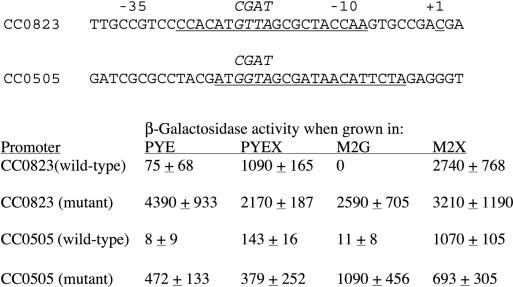Abstract
Microarray analysis was used to examine gene expression in the freshwater oligotrophic bacterium Caulobacter crescentus during growth on three standard laboratory media, including peptone-yeast extract medium (PYE) and minimal salts medium with glucose or xylose as the carbon source. Nearly 400 genes (approximately 10% of the genome) varied significantly in expression between at least two of these media. The differentially expressed genes included many encoding transport systems, most notably diverse TonB-dependent outer membrane channels of unknown substrate specificity. Amino acid degradation pathways constituted the largest class of genes induced in PYE. In contrast, many of the genes upregulated in minimal media encoded enzymes for synthesis of amino acids, including incorporation of ammonia and sulfate into glutamate and cysteine. Glucose availability induced expression of genes encoding enzymes of the Entner-Doudoroff pathway, which was demonstrated here through mutational analysis to be essential in C. crescentus for growth on glucose. Xylose induced expression of genes encoding several hydrolytic exoenzymes as well as an operon that may encode a novel pathway for xylose catabolism. A conserved DNA motif upstream of many xylose-induced genes was identified and shown to confer xylose-specific expression. Xylose is an abundant component of xylan in plant cell walls, and the microarray data suggest that in addition to serving as a carbon source for growth of C. crescentus, this pentose may be interpreted as a signal to produce enzymes associated with plant polymer degradation.
Caulobacter species are ubiquitous inhabitants of freshwater, marine, and subsurface environments and are sufficiently adaptable to low nutrient conditions that tapwater and even distilled water are ready sources for their isolation (19, 48, 49). The physiological properties that enable Caulobacter and other oligotrophic species to survive and reproduce in such environments are not well understood. In this work, we used whole-genome transcriptional profiling and genetic analysis to characterize metabolic pathways and transport systems that are differentially expressed by Caulobacter crescentus during growth in three standard laboratory media: a defined “minimal” inorganic salts medium supplemented with either glucose or xylose as the sole carbon source and a complex peptone-yeast extract medium (PYE), in which amino acids serve as the primary carbon source. These observations provide important baseline data on the genetic coordination of metabolism in this model organism.
The dimorphic life cycle of C. crescentus has served as a model for understanding prokaryotic development, cell cycle regulation, and asymmetric cell division (24). At each division, C. crescentus divides into two morphologically and physiologically distinct cells: motile swarmer progeny with a single polar flagellum, and sessile stalked progeny displaying an adhesive stalk. The swarmer progeny, in which DNA replication is initially inhibited, is specialized for dispersal and can seek new nutrient sources by chemotactic sensing. After about a third of their cell cycle, swarmer progeny cells eject their flagellum and grow a stalk from the same pole. The stalk enhances nutrient acquisition by increasing the effective membrane area available for transport systems without substantially increasing the cytoplasmic volume (20). Stalked cells initiate chromosome replication, grow a new flagellum, and divide, yielding new swarmer and stalked cells.
Genome-scale transcriptional profiling has shown that the expression of over 500 C. crescentus genes varies significantly over the course of the cell cycle (32). Among these are many genes involved in energy metabolism, nucleotide and amino acid biosynthesis, and protein synthesis. As shown here, a substantial fraction of these genes are also regulated in response to nutrients, raising intriguing questions as to how regulatory networks controlling metabolism and cell cycle progression might interact.
Caulobacter is obligately aerobic and heterotrophic and can utilize a variety of carbon sources. Previous studies have demonstrated the activity of Entner-Doudoroff pathway enzymes (14) during Caulobacter growth on glucose (48, 52), but there has been no quantitative analysis of glucose flow into the Entner-Doudoroff pathway versus alternatives such as the Embden-Meyerhof-Parnas glycolytic pathway. Related α-proteobacteria, including Rhizobium and Agrobacterium strains, use both the Entner-Doudoroff and Embden-Meyerhof-Parnas pathways, sometimes simultaneously (3, 42). The C. crescentus genome, however, lacks a clear homolog of phosphofructokinase, a key enzyme of the Embden-Meyerhof-Parnas pathway (45). Without this enzyme, any glucose-6-phosphate produced could serve as a substrate for glucose-6-phosphate dehydrogenase, directing carbon from glucose into the Entner-Doudoroff pathway. We show here that the Entner-Doudoroff pathway is essential in C. crescentus for growth with glucose as the sole carbon source.
Genome sequence analysis unexpectedly revealed that C. crescentus has the potential to degrade plant-derived biopolymers through the production of exoenzymes, including cellulases (endo-β-1,4-glucanases and β-glucosidases), xylanases and xylosidases, and polysaccharide deacetylases (45). In freshwater ponds, lakes, and rivers, Caulobacter species could use live or decaying aquatic vegetation as surfaces for biofilm formation. The major monomeric products released by degradative exoenzymes acting on lignocellulose would be glucose (from cellulose) and xylose (from xylan). The mechanism of xylose dissimilation in C. crescentus is unknown. The best-known bacterial pathway for xylose catabolism, isomerization to xylulose followed by phosphorylation to xylulose-5-phosphate and entry into the pentose phosphate pathway (23), is probably not operative in C. crescentus, given the absence of homologs for xylose isomerase and xylulose kinase in the genome. An inducible NADP-dependent xylose dehydrogenase activity has been reported in C. crescentus (48), but neither the gene encoding this activity nor the catabolic pathway in which it might participate is known. We identify here genes whose expression increases in the presence of xylose. These data suggest that C. crescentus uses xylose as an indicator of the availability of plant-derived polymers. We also identify a DNA sequence motif associated with many xylose-inducible genes and show that the motif is important for regulation.
The abundance of putative secreted peptidases in the C. crescentus genome suggests that extracellular degradation of proteins may also be important in natural habitats (45). Environmental amino acids support rapid growth of C. crescentus, leading us to compare gene expression in a complex amino acid-containing medium (PYE) to gene expression in defined minimal media. As expected, several amino acid degradation pathways are upregulated during growth in PYE, in contrast to induction of amino acid biosynthetic pathways (along with ammonia and sulfate assimilation systems) in minimal media. Interestingly, however, expression of many other genes encoding enzymes of key biosynthetic pathways does not change significantly between PYE and minimal media.
As a gram-negative bacterium, C. crescentus must acquire nutrients via transport across both outer and inner membranes, and high-affinity nutrient uptake is presumably necessary to support an oligotrophic lifestyle. The C. crescentus genome seems to lack homologs of the OmpF-type outer membrane porins characteristic of many gram-negative species (45) but is remarkably rich in TonB-dependent receptors, with at least 65 gene products containing the TonB box. As an alternative transport system, TonB-dependent receptors are thought to harness energy from the inner membrane proton motive force to the transport of macromolecular complexes, including vitamin B12 and siderophores, across the outer membrane (28, 41). With respect to transport across the inner membrane, over 80 ABC (ATP binding cassette) transporter components (61) are encoded in the Caulobacter genome. We show here that numerous TonB-dependent receptors and some ABC transporters are differentially expressed in each medium. Whether the deployment of such transporters is a key factor for Caulobacter and other oligotrophs to efficiently acquire nutrients in diverse nutrient-limited environments clearly warrants further investigation.
MATERIALS AND METHODS
Bacterial strains, media, and growth.
Cultures were grown in PYE (peptone-yeast extract) broth containing 0.2% Bacto Peptone (Difco) (0.1% yeast extract [Difco], 1 mM MgSO4, and 0.5 mM CaCl2) in M2 minimal salts medium (6.1 mM Na2HPO4, 3.9 mM KH2PO4, 9.3 mM NH4Cl, 0.5 mM MgSO4, 10 μM FeSO4 [EDTA chelate; Sigma Chemical Co.], 0.5 mM CaCl2) with 0.2% glucose as the sole carbon source (referred to subsequently as M2G) or in M2 medium with 0.2% xylose as the sole carbon source (referred to subsequently as M2X) (12). Liquid cultures were inoculated with C. crescentus colonies randomly picked off PYE agar plates (12). C. crescentus strain CB15N (NA1000) was used for microarray studies. The mutations and transcriptional fusions generated in this study were analyzed in the CB15N strain background. hex mutant strains (provided by Bert Ely, University of South Carolina) were derived from the CB15 strain background. Cultures from which RNA was extracted were grown in 125-ml flasks and shaken continuously at 275 rpm and incubated at 30°C.
RNA isolation.
For the microarrays, cells were harvested from a total of eight PYE cultures, eight M2G cultures, and five M2X cultures. The cultures were in exponential growth at the time of harvest, with mean densities as follows: PYE, OD600 = 0.37 ± 0.12; M2G, OD600 = 0.34 ± 0.13; and M2X, OD600 = 0.27 ± 0.05. (The cultures were not synchronized, and thus contained cells from all stages of the cell cycle.) Cells were isolated by centrifugation in a Beckman J2-21 M centrifuge at 7,740 × g at room temperature for 2 min. (In control experiments, approximately 85% of the cells were recovered in the pellet, and all stages of the cell cycle seemed to be recovered with similar efficiency based on phase contrast microscopy.)
The complete RNA extraction and purification protocol is described in the supplementary material (http://www.stanford.edu/group/caulobacter/metabolism) and described briefly here. Each cell pellet was resuspended in Trizol (Invitrogen) and incubated at 65°C for 10 min. Chloroform was added, and the samples were centrifuged in a microcentrifuge at 16,000 × g at 4°C for 15 min. The aqueous layer was transferred to a new tube, mixed with isopropanol, and frozen overnight at −80°C. The next day, RNA was pelleted by centrifugation at 16,000 × g at 4°C for 30 min. Supernatant was poured off, and the pellets were washed in 70% ethanol and resuspended in diethyl pyrocarbonate-treated water. Contaminating DNA was digested with DNase I (Ambion). RNA was extracted with acid phenol-chloroform, precipitated with ethanol and sodium acetate, and then resuspended in RNase-free water. RNA quality was visualized on agarose gels, and RNA concentration was determined spectrophotometrically.
PCR microarrays.
PCR products were spotted onto poly-l-lysine-coated glass microarray slides with an OmniGrid printer (GeneMachines) with 32 Telechem pins. The slides were postprocessed as described by Laub et al. (32). Primer sequences are available in the supplementary material and correspond largely to those used in Laub et al. (31). Slides from three printings were used.
Labeled indodicarbocyanine-dCTP (Cy3) and indocarbocyanine-dCTP (Cy5) cDNA probes were generated from 20 μg of total RNA by reverse transcription as described by Laub et al. (32), except that the amount of random hexamers was increased from 500 ng to 1 μg. Samples were hybridized competitively under coverslips to the microarray slides overnight at 65°C, then washed as described by Laub et al. (32). Hybridized arrays were scanned with a GenePix 4000B scanner (Axon Instruments), and scanned spots were converted to ratios (red/green) with GenePix Pro 4.0 software. Ratios and statistics on each spot were stored in an SQL Server database. The signal for each channel (red and green) was taken to be the mean of the foreground signal minus the median of the local background signal. Some spots were excluded by the criteria described in the online supplementary material. Array-wide normalization procedures are also described in the supplementary material.
Identifying genes differentially expressed between pairs of media.
Thirty-four microarrays were used to assay relative RNA abundance. Eighteen compared M2G and PYE, nine compared PYE and M2X, and seven compared M2X and M2G. To reduce dye bias, samples from each medium were labeled with Cy3 about half the time and with Cy5 about half of the time. We used least-squares techniques (59) to simultaneously estimate log2(M2G/M2X), log2(M2X/PYE), and log2(M2G/PYE) expression ratios for each gene. We used a correlated error model to account for dependencies introduced by hybridizing some RNA samples on multiple arrays. The variance of each estimate was modeled by assuming a normal error distribution.
For each gene, Student's t test was used to test the hypothesis that each log ratio, i.e., log2(M2G/M2X), log2(M2G/PYE), and log2(M2X/PYE), was zero. A two-sided P value, which accounted for the different number of samples available for each gene, was assigned to each ratio. For most genes, more arrays comparing M2G and PYE were done than were done for any other combination of media. Thus, the confidence regions for log2(M2G/PYE) tended to be smaller than those for log2(M2G/M2X) or for log2(M2X/PYE). To roughly equalize the number of false-negatives, significance levels were set so that, under the noise assumptions, 10 false-positives were expected for each of the log2(M2G/M2X) and the log2(M2X/PYE) comparisons and five false-positives were expected for the log2(M2G/PYE) comparisons. To minimize the effect of possible biases, to be considered significant, gene expression also needed to change at least 1.4-fold. Additionally, average changes of at least threefold were considered significant. Applying the above procedures to a randomized version of the data set yielded 18 false-positives (all comparisons combined). For more detail on the analysis procedure, see the supplementary material.
To aid in identifying patterns of coregulation, differentially expressed genes were sorted into several groups. For example, genes were assigned to the “up-in- PYE” set (equivalent to the “down-in-M2” set) if the increase in expression between PYE and both M2G and M2X was statistically significant or if the increase in expression between PYE and at least one of the M2 media was statistically significant, while the difference in expression between PYE and the other M2 medium was at least 1.75-fold. The down-in-PYE/up-in-M2, up-in- M2G, down-in-M2G, up-in-M2X, and down-in-M2X sets were constructed analogously.
Metabolic pathway analysis and identification of differentially regulated potential isozymes.
Functional analysis of microarray data was aided by CauloCyc (http://biocyc.org), a computationally derived pathway/genome database of C. crescentus. A pathway/genome database describes the genome of an organism, the product of each gene, the biochemical reaction(s), if any, catalyzed by each gene product, the substrates of each reaction, and the organization of those reactions into pathways. CauloCyc was computationally derived from the annotated genome of C. crescentus by the PathoLogic software, part of the Pathway Tools software suite (26, 54).
Each metabolic pathway in CauloCyc consists of a set of enzymatic functions (biochemical reactions) and the gene(s) that code for each function. When CauloCyc contained multiple genes for a given reaction, microarray data were used to construct 95% confidence intervals for each gene for each pairwise comparison with the noise model described above. This allowed pairs of genes whose products are annotated as catalyzing the same reaction, but which for one or more pairwise comparisons averaged at least a twofold difference in expression and had disjoint confidence intervals to be identified. Typically, we required at least two comparisons to yield distinct confidence intervals. For example, in a case where one gene in the pair was up in PYE and the other was unchanged, the confidence intervals for both PYE/M2G and PYE/M2X would need to be distinct. Also, if the PYE/M2G and PYE/M2X ratios for the first gene were 0.9 and 1.2, respectively, then the average PYE/M2G and PYE/M2X ratios for the second gene would need to be at least 1.8 and 2.4, respectively.
Gene annotations and categorization.
Most C. crescentus open reading frame (ORF) annotations were taken from GenBank accession number AE005673 (45). For genes annotated in GenBank as hypothetical or conserved hypothetical proteins, clusters of orthologous gene (COG) descriptions (57, 58) were used if they were more detailed. Gene names shown in the tables came from TIGR and generally refer to the homologous Escherichia coli gene (see www.tigr.org for more information). Annotations for CC2086 and CC1000 were obtained on the strength of BLAST matches to the NCBI NR database (2). BLAST was also used to identify homology between C. crescentus gene products and the gene products of other organisms. Classifications were taken from COG classifications when available; genes without COG classifications were categorized based on their GenBank annotations.
In order to create disjoint groups, genes assigned to multiple COG categories were placed into a single category (see supplementary materials). To facilitate comparisons, gene products involved in transport (based on either their GenBank or their COG annotation) were grouped together. Some genes coding for amino acid metabolism proteins were further classified as biosynthetic or degradative based on the metabolic pathways in CauloCyc. Nontransport genes responsible for assimilating sulfate into cysteine were classified in the amino acid metabolism and biosynthesis groups. Uncharacterized genes annotated simply as hypothetical proteins which did not have a COG number were placed in the hypothetical category; other uncharacterized genes were placed in the conserved hypothetical category.
Mapping of hex mutants.
C. crescentus mutants unable to use glucose as their sole carbon source (hex phenotype) were isolated by Ely and colleagues (13, 25). Mutations in eight such strains were mapped by φCr30 phage-mediated transduction, with a set of kanamycin-resistant marker strains (60). Mutations were further localized to single genes by complementation analysis. Candidate sets of genes (e.g., those expected to be involved in sugar metabolism) in the indicated genomic regions were amplified by PCR with AccuTaq Long-read Taq polymerase (Sigma-Aldrich) and the supplied reaction buffer plus 2% dimethyl sulfoxide. The products were cloned into plasmid pCR2.1 with the Topo Cloning System (Invitrogen), then subcloned into broad-host-range plasmid pMR20 in the E. coli S17.1 host. These plasmids were introduced into the hex mutant strains by conjugation, with selection on PYE containing tetracycline and nalidixic acid. Complementation of the hex phenotype was assessed by testing for colony formation on M2G agar plates, in which glucose serves as the sole carbon source.
Construction of mutant strains.
A nonpolar deletion removing nearly half of the coding region of CC2054, encoding glucokinase, was generated through a PCR-based strategy. Flanking primers were designed that annealed approximately 300 bp upstream and downstream of the CC2054 coding region. Complementary chimeric internal primers were designed that fused sequences from the 5′ and 3′ portions of the coding region, deleting codons 114 through 250 (out of 332 total) but maintaining a continuous reading frame. (The sequences of all primers used in this work are available on request.) Each of the chimeric internal primers was used in individual PCRs with the appropriate flanking primer. The products were isolated and combined in a subsequent PCR, with only the flanking primers for amplification.
Fusion of the initial PCR products in the second PCR was mediated by complementarity of the terminal chimeric regions, resulting in a single final product with much of the CC2054 coding region removed. This construct was cloned into plasmid pCR2.1 with the Topo Cloning System (Invitrogen), then subcloned into the kanamycin-resistant pNPTS138 vector for integration into the C. crescentus genome and sacB-mediated sucrose counterselection (60). Sucrose-resistant, kanamycin-sensitive isolates lacking the integrated plasmid were screened by colony PCR to determine whether the wild-type or mutant gene remained on the chromosome. Deletion mutants were streaked on M2G, M2X, and PYE to determine whether CC2054 is required for utilization of glucose as the sole carbon source.
lacZ reporter fusions to confirm microarray data.
The promoter regions of several genes were fused to a lacZ reporter to provide an alternative method of assessing regulation of expression of these promoters during growth on various media. The upstream regions of the CC0505, CC0823, and CC2057 genes were amplified by PCR, generally including approximately 300 bases upstream of the start codon for the indicated genes and up to several hundred bases downstream into the coding regions. PCR products were cloned into plasmid pCR2.1 with the Topo Cloning System (Invitrogen), then subcloned as EcoRI fragments into the pRKlac290 vector to generate a transcriptional fusion to the lacZ gene (56). Orientation in the pRKlac290 vector was verified by PCR with an insert-specific primer and a primer complementary to the coding sequence of the lacZ gene. pRK290 constructs containing the various promoters were delivered by conjugation from E. coli S17.1 into C. crescentus, with selection on PYE containing tetracycline and nalidixic acid. β-Galactosidase activity was assayed in log-phase cells as described previously (56).
Motif identification.
The MEME software (4) was used to identify matrix models describing DNA sequence motifs present upstream of genes whose expression is elevated in M2X or (separately) in M2G. The 475 bases upstream and the 125 bases downstream (600 bases total) of the start of each predicted C. crescentus open reading frame were searched. Bases interior to the predicted coding region were included in the search set to provide robustness against possibly misannotated start codons. Each possible motif location was scored with the motif matrix as well as with a third-order Markov model of C. crescentus intergenic regions (35). A location's score was the log of the probability that the motif was present minus the log of the probability that the sequence came from the background. Both strands of each sequence were searched. The cutoff score for the presence of the motif was taken as the score of the lowest scoring sequence used in generating the motif, except for glucose motif 2, where the second lowest score was used.
In Table 4, when overlapping instances of the xylose motif appeared on both DNA strands, only the highest scoring instance is shown. Both as a control and to estimate the number of false-positives, an equal number of 600-bp sequences were generated from the background distribution model and scored. One random sequence scored above the cutoff for the xylose motif, three were above the cutoff for glucose motif 1, and 22 were above the cutoff for glucose motif 2. Matrices describing the motifs are available in the supplementary material.
TABLE 4.
Occurrence of a putative xylose induction motif upstream of C. crescentus genes
| Gene diverging from intergenic region | Motif sequence | Distance from start codon (bases) |
|---|---|---|
| CC0337 | CTGCATGGGACCGGTAACAA | 197 |
| CC0505c | TAGAATGTTATCGCTACCAT | 24 |
| CC0670 | CGAAATGGTACCGCTAACAT | 60 |
| CC0785 | AGAAATGATAGCGCTATCAA | 211 |
| CC0788c | ATAGCTGGTAGCGCTACCAA | 36 |
| CC0801e | CGCGTTGATAGCGCTAACAA | 68 |
| CC0814a,c | CGCGCTGATACCGGTAACAC | −23 |
| CC0821b | AGACTTCGTACCGCTATATC | 454 |
| CC0823a,c,d | CCACATGTTAGCGCTACCAA | 88 |
| CC0979c/CC0980b | AAAAAAGTGACCGCTACCAT | 102/66 |
| CC0988e | CTTTCTGGTACCGCTACCGC | −3 |
| CC0999c | ACCGATGTTACCGCTAACTA | 130 |
| CC1534c/CC1535c | ACAAATGGTAGCGGTATCTT | 183/100 |
| CC2152a,c | AGAAATGTTACCGCTAACGC | 51 |
| CC2357 | ACAATTGATAGCGCTATCAT | 110 |
| CC2586c | CACGATGTTACCGGTCACAT | 105 |
| CC2620b | CCAATTGGTACCGCTACCAA | 207 |
| CC2622e | CCAATTGGTACCGCTACCAA | 148 |
| CC2790b/CC2791c | CTATATGGGAGCGCTAACAT | 120/50 |
| CC2804c/CC2805c | ATCAATGGTAGCGCTACCTT | 51/426 |
| CC2811c/CC2812c | CCTAATGGTACCGGTACATC | 117/120 |
| CC2832c | CAATCTGGTAGCGCTACCAA | 82 |
| CC3054c | AGAAATGGTACCGGTCTCAT | 114 |
| CC3161a,c | AGGGATGATATCGCTAAGAT | 299 |
| CC3323a,e | CCTCATGGTAGCGGTCTCAC | 79 |
| Multilevel consensus sequence | AAAAATGGTACCGCTACCAT | |
| CCCGTGGATA | ||
| GTC | ||
| T |
Two promoters diverge from the intergenic region containing the motif. Only the gene shown was up in M2X.
No microarray data available.
Gene was up in M2X and met significance criteria.
Gene was up in M2X but did not meet significance criteria.
When searching for the xylose motif upstream of Xanthomonas campestris and Xanthomonas citri genes, background models of X. campestris and X. citri intergenic regions were used. Gene predictions and coordinates for X. campestris and X. citri came from GenBank accession numbers AE008922 and AE008923, respectively (10). Genome sequences were downloaded from GenBank (ftp://ftp.ncbi.nlm.nih.gov/) and processed locally with the Genome-tools software suite (33). Genes were predicted to be in the same transcription unit (operon) if they are next to each other on the same chromosome strand and if their intergenic spacing (predicted stop codon to predicted start codon) is less than or equal to 70 bp.
To test the functionality of the motif associated with xylose-regulated genes, the sequences upstream of the CC0823 and CC0505 genes were altered by site-directed mutagenesis (60). The templates for mutagenesis were the same promoter-containing fragments used to assay regulation of the wild-type promoters. The mutated promoter regions, with identical flanking sequences, were similarly subcloned into the pRKlac290 lacZ transcriptional fusion vectors and introduced into C. crescentus for assay.
RESULTS
Growth of C. crescentus on various carbon sources.
C. crescentus can use diverse carbon sources for growth, including various carbohydrates (48), fatty acids (46), amino acids (15, 48), and aromatic compounds (6). We examined the growth of C. crescentus strain CB15, the strain used to determine the genome sequence, as well as the CB15N (NA1000) derivative used routinely for Caulobacter cell cycle synchronization experiments, on M2 minimal salts agar (12) with various carbon sources. Carbon sources tested included hexoses and hexose derivatives (glucose, fructose, galactose, mannose, glucosamine, gluconate, glucuronic acid, and mannitol), pentoses (xylose, arabinose, and ribose), disaccharides (sucrose, lactose, and trehalose), and other carbon compounds (acetate, glycerol, and glutamate).
Only glucose, xylose, and glutamate produced colonies of at least 1 mm in diameter within 72 h, although colony growth was only slightly slower on sucrose. Trehalose generated 1-mm colonies within 5 days, while galactose, mannose, mannitol, arabinose, and ribose supported growth of 1-mm colonies within 6 to 10 days at 30°C. Growth on M2 medium containing 0.2% fructose, acetate, glycerol, gluconate, glucuronic acid, or glucosamine was barely detectable after 10 days at 30°C. No significant differences were observed between CB15 and CB15N during growth on these carbon sources.
To more quantitatively assess growth rates, CB15N was grown in liquid batch culture on PYE (an amino acid-rich medium typically used for laboratory culture of C. crescentus), M2 with glucose (M2G), and M2 with xylose (M2X). During the exponential growth phase of cultures incubated aerobically at 30°C, CB15N cultures exhibited a doubling time of 89 min on PYE, 114 min on M2G, and 127 min on M2X.
Transcriptional profiling of C. crescentus grown on PYE and minimal media.
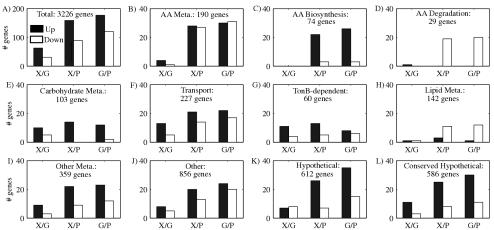
To assess gene expression in cultures grown in PYE, M2G, and M2X, RNA was prepared from log-phase C. crescentus CB15N cultures. RNA samples were fluorescently labeled and competitively hybridized on spotted arrays displaying PCR product probes representing 3,528 predicted C. crescentus genes (93% of the 3,761 open reading frames predicted in the C. crescentus genome) (31, 32). Analysis of the data produced M2X/M2G expression ratios for 3,074 genes, PYE/M2G ratios for 3,179 genes, and M2X/PYE ratios for 3,089 genes. (Due to space constraints, only selected data are presented and discussed here. The entire data set, as well as all tables numbered with an S prefix, are presented in the supplementary materials, which are available online at http://www.stanford.edu/group/caulobacter/metabolism. Additional information on probes, data collection, data processing, and statistical analysis can be found at the same site.) Expression of 390 genes was found to vary significantly and reproducibly. Genes that were found to be differentially expressed between each pair of media are shown by functional categories in Fig. 1.
FIG. 1.
Gene expression changes between M2G, M2X, and PYE media by functional category. Classifications are based on NCBI COG classifications (57, 58) and CauloCyc. Shown are the number of genes found to be significantly increased (dark bars) or decreased (light bars) between each pair of media (X/G, M2X/M2G; X/P, M2X/PYE; G/P, M2G/PYE). The number of genes in each category for which at least one comparison could be made is shown. AA, amino acid. Panel F shows all genes coding for transport proteins, while panel G shows only those genes coding for TonB-dependent receptors. The other metabolic functions category (Other Meta.) shown in panel I includes coenzyme metabolism, energy production and conservation, inorganic ion metabolism, nucleotide metabolism, and secondary metabolite metabolism. In panel J, the other functions category includes cell cycle control, motility, stress responses and defense mechanisms, intracellular trafficking and secretion, posttranslational modification, protein turnover, chaperones, replication, recombination, DNA repair, signal transduction, transcription, and translation.
Sets of genes that appear to be coordinately regulated in each medium were identified, perhaps revealing information as to their function. For example, there were 88 up-in-PYE genes, whose expression is elevated in PYE compared to both M2G and M2X. (Up-in-PYE genes can also be said to be down in M2.) Similarly, we found 119 up-in-M2/down-in-PYE genes, 26 up-in-glucose (M2G) genes, eight down-in-glucose genes, 51 up-in-xylose (M2X) genes, and five down-in-xylose genes. Not surprisingly, given the differences in carbon, nitrogen, sulfur, and phosphate sources, the greatest differences in gene expression were observed between PYE and M2 media, irrespective of the carbon source in the M2 cultures. Examination of those with known or predicted functions revealed some trends. For example, many genes encoding transporters or components of transport systems were differentially regulated in the three media. Also, the expression of genes encoding components of amino acid degradation pathways tended to be elevated in PYE, whereas biosynthetic pathways were frequently elevated in M2 medium (Fig. 1C and D and Fig. 2). These observations are discussed in more detail in the following sections.
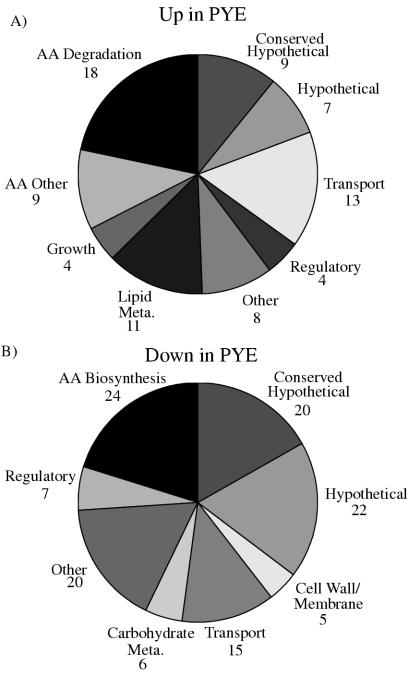
FIG. 2.
Medium-specific expression changes. (A) Function of genes whose expression is increased in PYE compared to both M2G and M2X. The growth category includes several genes whose expression may have increased due to the faster growth rate in PYE than in M2G or M2X. (B) Function of genes whose expression is increased in M2G and M2X compared to PYE. Genes whose products incorporate sulfate into cysteine or ammonia into glutamine or glutamate are in the amino acid (AA) biosynthesis class. Sulfate transporters are in the transport category.
Nutrient transport.
The differential gene expression data suggest that C. crescentus modulates the transporters it synthesizes in response to the growth medium (Fig. 1F and G; Table S1 in supplementary materials). Twenty-five TonB-dependent receptors showed differential regulation in at least one pairwise medium comparison, with the largest coregulated set being the nine specifically induced by xylose (i.e., elevated in M2X relative to both M2G and PYE) (Tables 1, 2, and 3, Table S1). Six ABC transport components were repressed in PYE, including three predicted to be involved in sulfate transport; five were induced in PYE, including components similar to those used to transport cysteine, sugar(s), and phosphonates in other bacteria. No ABC transport components responded to growth on xylose, and only one was induced by glucose.
TABLE 1.
Genes involved in amino acid biosynthesis or degradation that show differential expression between PYE and M2 minimal media
| Pathway or gene | Relative mRNA ratio
|
|
|---|---|---|
| PYE/M2G | PYE/M2X | |
| Alanine degradation | ||
| CC3574, alanine dehydrogenase, ald | 8.64 | 5.32 |
| Arginine degradation | ||
| CC0581, arginine N-succinyltransferase, astA | 3.11 | 3.41 |
| CC1606, arginine N-succinyltransferase, astA | 2.96 | 3.26 |
| CC1607, succinylglutamic semialdehyde dehydrogenase, astD | 2.56 | 2.63 |
| CC1608, succinylarginine dihydrolase, astB | 2.80 | 3.03 |
| Arginine and glutamate degradation | ||
| CC0584, succinylornithine transaminase, putative | 2.89 | 3.50 |
| Aromatic amino acid and histidine synthesis | ||
| CC2300, phospho-2-dehydro-3-deoxyheptonate aldolase, aroG | 0.34 | 0.33 |
| CC2534, histidinol-phosphate aminotransferase, hisC | 3.99 | 3.02 |
| Aromatic amino acid synthesis | ||
| CC1116, chorismate mutase, putative | 1.55 | 1.78 |
| CC2222, chorismate mutase, putative | 0.52 | 0.52 |
| CC2223, histidinol-phosphate aminotransferase, hisC | 0.67 | 0.69 |
| Glutamate degradation | ||
| CC0088, NAD-specific glutamate dehydrogenasea | 3.35 | 3.25 |
| Glycine degradation | ||
| CC3352, glycine cleavage system P protein, subunit 2, gcvP | 3.56 | 4.10 |
| CC3353, glycine cleavage system P protein, subunit 1, gcvP | 4.34 | 5.27 |
| CC3354, glycine cleavage system H protein, gcvH | 4.76 | 5.32 |
| CC3355, glycine cleavage system T protein, gcvT | 4.36 | 5.09 |
| Histidine degradation | ||
| CC0957, urocanate hydratase, hutU | 3.67 | 3.56 |
| CC0958, formiminoglutamase, hutG | 3.90 | 4.22 |
| CC0959, histidine ammonia-lyase, hutH | 3.54 | 4.80 |
| CC0960, imidazolonepropionase, hutI | 2.76 | 2.49 |
| Incorporation of ammonia into glutamate | ||
| CC1969, glutamine synthetase, class I, glnA | 0.36 | 0.52 |
| CC3606, glutamate synthase, small subunit, gltD | 0.39 | 0.33 |
| CC3607, glutamate synthase, large subunit, gltB | 0.25 | 0.21 |
| Isoleucine and valine synthesis | ||
| CC2100, acetolactate synthase, large subunit, ilvB | 0.21 | 0.20 |
| CC2120, ketol-acid reductoisomerase, ilvC | 0.39 | 0.40 |
| Leucine synthesis | ||
| CC0193, 3-isopropylmalate dehydrogenase, leuB | 0.31 | 0.32 |
| CC0195, 3-isopropylmalate dehydratase, small subunit, leuD | 0.28 | 0.32 |
| CC0196, 3-isopropylmalate dehydratase, large subunit, leuC | 0.28 | 0.28 |
| CC1541, 2-isopropylmalate synthase, leuA | 0.23 | 0.21 |
| Methionine synthesis | ||
| CC0050, S-adenosylmethionine synthetase | 0.56 | 0.58 |
| CC0482, 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase, metF | 0.23 | 0.24 |
| CC2138, 5-methyltetrahydrofolate-homocysteine methyltransferase | 0.29 | 0.29 |
| Phenylalanine degradation | ||
| CC2533, 4-hydroxyphenylpyruvate dioxygenase, hpd | 3.82 | 3.22 |
| Proline degradation | ||
| CC0804 proline dehydrogenase/delta-1-pyrroline-5-carboxylate dehydrogenase, putA | 3.74 | 3.78 |
| Serine synthesis | ||
| CC3215, d-3-phosphoglycerate dehydrogenase, serA | 0.23 | 0.22 |
| CC3216, phosphoserine aminotransferase, serC | 0.27 | 0.24 |
| Sulfate acquisition and cysteine synthesis | ||
| CC1119, sulfite reductase (NADPH) hemoprotein, cysI | 0.21 | 0.21 |
| CC1121, phosphoadenylylsulfate reductase, cysH | 0.27 | 0.26 |
| CC1426, cysteine synthase, cysB | 0.69 | 0.63 |
| CC1482, sulfate adenylate transferase, subunit 1/ adenylylsulfate kinase, cysN/C | 0.20 | 0.19 |
| CC1483, sulfate adenylate transferase, subunit 2, cysD | 0.27 | 0.25 |
| CC1596, sulfate ABC transporter, permease protein, cysT | 0.33 | 0.34 |
| CC1597, sulfate ABC transporter, permease protein, cysW | 0.39 | 0.49 |
| CC1598, sulfate ABC transporter, ATP-binding protein, cysA | 0.34 | 0.40 |
| CC3625, cysteine synthase, cysK | 0.39 | 0.46 |
| Threonine synthesis | ||
| CC3399, threonine synthase, thrC | 0.41 | 0.39 |
| Valine degradation | ||
| CC2274, methylmalonate-semialdehyde dehydrogenase, putative | 3.08 | 2.80 |
TABLE 2.
Genes whose expression increased in M2G relative to both M2X and PYE
| Gene or functional category | Relative mRNA ratio
|
|
|---|---|---|
| M2G/M2X | M2G/PYE | |
| Carbohydrate metabolism | ||
| CC2054, glucokinase, glk | 2.73 | 3.27 |
| CC2055, phosphogluconate dehydratase, edd | 2.64 | 4.26 |
| CC2056, 6-phosphogluconolactonase, pgl | 2.50 | 3.78 |
| CC2057, glucose-6-phosphate 1-dehydrogenase, zwf | 2.55 | 3.51 |
| Transport | ||
| CC0028, TonB-dependent receptor | 2.04 | 6.23 |
| CC0214, TonB-dependent receptor | 1.91 | 7.57 |
| CC1103, sugar transporter, fucose permease family | 4.14 | 5.28 |
| CC1517, TonB-dependent receptor | 3.27 | 8.40 |
| CC1518, ABC transporter, ATP-binding protein | 3.84 | 4.69 |
| CC2928, TonB-dependent receptor | 2.11 | 3.41 |
| Other categories | ||
| CC1493, phosphoenolpyruvate carboxylase, ppc | 3.14 | 3.53 |
| CC2053, transcriptional regulator, LacI family | 2.38 | 1.92 |
| CC2226, EF hand domain protein | 1.67 | 1.84 |
| CC2355, sal operon transcriptional repressor, salR | 1.92 | 2.01 |
| CC3062, thiamine biosynthesis protein, apbE | 1.91 | 2.11 |
| CC3086, beta-lactamase, putativea | 1.88 | 6.19 |
| Uncharacterized | ||
| CC0027, CC0669, CC0846, CC1520, CC2160, CC2193, CC3059, CC3060, CC3061, CC3163 | >1.4 | >1.4 |
TABLE 3.
Genes whose expression increased in M2X relative to both M2G and PYE
| Functional category or genef | Relative mRNA ratio
|
|
|---|---|---|
| M2X/M2G | M2X/PYE | |
| Degradation | ||
| CC0788, β-galactosidase, putativea,b,c | 3.46 | 3.66 |
| CC0819, dehydratase, IlvD/Edd familyd | 4.99 | 5.21 |
| CC0822, aldehyde dehydrogenase, aldAd | 11.6 | 11.9 |
| CC0945, oxidoreductase, glucose-methanol-choline family | 1.56 | 2.03 |
| CC1534, aspartate aminotransferase, aspc | 1.51 | 2.77 |
| CC2152, secreted protein, putativec | 8.75 | 9.25 |
| CC2153, polysaccharide deacetylased | 5.39 | 6.36 |
| CC2791, fatty aldehyde dehydrogenasec | 3.14 | 3.56 |
| CC2802, xylosidase/arabinosidase, xylB | 3.78 | 3.86 |
| CC2811, α-glucuronidasea,c | 2.60 | 2.99 |
| CC2812, mandelate racemase/muconate- lactonizing enzyme familyc | 2.58 | 2.75 |
| CC3054, xylosidase/arabinosidase, xarBa,c | 7.11 | 9.32 |
| CC3250, fructose-bisphosphate aldolase, class II, fbaA | 3.03 | 1.65 |
| Transport | ||
| CC0442, TonB-dependent receptora | 2.68 | 3.46 |
| CC0814, major facilitator family transporterc | 3.58 | 6.28 |
| CC0991, TonB-dependent receptora | 1.92 | 1.75 |
| CC0999, TonB-dependent receptor, putativea,c | 4.99 | 5.66 |
| CC1000, peptide transport system permease protein, SapC familye | 2.03 | 1.07 |
| CC2804, TonB-dependent receptora,c | 8.63 | 9.85 |
| CC2832, TonB-dependent receptora,c | 3.84 | 3.73 |
| CC3127, TonB-dependent receptora | 1.59 | 1.80 |
| CC3161, TonB-dependent receptora,c | 3.63 | 4.41 |
| CC3336, TonB-dependent receptor, putative | 2.46 | 3.97 |
| CC3461, TonB-dependent receptor | 3.16 | 4.38 |
| Other | ||
| CC0247, response regulator | 1.66 | 2.33 |
| CC0820, Smp-30/Cgr1 family proteind | 2.81 | 3.36 |
| CC0878, ATP-dependent Clp protease, ATP-binding subunit, clpB | 1.58 | 1.80 |
| CC0979, peptidyl-prolyl cis-trans isomerase A, ppiAa,c | 1.47 | 1.75 |
| CC1294, sensor histidine kinase | 2.17 | 2.23 |
| CC1363, proton pump, putative | 3.46 | 3.61 |
| CC1764, isocitrate lyase, aceA | 2.83 | 3.73 |
| CC1770, ubiquinol oxidase subunit IV, qoxD | 2.91 | 2.60 |
| CC1771, ubiquinol oxidase subunit III, qoxC | 3.92 | 2.57 |
| Uncharacterized or poorly characterized | ||
| CC0127, CC0505,a,c CC0553, CC0823,c CC0925,a CC1535,c CC1536, CC2519, CC2585,d CC2586,c CC2626, CC2645, CC2805,c CC2806,a,d CC2807, CC3055, CC3190, CC3335 | >1.4 | >1.4 |
Predicted to have a signal peptide.
Annotation came from COGs.
Contains a xylose induction motif upstream.
In a putative transcription unit that has a xylose induction motif upstream.
Annotation came from BLASTP.
Annotations are from GenBank (45) unless otherwise noted.
Previous examination of the membrane proteome of C. crescentus found 12 outer membrane proteins (11 of which are TonB-dependent receptors) to be more abundant in cells grown in M2G compared to PYE (47). The microarray data presented here are generally consistent with that analysis, suggesting that differences in transporter transcript levels are positively correlated with differences in the respective protein levels. Genes encoding three of the TonB-dependent receptors identified in the membrane proteome analysis (CC1517, CC0214, and CC0028) showed elevated expression in both M2 media in the microarray analysis, with notably higher expression in M2G than M2X (Table 2). CC1666 showed a fourfold increase in expression in both M2 media relative to PYE. Expression of CC1750 also increased significantly between M2G and PYE, though little change was observed in M2X. There were, however, some discrepancies between the microarray and proteome data. Expression of CC3461 was strongly induced in M2X (Table 3), but the difference in expression in M2G versus PYE was not statistically significant. Expression of CC1915 and CC1970 did not change significantly in M2 media compared to PYE, and expression of CC1099 actually decreased in M2G relative to PYE, contrary to the proteomic data. Microarray data comparing expression in M2G and PYE were not obtained for CC1781, CC2194, and CC3500.
Degradation pathways induced in PYE.
PYE supplies amino acids that can be employed in protein synthesis, used to fuel energy production, or transformed into other necessary metabolites. The expression of a number of genes encoding components of amino acid degradation pathways was elevated in PYE (Fig. 1D and 2A). Particularly notable are components of pathways involved in degrading arginine, histidine, alanine, glycine, glutamate, and proline, which are abundant in peptone and in yeast extract, the main components of PYE (BD Diag-notic Systems, http://www.bd.com/diagnostics/microservices/regulatory/typicalanalysis/index.asp) (Table 1). Arginine catabolism appears to occur via the arginine succinyltransferase pathway, with enzymes from two potential transcription units (CC0581-0584 and CC1606-1608), yielding CO2, NH3, NADH, glutamate, and succinate.
Histidine degradation in C. crescentus (15) has been shown to proceed through the initial steps of the classical Hut (histidine utilization) pathway defined in Aerobacter (Klebsiella) aerogenes (36). That pathway yields glutamate and formamide, but C. crescentus produces formate rather than formamide, suggesting that the terminal steps of the pathway resemble that of Pseudomonas putida (34). The known C. crescentus Hut enzymes are products of a single PYE-induced locus (CC0957-0960). CC0958 is annotated in GenBank as formiminoglutamase, which catalyzes the terminal step of the K. aerogenes Hut pathway, yielding formamide. The annotation may be erroneous in that the CC0958 polypeptide sequence aligns closely (BLAST E value, 3 × 10−48) with the P. putida N-formylglutamate amidohydrolase (GenBank accession number AAB86969), which catalyzes the terminal step of the pathway version yielding formate. Conservation of this gene product in other α-proteobacterial genomes (Agrobacterium tumefaciens, Sinorhizobium meliloti, Mesorhizobium loti, Brucella melitensis, and Rhodospirillum rubrum) suggests that this pathway for histidine utilization is standard for this group.
The induced pathways suggest mechanisms by which C. crescentus generates energy from amino acid degradation during growth in PYE. For example, induction of alanine dehydrogenase (CC3574) allows the conversion of alanine to pyruvate and generates NADH. The pyruvate is presumably fed into the tricarboxylic acid cycle, while NADH feeds respiratory electron transport. Similarly, a bifunctional proline dehyrogenase/pyrroline-carboxylate dehydrogenase (CC0804, homologous to E. coli putA) is also induced, which generates FADH2 and NADH while producing glutamate from proline.
Glutamate can serve as an excellent source of carbon and nitrogen for C. crescentus (13). Expression of CC0088, which encodes a member of a recently identified class of glutamate dehydrogenases (40), increased threefold during growth in PYE relative to M2 media. Glutamate dehydrogenase carries out oxidative deamination of glutamate, yielding α-ketoglutarate, ammonia, and NADH. The activity of the related glutamate dehydrogenase in Streptomyces clavuligerus increases significantly in cultures grown with glutamate as the sole nitrogen and carbon source compared to cultures grown with ammonia and glycerol (40). Ely et al. (13) previously reported that C. crescentus has no detectable glutamate dehydrogenase activity, but their assay used NADP+ as the electron acceptor, whereas Minambres et al. (40) found the S. clavuligerus homolog to be NAD+ specific.
The yeast extract in PYE may contain significant levels of fatty acids, which C. crescentus utilizes as an energy source, based on the observation that growth on PYE induced expression of several genes encoding enzymes in the β-oxidation pathway for fatty acids. These include acyl-coenzyme A dehydrogenase (CC1350: PYE/M2G = 3.55, PYE/M2X = 3.55), enoyl-coenzyme A hydratase/isomerase (CC1352: PYE/M2G = 2.33, PYE/M2X = 2.34; CC2169: PYE/M2G = 3.54, PYE/M2X = 3.65), and acetyl/propionyl-coenzyme A carboxylase (CC2168: PYE/M2G = 2.17, PYE/M2X = 2.10). The fatty acid oxidation genes induced in PYE are a subset of a large family of similarly annotated genes in the C. crescentus genome (Table S2). Whether the induced gene products are functionally distinct from the uninduced ones is unknown, as is the mechanism of their differential regulation.
Assimilation and biosynthesis in minimal media.
During growth in M2 medium, C. crescentus must synthesize each amino acid. As expected, this leads to elevated expression of genes encoding components of several amino acid biosynthetic pathways, including those for leucine, isoleucine, valine, serine, threonine, cysteine, methionine, glutamine, and glutamate (Fig. 2B and Table 1). Interestingly, several pathways showed no apparent difference in expression compared to growth on PYE, and in some only a subset of the genes showed increased expression. For example, among the many enzymes included in the pathways for synthesis of the aromatic amino acids tryptophan, tyrosine, and phenylalanine, only CC2300 (encoding 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase, the first enzyme of the chorismate pathway) and CC2222, encoding chorismate mutase, showed significant induction in M2. The observed lack of induction of other aromatic amino acid biosynthesis genes in M2 is consistent with previous S1 nuclease mapping data (55), which found that trpF (CC3545), trpB (CC3544), and trpA (CC3543) mRNA levels did not change between cells grown on PYE and M2-sucrose. In contrast, microarray studies found that mRNA levels for genes in the E. coli trpEDCBA operon increased two- to fivefold in minimal media compared to the same media supplemented with tryptophan (27).
In M2 medium, sulfate supplies sulfur to the cells, and genes encoding components of the sulfate assimilation pathway are strongly induced relative to PYE (Table 1). These include a sulfate-specific ABC transport system (encoded by CC1596-1598, whose products are homologous to the E. coli CysTWA sulfate transporter), a periplasmic binding protein that probably delivers sulfate to the ABC transport system (CC0286), and the enzymes that incorporate sulfate-derived sulfur into cysteine: sulfate adenylate transferase/adenylylsulfate kinase (CC1482-1483), phospho-adenylylsulfate reductase (CC1121), sulfite reductase (CC1119), and cysteine synthase (CC1426 and CC3625). Expression of CC3063 (cysJ, the α subunit of sulfite reductase) did not change significantly.
During growth in M2, ammonia serves as the sole nitrogen source. Ely et al. (13) showed that ammonia assimilation in C. crescentus occurs via the combined action of glutamine synthetase and glutamate synthase. They observed fivefold induction of glutamate synthase activity and fourfold induction of glutamine synthetase activity in M2G versus PYE medium. Similarly, our microarray data show elevated expression in M2 media (compared to PYE) for the genes encoding glutamine synthetase (CC1969: M2G/PYE = 2.78, M2X/PYE = 1.93) and glutamate synthase subunits (CC3606: M2G/PYE = 2.54, M2X/PYE = 3.07; CC3607: M2G/PYE = 4.06, M2X/PYE = 4.75).
Very few genes encoding enzymes involved in nucleotide biosynthesis showed significant changes in expression in M2 versus PYE. Some enzymes involved in the synthesis of other relevant metabolites and cofactors, however, showed significant induction in M2. De novo purine synthesis and some reactions in amino acid synthesis depend on formyl-tetrahydrofolate as a donor of one-carbon units. The formyl-tetrahydrofolate synthesis pathway in C. crescentus has not been clearly elucidated, but expression of genes encoding several enzymes that could be relevant, including CC2138 (5-methyl-tetrahydrofolate-homocysteine methyltransferase), CC2140(5,10-methylenetetrahydrofolate reductase), and CC3630(formyltetrahydrofolate deformylase), is induced during growth in M2 medium. With respect to one-carbon unit metabolism, expression of S-adenosylmethionine synthetase (CC0050), which produces the key methyl donor S-adenosylmethionine, was also increased in M2.
Glucose catabolism.
C. crescentus expresses enzymes of the Entner-Doudoroff pathway during growth on glucose, and it has been proposed that this is the primary route of glucose catabolism in this organism (52). Our microarray results show that the expression of CC1495, which codes for 2-dehydro-3-deoxyphosphogluconate aldolase (commonly referred to as KDPG aldolase), which carries out the last step in the Entner-Doudoroff pathway, was increased in both M2G and M2X compared to PYE but did not vary significantly between M2G and M2X. The microarray data also show that the genes encoding the first four enzymes in the Entner-Doudoroff pathway (CC2054 to CC2057) are more highly expressed in M2G than in either M2X or PYE, although there is some elevation of expression in M2X relative to PYE.
The CC2057 to CC2054 coding regions are closely spaced (3, 7, and 15 bp of separation, respectively) and probably form an operon. To independently verify the regulation of this operon, the CC2057 promoter region (including 287 bases upstream of the predicted start codon) and coding sequence were cloned into a low-copy-number lacZ fusion vector. Expression of the CC2057 promoter fusion in strain CB15 grown in M2G (1,460 ± 445 Miller units) and in PYE supplemented with glucose (1,460 ± 430 units) was higher than in M2X (917 ± 90 units) or PYE (693 ± 185 units). The glucose-dependent increase in expression was lower than observed in the microarray experiments (Table 2) but qualitatively consistent.
C. crescentus hex (hexose) mutant strains that are unable to use glucose as their sole carbon source have been isolated (25). We examined eight hex mutant strains to further characterize the phenotype. All of the strains grew normally on PYE and M2X agar plates. Most of these strains appeared completely incapable of growth on M2G, the exceptions being two strains that formed colonies very slowly on M2G (see below). To identify the basis for this inability to utilize glucose, the hex mutations were mapped by transductional linkage to kanamycin resistance markers (60). The specific genes in which the mutations resided were then identified by complementation, based on restoration of the ability to grow on M2G. Three strains (SC333, SC337, and SC414) contained mutations that mapped to CC2055, a homolog of the E. coli edd gene encoding phosphogluconate dehydratase, which catalyzes the Entner-Doudoroff pathway reaction in which phosphogluconate is converted to 2-keto-3-deoxy-6-phosphogluconate. Another mutation (SC144) was mapped to CC1495, a homolog of the E. coli eda gene encoding KDPG aldolase, which cleaves 2-keto-3-deoxy-6-phosphogluconate to yield glyceraldehyde-3-phosphate and pyruvate.
Four other hex mutant strains were examined, including two (SC114 and SC481) that showed no growth on M2 with glucose or galactose as the carbon source and two (SC155 and SC162) that grew very slowly on glucose or galactose. All four mutations mapped to CC1493 (ppc), which encodes phosphoenolpyruvate carboxylase, an enzyme that catalyzes the addition of CO2 to phosphoenolpyruvate to generate oxaloacetic acid. Expression of CC1493 was elevated in M2G compared to both M2X and PYE. During growth on minimal media such as M2G, tricarboxylic acid cycle intermediates are siphoned off for use in various biosynthetic pathways. In order for the cycle to continue, oxaloacetate levels must be replenished (17). The difficulty experienced by ppc mutants in growing on M2 with glucose or galactose as the carbon source demonstrates the critical role of this replenishment process. We suspect that the mutations in SC155 and SC162 compromise the function of the phosphoenolpyruvate carboxylase, reducing (but not eliminating) oxaloacetate production.
Entry of glucose into the Entner-Doudoroff pathway typically requires the sugar to be phosphorylated, though exceptions in some archaea have been noted (9). In E. coli, glucose is phosphorylated as it enters the cell through a phosphoenolpyruvate-dependent phosphotransferase transporter system (50). Interestingly, though the C. crescentus genome encodes several putative phosphotransferase transporter system-type transport systems, none were induced n response to glucose in our data. However, the putative operon encoding several other components of the Entner-Doudoroff pathway also includes a glucokinase homolog (CC2054), which could account for production of glucose-6-phosphate. Since no mutations with the hex phenotype were identified in this gene, a knockout of CC2054 was constructed. The mutant strain showed no detectable growth with glucose as the sole carbon source, a phenotype consistent with its being required for phosphorylation of glucose prior to entry into the Entner-Doudoroff pathway. This suggests that in C. crescentus, glucose is not transported through or phosphorylated by a phosphotransferase transporter system-type system. Based on the microarray data, one candidate for transporting glucose across the inner membrane is encoded by CC1103, whose product exhibits significant similarity to the E. coli fucose permease.
The MEME software (4) was used to identify conserved DNA motifs associated with glucose-induced genes. Two palindromic motifs were found, associated with nonoverlapping sets of genes (Tables S3 and S4, supplementary materials). The first motif appears in 13 intergenic regions in the C. crescentus genome (Table S3). Genes with the first motif upstream include CC2057 (the first gene of the operon encoding enzymes of the Entner-Doudoroff pathway), four genes encoding transporters (CC0970, CC1103, CC1500, and CC1754), two genes coding for 1,4-β-d-glucan glucohydrolase homologs (CC0797 and CC2052), and a gene for a putative glucokinase family protein (CC3167). The second motif, albeit less statistically significant (see above), appears in 14 intergenic regions, including probable promoter regions for 11 genes showing elevated expression in glucose (Table S4). These include CC1493 (encoding phosphoenolpyruvate carboxylase) and four genes encoding transporters (CC0214, CC1517, CC1518, and CC2928). Though intriguing, the significance of these two motifs with respect to expression and regulation of these genes remains to be investigated.
Xylose regulon.
A substantial fraction of the genes whose expression increased in xylose encode enzymes predicted to contribute to the degradation of extracellular polymers (Table 3). This class includes two xylosidase/arabinosidases (CC2802, CC3054), an α-glucouronidase (CC2811), a putative β-galactosidase (CC0788), a putative polysaccharide deacetylase (CC2153), and a gene annotated as encoding a secreted protein (CC2152). Expression of CC0989 and CC2357, which are annotated as encoding xylosidases, did not increase significantly in M2X in the microarray data. Expression of a fructose-bisphosphate aldolase (CC3250), which could act in degradation or in gluconeogenesis, was also increased in xylose.
With SignalP (43, 44), four additional nontransport, xylose-induced genes encoding proteins with probable signal peptides for export were identified. Based on homology, probable functions could be identified for only one of these (CC0979, peptidyl-prolyl cis-trans isomerase A). Another one (CC0925) is annotated as an OmpA-related protein, suggesting that it may be targeted to the outer membrane. Also targeted to the outer membrane are a set of nine TonB-dependent receptors whose expression is induced by xylose. As noted previously, these proteins could support transport across the outer membrane. Xylose also induced expression of a gene (CC0814) encoding a member of the major facilitator superfamily of inner membrane proteins. The CC0814 product is strikingly similar to the E. coli XylE xylose/H+ symporter (BLASTP E value, 2 × 10−70), suggesting a role in xylose import into the cytoplasm. Given that E. coli, as an example, utilizes at least one other xylose transport system in addition to XylE, it is possible that other inner membrane transporters aid in acquiring xylose in C. crescentus as well.
The CC0823 through CC0819 genes probably form an operon, based on the close spacing of the coding regions and the coordinated induction by xylose. The extent of induction observed for these genes ranged from a low of 2.8-fold (M2X/M2G) for CC0820 to a high of 11.6-fold for CC0822. (No expression data were obtained for CC0821.) A chromosomal Tn5-lacZ insertion previously isolated in the CC0823 coding region showed strong induction of β-galactosidase activity by xylose (≈40-fold upon addition of xylose to PYE) (39). Plasmid-borne transcriptional fusions of CC0823 to lacZ displayed 10-fold-higher activity in M2X than in M2G (39), similar to the sevenfold induction observed for CC0823 mRNA in M2X versus M2G in the microarray data. The Tn5-lacZ insertion in CC0823 prevented growth with xylose as the sole carbon source (39), implying that CC0823 or a downstream gene in this operon is critical for metabolism of xylose.
Regions upstream of the genes in the C. crescentus xylose regulon were scanned with MEME (4) to identify conserved sequences potentially representing transcription factor binding sites. A conserved 20-bp palindromic motif strongly associated with xylose-regulated genes was identified (Table 4). The motif appeared 25 times in intergenic regions in the C. crescentus genome. Fifteen of these sites were adjacent to at least one gene whose expression is significantly increased in xylose (Table 4), including CC0823, where it overlapped the predicted −10 region of the promoter (Fig. 3). Transcription factors controlling gene expression in response to xylose have been identified in several bacterial species, and binding sites for such factors are sometimes conserved between species (38). However, the motif identified here differs substantially from the consensus binding site of the XylR repressor found in the Bacillus/Clostridium group (53), and it does not resemble the recognition sequence for the XylR activator of some γ-proteobacteria (30).
FIG. 3.
Mutational analysis of a conserved motif associated with xylose-induced genes. The wild-type sequences of the regions upstream of the CC0823 and CC0505 genes are shown above the tabulated data. The italicized segments of each wild-type promoter were altered by site-directed mutagenesis to the sequences shown above the wild-type sequences. The numbering refers specifically to the CC0823 promoter (39). The portions of the sequences most similar to the motif associated with other xylose-regulated genes (Table 4) are underlined. β-Galactosidase activities are for CB15N cultures containing plasmid-borne transcriptional fusions of each promoter to lacZ (see text). Cultures were grown to mid-log phase in the indicated medium for each assay. Background β-galactosidase activity attributable to the vector has been subtracted. At least three replicates of each culture were assayed to obtain the averages and standard deviations shown.
The only xylose-regulated gene whose transcription start site has been identified is CC0823 (39). Overlap of the conserved motif with the −10 region (Fig. 3) of this promoter is consistent with a repressor binding site designed to inhibit transcription initiation by interfering with RNA polymerase binding. It should be noted that this motif is distinct from the dyad symmetric element suggested by Meisenzahl et al. (39) as a potential regulatory site for the CC0823 promoter.
To experimentally examine the role of this motif in xylose-dependent regulation, the motifs associated with CC0823 and CC0505 were mutationally altered (Fig. 3). CC0505 encodes a signal peptide-containing protein, possibly secreted, whose function is unknown. CC0505 is not necessary for xylose utilization (data not shown), but its expression showed substantial xylose induction in microarray experiments (M2X/M2G = 5.8, M2X/PYE = 6.2). Wild-type and mutant promoters were fused to the lacZ reporter gene on a low-copy-number plasmid, and β-galactosidase expression was assayed in the same media used in the microarray experiments as well as PYE supplemented with xylose (Fig. 3). There were clear differences between the two promoters—the CC0823 promoter yielded notably higher expression under all conditions—but the consequences of altering the conserved motif were similar. In both cases, the mutant promoters showed strong constitutive expression in media lacking xylose. These data support a model in which the conserved motif functions as a binding site for a repressor protein that obstructs these promoters in the absence of xylose and releases in the presence of xylose, allowing access to RNA polymerase. Definition of this regulatory motif is particularly significant in that the CC0823 promoter is widely used as a heterologous promoter directing inducible gene expression in C. crescentus (for example, see references 11 and 39); defining the target site for regulation should aid in identifying the regulator itself and in manipulating the system to improve its utility.
Possible regulatory proteins.
A number of putative transcription factor or proteins involved in signal transduction pathways were differentially expressed in the media examined here. Regulators induced in M2 media relative to PYE included a homolog of ArsR (CC2141: M2G/PYE = 2.23, M2X/PYE = 2.15), a response regulator (CC3477: M2G/PYE = 2.81, M2X/PYE = 2.40), a histidine kinase (CC0285: M2G/PYE = 1.88, M2X/PYE = 1.82) a hybrid histidine kinase/response regulator (CC0026: M2G/PYE = 2.09, M2X/PYE = 1.99), a LuxR-related protein (CC0782: M2G/PYE = 2.53, M2X/PYE = 3.08), and extracytoplasmic function sigma factors CC2883 (M2G/PYE = 2.38, M2X/PYE = 2.07) and CC3475 (M2G/PYE = 2.03, M2X/PYE = 1.89). Two putative transcription factors, both members of the LacI family (CC2053 and CC2355), appeared to be specifically induced by glucose (expression significantly greater on M2G than M2X or PYE, Table 2). Expression of a response regulator (CC0247) and a sensor histidine kinase (CC1294) was induced during growth on xylose (Table 3). A few potential transcriptional regulators showed relatively minor (less than twofold) but statistically significant increases in expression in PYE relative to the M2 media, including a homolog of the PupR anti-sigma factor (CC0982), a response regulator (CC1610), a putative cyclic AMP binding protein (CC2171), and an AsnC family member (CC3573) (see supplementary materials).
Cell cycle regulation of metabolic genes.
The expression of several C. crescentus genes associated with routine metabolic functions has been shown to be cell cycle regulated during growth in M2G medium (32). A subset of those cell cycle-regulated genes were differentially regulated in the growth media tested here (supplementary material). Twelve cell cycle-regulated genes were up and one was down in xylose, two were down and six were up in glucose, and 37 were down and 21 were up in PYE. The genes regulated by both the growth media and the cell cycle did not seem to have a common time of peak expression during the cell cycle. Cell cycle-regulated genes could appear to be medium regulated in microarray experiments with mixed cultures if the cultures grown in various media had different proportions of swarmer and stalk cells, but we saw no obvious evidence for that to be the case here.
The faster growth rate of C. crescentus in PYE compared to the M2 media likely explains why some cell cycle-regulated genes, including dnaA (CC0008) and recA (CC1087), appear to be increased in PYE. Some of the cell cycle-regulated genes whose expression was elevated in PYE, including phnC, phnD, and phnE (CC0361 to CC0363), are involved in transport, while others, such as gcvT (CC3335) and gcvH (CC3334), are involved in degradation. Conversely, many of the cell cycle-regulated genes whose expression was down in PYE are involved in biosynthesis. For promoters regulated by both the cell cycle and nutrient composition, it remains to be determined whether the same transcription factors are used in both forms of regulation.
DISCUSSION
Past studies of bacterial metabolism have primarily focused on cells adapted to high nutrient conditions. In this work, in contrast, we analyzed the metabolism of an oligotrophic organism, Caulobacter crescentus. Oligotrophs thrive where nutrients are perpetually scarce, the usual situation in most aquatic and terrestrial habitats on earth. An improved understanding of oligotrophs' metabolic activities is essential to understanding biogeochemical processes that have major impact on the environment (20). We describe here a comparison of gene expression during growth of C. crescentus on common laboratory media. These data provide insights into C. crescentus metabolic pathways and regulatory strategies and establish a baseline for future experiments with these media. We assessed expression patterns for over 80% of the known genes in the C. crescentus genome in each pairwise medium comparison and found that over 12% of the genes examined showed significant differences in expression in at least one medium comparison. Not surprisingly, most of the differences occurred between the complex PYE medium and the M2 minimal salts media regardless of the carbon source used in the M2 medium. Fewer differences were seen between the two M2 media, but the differences that were observed may identify transporters, enzymes, and regulators specifically relevant to glucose and xylose utilization.
The Entner-Doudoroff pathway has been shown to be active in C. crescentus during growth on glucose (48, 52). In the data presented here, all of the genes encoding enzymes of the Entner-Doudoroff pathway were found to be significantly induced during growth on glucose-containing media compared to PYE. Mutations in three genes (CC1495, CC2054, and CC2055) encoding enzymes of the Entner-Doudoroff pathway rendered C. crescentus incapable of growth with glucose as the sole carbon source. The Entner-Doudoroff pathway is thus essential for glucose utilization in C. crescentus, a situation previously seen only in Pseudomonas aeruginosa (5) and Burkholderia cepacia (1). In contrast, in several free-living α-proteobacteria in which glucose metabolism has been examined, both the Embden-Meyerhof-Parnas and Entner-Doudoroff pathways are used (3, 8, 42).
The apparent absence of the enzyme phosphofructokinase in C. crescentus would preclude use of the upper branch of the Embden-Meyerhof-Parnas pathway. Thus, instead of generating glyceraldehyde-3-phosphate and dihydroxyacetone phosphate, glucose is processed into glyceraldehyde-3-phosphate and pyruvate. There appears to be little transcriptional regulation of the genes encoding enzymes for the latter steps of glycolysis (taking glyceraldehyde-3-phosphate to pyruvate), as mRNA levels showed minimal differences between media. In contrast, in E. coli, expression of all genes in the lower branch of the Embden-Meyerhof-Parnas pathway except pykA and pykF (pyruvate kinases) increases during fermentation on glucose compared to fermentation on xylose (16). The E. coli gene expression data agree with metabolic flux estimates in that all steps in the lower part of the glycolytic pathway are expected to have higher flux during fermentation of glucose compared to fermentation of xylose except for the conversion of phosphoenolpyruvate to pyruvate (16). Whether similar expression of glycolytic genes during growth of C. crescentus on xylose and glucose likewise reflects comparable carbon flux through the lower part of the glycolytic pathway during growth on glucose and xylose is uncertain, given that the route of xylose catabolism is unknown.
There are many genes whose products have the same predicted enzymatic function in the current C. crescentus genome annotation but which exhibited distinct expression patterns in the microarray data (Table S2). Some differentially regulated isozymes may participate in distinct pathways that are differentially induced depending on the medium. For example, in an oligotrophic Spirillum sp., the relative activities of three forms of lactate dehydrogenase vary as a function of the lactate concentration in the growth medium (37). The exact roles of many of the C. crescentus isozymes remain to be determined. In such cases, differential regulation may provide clues to distinct functions.
One example is particularly relevant to glucose catabolism. Two genes (CC0784 and CC1495) are annotated as encoding KDPG aldolase, which catalyzes the final step in the Entner-Doudoroff pathway. Expression of CC1495 is elevated in M2 minimal media with either xylose or glucose as the carbon source compared to PYE, whereas expression of CC0784 is unchanged. Subsequent mutational results demonstrated that CC1495 is essential for growth on glucose, implying that CC0784 (assuming that it is expressed) is not capable of functionally substituting for CC1495 in glucose catabolism. On the other hand, the CC1495 mutant strains could still form colonies with galactose as the carbon source. The CC0784 polypeptide sequence resembles gene products annotated as 2-dehydro-3-deoxyphosphogalactonate aldolases from several organisms, including Ralstonia solanacearum, Bradyrhizobium japonicum, and Brucella species; this resemblance is considerably stronger than that shown by the CC1495 polypeptide. It has been reported previously that C. crescentus may use some version of the Entner-Doudoroff pathway for galactose catabolism (29). Our results suggest that the CC0784 product, rather than participating in the glucose Entner-Doudoroff pathway, may be involved in a version of the Entner-Doudoroff pathway dedicated to galactose.
The pathway of xylose degradation in C. crescentus may be distinct from known pathways in other microbes, as genes encoding key enzymes for those pathways (particularly xylose isomerase and xylulose kinase) are not apparent in the C. crescentus genome. An inducible xylose dehydrogenase activity has been demonstrated in C. crescentus, which may serve as the initial step in xylose catabolism (48). The only known C. crescentus mutant strain that is unable to grow on xylose resulted from a Tn5-lacZ insertion in the xylX (CC0823) gene (39). This insertion showed strongly xylose-inducible β-galactosidase activity. Microarray data presented here confirm xylose induction of CC0823, which turns out to be the first gene in a potential five-gene operon (CC0823 to CC0819), the largest transcription unit whose expression responds to xylose.
The CC0823 product does not resemble any gene of known function, but it is homologous to genes in other genomes, including Mesorhizobium loti, Streptomyces coelicolor, Agrobacterium tumefaciens, Pseudomonas putida, Bradyrhizobium japonicum, and Sinorhizobium meliloti. The xyl mutant phenotype of the Tn5 insertion could reflect a critical role for the CC0823 product in xylose metabolism (39); alternatively, the xyl mutant phenotype could result from polar effects of the insertion on expression of critical downstream genes in the operon. Two putative dehydrogenases (CC0822 and CC0821) of unknown substrate specificity are encoded in this operon, either of which could potentially be responsible for inducible xylose dehydrogenase activity.
The ability to replenish intermediates of the tricarboxylic acid cycle can be critical for growth of microbes on a single carbon source (17). Under such conditions, tricarboxylic acid cycle intermediates are continually siphoned off for various biosynthetic processes, compromising continued flux through the pathway. Expression data and genetic analysis suggest that carbon flux through the tricarboxylic acid cycle is managed differently during growth on glucose and xylose. The expression of phosphoenolpyruvate carboxylase is induced during growth on glucose but not xylose, and genetic evidence is presented above that supplementation of the tricarboxylic acid cycle with oxalacetate via phosphoenolpyruvate carboxylase is critical for growth on glucose but not xylose.
In contrast, expression of isocitrate lyase (CC1764) is specifically elevated during growth on xylose. Isocitrate lyase catalyzes the conversion of isocitrate to glyoxylate and succinate, initiating the glyoxylate bypass. The succinate generated by this process can be drawn off for other biosynthetic pathways. Glyoxylate, meanwhile, can be combined with acetyl-coenzyme A by malate synthase to produce malate, allowing a modified tricarboxylic acid cycle to continue. Interestingly, expression of malate synthase, encoded by the gene (CC1765) adjacent to that encoding isocitrate lyase, was not observed to increase coordinately in response to xylose in the microarrays. Future studies are warranted to examine the role of the glyoxylate bypass during growth on xylose and other carbon sources.
The ability to simultaneously import a variety of growth substrates could be critical for survival in oligotrophic environments (21). As such, it seems unlikely that all of the putative transporters and exoenzymes whose expression is induced by xylose are utilized solely, or are even necessary, for the import and utilization of this sugar. The CC0505 gene, for example, is strongly induced by xylose, but a CC0505 knockout strain shows no defects in growth on xylose (data not shown). We hypothesize that C. crescentus uses xylose as an indicator that metabolites derived from the breakdown of plant cell walls are available. Other microorganisms, particularly soil microbes such as Streptomyces species, are known to use small molecules resulting from extracellular polymer breakdown as a positive feedback signal to induce degradative enzymes (22). Similarly, growth of Thermotoga maritima on xylose induces genes for two xylanases and an α-glucosidase, and growth of T. maritima on xylan additionally increases expression of genes for an α-glucuronidase and a β-d-galactosidase (7).
Since xylose in natural habitats would presumably be acquired by breakdown of xylan, xylose may be used to trigger the expression of extracellular hydrolytic enzymes relevant to breakdown of lignicellulose (of which xylan is a major constituent) as well as the enzymes needed for the catabolism of xylose itself, as in the fungus Aspergillus niger (18). Environments rich in xylan should contain cellulose as well; interestingly, however, xylose does not induce expression of endoglucanases or β-glucosidases that could (at least theoretically) be used by C. crescentus for cellulose degradation.
The abundance of TonB-dependent receptors induced by xylose could provide cells with the ability to import products of extracellular lignocellulose degradation, but it remains to be determined what metabolites each transporter is capable of bringing into the periplasm. In an effort to further characterize the transporters induced by xylose, we used BLASTP to compare xylose-induced C. crescentus transporters to the NCBI database. Somewhat surprisingly, many of the strongest homologies were to proteins encoded by Xanthomonas campestris, a plant pathogen that secretes exoenzymes to attack plant cell walls. The virulence of at least one Xanthomonas species (X. oryzae, which causes rice blast disease) is dependent on the ability to secrete xylanase (51). The X. campestris genome contains bidirectional best hits to four of the xylose-induced TonB-dependent receptors (CC0442, CC0999, CC2832, and CC3336) as well as the putative inner membrane xylose transporter (CC0814) and the SapC peptide permease homolog (CC1000).
Although Caulobacter and Xanthomonas are not close phylogenetically, their genomic contents bear intriguing similarities. In addition to having sets of putative exoenzymes similar to C. crescentus, the sequenced genomes of the Xanthomonas species X. campestris and X. citri also display an abundance (nearly 50) of TonB-dependent receptors (10). There are also hints that regulation by xylose may be similar in X. campestris and C. crescentus. The X. campestris genome has several close matches to the DNA sequence motif associated with xylose regulation in C. crescentus (Table 4), with three of the four strongest instances of this motif in the X. campestris genome located upstream of putative transcription units containing xylose-associated genes (xylose isomerase, xylosidase, and xylanase) (Table S5). Thus, although C. crescentus has not been reported to act as a plant pathogen, it may have mechanisms for acquiring plant polymer-derived metabolites similar to those found in Xanthomonas species.
Environmentally responsive signal transduction pathways in C. crescentus have not been extensively explored. The collection of large microarray data sets, as presented here, provides some initial insight into regulatory responses to nutrient levels in this organism. A set of genes whose expression responds to the availability of xylose have been identified, along with a cis-acting site that appears to represent the target for a xylose-responsive repressor. Two potential regulatory sites have also been identified for glucose-responsive genes; these need to be tested, and the trans-acting factors controlling xylose and glucose regulation will be pursued in future work. Coregulation of many genes and potential operons encoding amino acid synthesis or degradation functions has been observed.
Future work will continue to characterize the traits that allow C. crescentus to be a successful oligotroph by exploring this organism's metabolism in more dilute media, in mixtures of substrates, and in continuous cultures. Finally, the stage is set for exploring the interface between environmental and cell cycle regulation. What environmental cues influence cell cycle progression? Are there cell cycle checkpoints that determine if a cell has sufficient resources to proceed with division? How are individual promoters influenced by both the environmental and cell cycle state? As just one example, expression of the glycine cleavage system (gcv) genes (CC3355 to CC3352) is strongly induced in PYE medium versus M2, presumably due to relatively high levels of exogenous glycine. During the cell cycle, expression of the gcv genes peaks in swarmer cells and then is repressed dramatically in predivisional cells (32). Is the same transcription factor used in both forms of regulation? What controls cell cycle regulation of this promoter, and why? Further application of molecular and classical genetic approaches with microarray analysis can provide insight into such questions.
Acknowledgments
A.K.H., M.M., P.R., and H.H.M. were supported by grant DE-FG03-01ER63219-A001 from the Department of Energy. A.K.H., M.M., and H.H.M. were supported by grant MDA972-00-1-0032-P00005 from the Defense Advanced Research Projects Agency Defense Sciences Office. A.K.H. was also supported by NIH HG00044 from the National Institutes of Health. P.R. was also supported by grant N66001-01-C-8011 from the Defense Advanced Research Projects Agency. The work of C.S., D.Y., and N.A. was supported by a grant from Santa Clara University and by the Community of Science Scholars Initiative program at SCU, made possible by a grant from the Howard Hughes Medical Institute to support undergraduate research. C.S. is also supported by grant MCB-0317037 from the National Science Foundation.
We thank Bert Ely (University of South Carolina), who kindly provided us with several hex mutant strains for analysis, and Lisandra West, for support in generating the microarray data set.
REFERENCES
- 1.Allenza, P., and T. G. Lessie. 1982. Pseudomonas cepacia mutants blocked in the Entner-Doudoroff pathway. J. Bacteriol. 150:1340-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, L. O., L. K. Nakamura, G. Julian, and L. A. Bulla, Jr. 1975. Carbohydrate catabolism of selected strains in the genus Agrobacterium. Appl. Microbiol. 30:731-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 5.Blevins, W. T., T. W. Feary, and P. V. Phibbs, Jr. 1975. 6-Phosphogluconate dehydratase deficiency in pleiotropic carbohydrate-negative mutant strains of Pseudomonas aeruginosa. J. Bacteriol. 121:942-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee, D. K., and A. W. Bourquin. 1987. Metabolism of aromatic compounds by Caulobacter crescentus. J. Bacteriol. 169:1993-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chhabra, S. R., K. R. Shockley, S. B. Conners, K. L. Scott, R. D. Wolfinger, and R. M. Kelly. 2003. Carbohydrate-induced differential gene expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J. Biol. Chem. 278:7540-7552. [DOI] [PubMed] [Google Scholar]
- 8.Conrad, R., and H. G. Schlegel. 1977. Different degradation pathways for glucose and fructose in Rhodopseudomonas capsulata. Arch. Microbiol. 112:39-48. [DOI] [PubMed] [Google Scholar]
- 9.Dandekar, T., S. Schuster, B. Snel, M. Huynen, and P. Bork. 1999. Pathway alignment: application to the comparative analysis of glycolytic enzymes. Biochem. J. 343:115-124. [PMC free article] [PubMed] [Google Scholar]
- 10.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 11.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415-424. [DOI] [PubMed] [Google Scholar]
- 12.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 13.Ely, B., A. B. Amarasinghe, and R. A. Bender. 1978. Ammonia assimilation and glutamate formation in Caulobacter crescentus. J. Bacteriol. 133:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Entner, N., and M. Duodoroff. 1952. Glucose and gluconic acid oxidation of Pseudomonas saccharophila. J. Biol. Chem. 196:853-862. [PubMed] [Google Scholar]
- 15.Ferber, D. M., F. Khambaty, and B. Ely. 1988. Utilization of histidine by Caulobacter crescentus. J. Gen. Microbiol. 134:2149-2154. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez, R., H. Tao, K. T. Shanmugam, S. W. York, and L. O. Ingram. 2002. Global gene expression differences associated with changes in glycolytic flux and growth rate in Escherichia coli during the fermentation of glucose and xylose. Biotechnol. Prog. 18:6-20. [DOI] [PubMed] [Google Scholar]
- 17.Gottschalk, G. 1978. Bacterial metabolism. Springer-Verlag, New York, N.Y.
- 18.Hasper, A. A., J. Visser, and L. H. de Graaff. 2000. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol. Microbiol. 36:193-200. [DOI] [PubMed] [Google Scholar]
- 19.Henrici, A. T., and D. E. Johnson. 1935. Studies on freshwater bacteria. II. Stalked bacteria, a new order of schizomycetes. J. Bacteriol. 30:61-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch, P. 1986. Microbial life at extremely low nutrient levels. Adv. Space Res. 6:287-298. [DOI] [PubMed] [Google Scholar]
- 21.Hirsch, P., M. Bernhard, S. S. Cohen, J. C. Ensign, H. W. Jannasch, A. L. Koch, K. C. Marshall, A. Matin, J. S. Poindexter, S. C. Rittenberg, D. C. Smith, and H. Veldkamp. 1979. Life under conditions of low nutrient concentrations, group report, p. 357-372. In M. Shilo (ed.), Strategies of microbial life in extreme environments. Dahlem Konferenzen, Berlin, Germany.
- 22.Hodgson, D. A. 2000. Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv. Microb. Physiol. 42:47-238. [DOI] [PubMed] [Google Scholar]
- 23.Jackson, S., and S. W. Nicolson. 2002. Xylose as a nectar sugar: from biochemistry to ecology. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 131:613-620. [DOI] [PubMed] [Google Scholar]
- 24.Jenal, U., and C. Stephens. 2002. The Caulobacter cell cycle: timing, spatial organization and checkpoints. Curr. Opin. Microbiol. 5:558-563. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, R. C., and B. Ely. 1977. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics 86:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karp, P. D., S. Paley, and P. Romero. 2002. The Pathway Tools software. Bioinformatics 18(Suppl. 1):S225-S232. [DOI] [PubMed] [Google Scholar]
- 27.Khodursky, A. B., B. J. Peter, N. R. Cozzarelli, D. Botstein, P. O. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:12170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 29.Kurn, N., I. Contreras, and L. Shapiro. 1978. Galactose catabolism in Caulobacter crescentus. J. Bacteriol. 135:517-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laikova, O. N., A. A. Mironov, and M. S. Gelfand. 2001. Computational analysis of the transcriptional regulation of pentose utilization systems in the gamma subdivision of Proteobacteria. FEMS Microbiol. Lett. 205:315-322. [DOI] [PubMed] [Google Scholar]
- 31.Laub, M. T., S. L. Chen, L. Shapiro, and H. H. McAdams. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA 99:4632-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 33.Lee, W., and S. L. Chen. 2002. Genome-tools: a flexible package for genome sequence analysis. BioTechniques 33:1334-1341. [DOI] [PubMed] [Google Scholar]
- 34.Leidigh, B. J., and M. L. Wheelis. 1973. Genetic control of the histidine dissimilatory pathway in Pseudomonas putida. Mol. Gen. Genet. 120:201-210. [DOI] [PubMed] [Google Scholar]
- 35.Liu, X., D. L. Brutlag, and J. S. Liu. 2001. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 127-138. [PubMed]
- 36.Magasanik, B., and H. R. Bowser. 1955. The degradation of histidine by Aerobacter aerogenes. J. Biol. Chem. 213:517-580. [PubMed] [Google Scholar]
- 37.Matin, A. 1979. Microbial regulatory mechanisms at low nutrient concentrations as studied in chemostat, p. 323-339. In M. Shilo (ed.), Strategies of microbial life in extreme environments. Dahlem Konferenzen, Berlin, Germany.
- 38.McGuire, A. M., J. D. Hughes, and G. M. Church. 2000. Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res. 10:744-757. [DOI] [PubMed] [Google Scholar]
- 39.Meisenzahl, A. C., L. Shapiro, and U. Jenal. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minambres, B., E. R. Olivera, R. A. Jensen, and J. M. Luengo. 2000. A new class of glutamate dehydrogenases (GDH). Biochemical and genetic characterization of the first member, the AMP-requiring NAD-specific GDH of Streptomyces clavuligerus. J. Biol. Chem. 275:39529-39542. [DOI] [PubMed] [Google Scholar]
- 41.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675-681. [DOI] [PubMed] [Google Scholar]
- 42.Mulongoy, K., and G. H. Elkan. 1977. Glucose catabolism in two derivatives of a Rhizobium japonicum strain differing in nitrogen-fixing efficiency. J. Bacteriol. 131:179-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen, H., and A. Krogh. 1998. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:122-130. [PubMed] [Google Scholar]
- 45.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, C. M. Fraser, and J. Eisen. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Connell, M., S. Henry, and L. Shapiro. 1986. Fatty acid degradation in Caulobacter crescentus. J. Bacteriol. 168:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phadke, N. D., M. P. Molloy, S. A. Steinhoff, P. J. Ulintz, P. C. Andrews, and J. R. Maddock. 2001. Analysis of the outer membrane proteome of Caulobacter crescentus by two-dimensional electrophoresis and mass spectrometry. Proteomics 1:705-720. [DOI] [PubMed] [Google Scholar]
- 48.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poindexter, J. S. 1981. The caulobacters: ubiquitous unusual bacteria. Microbiol. Rev. 45:123-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray, S. K., R. Rajeshwari, and R. V. Sonti. 2000. Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Mol. Plant-Microbe Interact. 13:394-401. [DOI] [PubMed] [Google Scholar]
- 52.Riley, R. G., and B. J. Kolodziej. 1976. Pathway of glucose catabolism in Caulobacter crescentus. Microbios 16:219-226. [PubMed] [Google Scholar]
- 53.Rodionov, D. A., A. A. Mironov, and M. S. Gelfand. 2001. Transcriptional regulation of pentose utilisation systems in the Bacillus/Clostridium group of bacteria. FEMS Microbiol. Lett. 205:305-314. [DOI] [PubMed] [Google Scholar]
- 54.Romero, P., and P. Karp. 2003. PseudoCyc, a pathway-genome database for Pseudomonas aeruginosa. J. Mol. Microbiol. Biotechnol. 5:230-239. [DOI] [PubMed] [Google Scholar]
- 55.Ross, C. M., and M. E. Winkler. 1988. Regulation of tryptophan biosynthesis in Caulobacter crescentus. J. Bacteriol. 170:769-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephens, C. M., and L. Shapiro. 1993. An unusual promoter controls cell-cycle regulation and dependence on DNA replication of the Caulobacter fliLM early flagellar operon. Mol. Microbiol. 9:1169-1179. [DOI] [PubMed] [Google Scholar]
- 57.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 58.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weisberg, S. 1985. Applied linear regression, 2nd ed. John Wiley & Sons, Inc., New York, N.Y.
- 60.West, L., D. Yang, and C. Stephens. 2002. Use of the Caulobacter crescentus genome sequence to develop a method for systematic genetic mapping. J. Bacteriol. 184:2155-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young, J., and I. B. Holland. 1999. ABC transporters: bacterial exporters—revisited five years on. Biochim. Biophys. Acta 1461:177-200. [DOI] [PubMed] [Google Scholar]