. position paper. Wkly. Epidemiol. Rec. 74:137-144. [Google Scholar]
- 53.Ziebuhr, W., K. Ohlsen, H. Karch, T. Korhonen, and J. Hacker. 1999. Evolution of bacterial pathogenesis. Cell. Mol. Life Sci. 56:719-728. [DOI] [PMC free article] [PubMed] [Google Scholar]