Abstract
Over 30 polypeptides are synthesized at various times during sporulation in Bacillus subtilis, and they are assembled at the surface of the developing spore to form a multilayer protein structure called the coat. The coat consists of three main layers, an amorphous undercoat close to the underlying spore cortex peptidoglycan, a lamellar inner layer, and an electron-dense striated outer layer. The product of the B. subtilis oxdD gene was previously shown to have oxalate decarboxylase activity when it was produced in Escherichia coli and to be a spore constituent. In this study, we found that OxdD specifically associates with the spore coat structure, and in this paper we describe regulation of its synthesis and assembly. We found that transcription of oxdD is induced during sporulation as a monocistronic unit under the control of σK and is negatively regulated by GerE. We also found that localization of a functional OxdD-green fluorescent protein (GFP) at the surface of the developing spore depends on the SafA morphogenetic protein, which localizes at the interface between the spore cortex and coat layers. OxdD-GFP localizes around the developing spore in a cotE mutant, which does not assemble the spore outer coat layer, but it does not persist in spores produced by the mutant. Together, the data suggest that OxdD-GFP is targeted to the interior layers of the coat. Additionally, we found that expression of a multicopy allele of oxdD resulted in production of spores with increased levels of OxdD that were able to degrade oxalate but were sensitive to lysozyme.
Bacterial endospores are designed to withstand long periods of dormancy and to resist physical and chemical conditions that would rapidly destroy vegetative cells. This extreme endurance can be attributed to several factors, including the composition and structural organization of the layers that surround the mature spore core (10, 19, 36). In all endospore formers, the spore core is surrounded by a thick modified peptidoglycan called the cortex, which is a key element in heat resistance (36). The cortex is covered by a multilayer protein coat, which confers resistance to noxious chemicals and to peptidoglycan-breaking enzymes, such as lysozyme (10, 19, 36). In addition, the coat contributes to the ability of the spore to monitor its environment and to initiate germination upon proper stimulation (10, 19, 36). In Bacillus subtilis, the spore coat is composed of over 30 polypeptides, which are organized into three main layers, an amorphous undercoat, a lamellar inner coat, and an electron-dense striated outer coat (2, 10, 13, 19, 28, 29). Assembly of the coat is initiated soon after the asymmetric division that partitions the sporulating cell into a larger mother cell and a smaller forespore. The early events in coat assembly are controlled by the mother cell-specific RNA polymerase sigma factor σE (16, 26, 42). Several of the proteins whose synthesis is driven by σE have morphogenetic roles; i.e., irrespective of their association with the final structure they act by laying down an imprint that prepares the surface of the developing spore for the ordered assembly of the coat structural components (10, 19).
SpoIVA, CotE, SpoVID, and SafA are morphogenetic proteins whose synthesis is under the control of σE (5, 39, 44, 48, 50, 57). SpoIVA localizes along the asymmetric division septum, and after the engulfment of the forespore by the mother cell, it encircles the forespore protoplast close to its outer membrane (11). SpoIVA is required for assembly of CotE as a ring-like structure about 75 nm from the spore outer membrane. The space between SpoIVA and CotE is presumably filled with a scaffold or matrix and later becomes the inner coat region, whereas the CotE ring itself appears to serve as the nucleation site for outer coat assembly (11). Accordingly, a cotE mutant forms spores that retain some inner coat but are devoid of an outer coat and are lysozyme sensitive (11, 56). The localization of SpoVID at the surface of the developing spore also requires SpoIVA, but it is CotE independent (11, 40). SpoVID is not required for formation of the CotE ring, but it is needed for maintenance of this ring around the forespore at later stages of coat assembly (11). An absence of SpoVID leads to misassembly of the coat as swirls of material dispersed throughout the mother cell cytoplasm and in lysozyme-sensitive spores (5). SpoVID, but not CotE, is also required for the targeting of SafA to the spore surface (40). SafA has a cell wall-binding motif at its N terminus and has been shown by immunogold labeling to localize to the cortex-coat interface (39). A safA mutant forms spores that are deficient in lysozyme resistance and germination (39, 50). Since SafA and SpoVID directly interact, it has been proposed that SafA may act as a bridge between the cortex and coat structures (39, 40).
It is only at a later developmental stage, after engulfment of the forespore by the mother cell, that assembly of the coat structure becomes apparent by electron microscopy (10, 19). Conclusion of the engulfment process triggers activation of the late mother cell regulator σK, which replaces σE in the mother cell line of gene expression (16, 26, 42) and drives expression of most of the genes that code for coat structural components (10, 19). Expression of the coat structural genes is additionally modulated by the action of the transcriptional regulator GerE (3, 17, 22, 23, 45, 49, 54, 55, 57). Spores of a gerE mutant lack the ultrastructural features normally associated with the inner coat layers (10, 33). In addition, they are deficient in expression of several genes encoding prominent components of the outer coat, such as the CotC and CotG proteins (9, 45, 57).
The roles of most individual coat structural proteins in the assembly and function of the spore coat are unclear, as null mutations often do not have a measurable phenotypic effect (10, 19). This suggests that there is extensive redundancy or that the various components make minor contributions to the structure and function of the coat layers (10, 19). Nevertheless, some of the coat proteins are enzymes or exhibit sequence similarity to enzymes that suggest that they are involved in the assembly process or in the final spore attributes. Ultimately, a description of the assembly process that also accounts for the coat properties will require detailed functional and structural characterization of selected components. For example, the CotA protein (9), a component of the spore outer coat layers (56), was recently shown to be a highly thermostable laccase involved in spore resistance to UV light and hydrogen peroxide (21, 32). The crystal structure of CotA was determined, in anticipation of the possibility that it could serve as a platform for detailed analysis of the mechanism underlying the assembly and function of CotA in the spore coat (14).
In this study, we were concerned with the regulation of expression and assembly of the product of the yoaN (oxdD) gene, which, when overproduced in Escherichia coli, was shown to have Mn-dependent oxalate decarboxylase activity (52). Oxalate decarboxylases (EC 4.1.1.2) convert oxalate to formate and CO2 (12, 51, 52). The best-characterized enzymes are enzymes that have a fungal origin, are induced by oxalate, and appear to control excessive concentrations of oxalate (12). The first oxalate decarboxylase to be identified in a prokaryote was the acid-inducible OxdC enzyme from B. subtilis, which may have a role in proton consumption within the cytoplasm (51). OxdC belongs to the cupin superfamily, whose members contain a β-sandwich domain consisting of one six-strand β-sheet and one five-strand β-sheet (1, 12, 51). OxdC is a homohexameric enzyme in which each monomer has two cupin β-barrel domains, and hence it belongs to the bicupin subclass of the cupin superfamily (1, 12, 51). OxdD is very similar to OxdC and to known oxalate decarboxylases (51). No role has been reported for OxdD. However, both OxdD and OxdC were found in a recent proteomics-based study to be spore-associated proteins (28). Here, we show that OxdD is specifically associated with the spore coat. We found that transcription of oxdD is under the control of σK, is monocistronic, and is negatively regulated by GerE. An enzymatically active OxdD-green fluorescent protein (GFP) fusion protein localized to the coat layers in a safA-dependent manner. Mutations in gerE also interfered with the assembly of OxdD-GFP. In contrast, cotE was not required for the assembly of OxdD-GFP but determined its stable association with the coat. The data suggest that OxdD is targeted to the inner layers of the coat. A multicopy allele of oxdD resulted in spores with increased levels of OxdD and with oxalate decarboxylase activity.
MATERIALS AND METHODS
Bacterial strains, media, and general techniques.
The bacterial strains used in this study are listed in Table 1. Difco sporulation medium (DSM) was used to induce sporulation by nutrient exhaustion (37). Genetic manipulations of E. coli and B. subtilis were performed and spore resistance and germination properties were assessed as previously described (8). The high-fidelity Pfu polymerase (Stratagene, La Jolla, Calif.) was used to generate PCR fragments for cloning. These fragments were sequenced, whenever required, to ensure that no mutations were introduced.
TABLE 1.
Bacterial strains
| Strain | Genotype and phenotype | Origin or reference |
|---|---|---|
| JH642 | trpC2 pheA12, wild type | Laboratory stock |
| MO1027 | trpC2 pheA12, ΔsigK::erm, Emr | P. Stragier |
| MB24 | trpC2 metC3, wild type | P. Stragier |
| IS105 | trpC2 ΔcotE::cm, Cmr | BGSCa |
| AOB68 | trpC2 metC3 ΔsafA::sp, Spr | 39 |
| AH77 | trpC2 metC3 ΔsigK::erm, Emr | Laboratory stock |
| AH94 | trpC2 metC3 gerE36 | Laboratory stock |
| AH2721 | trpC2 metC3 ΔgerE::km, Kmr | Laboratory stock |
| AH2835 | trpC2 metC3 ΔcotE::cm, Cmr | Laboratory stock |
| AH2873 | trpC2 metC3 ΔoxdD::oxdD-gfp, Spr | This study |
| AH2883 | trpC2 metC3 ΔoxdD::oxdD-gfp ΔcotE::cm, Spr Cmr | This study |
| AH2884 | trpC2 metC3 gerE36 ΔcotE::cm, Cmr | This study |
| AH2886 | trpC2 metC3 ΔamyE::PoxdD-lacZ, Cmr | This study |
| AH2890 | trpC2 metC3 ΔsigK::erm ΔamyE::PoxdD-lacZ, Cmr Emr | This study |
| AH2891 | trpC2 metC3 ΔgerE::km ΔamyE::PoxdD-lacZ, Cmr Kmr | This study |
| AH2892 | Tuner(DE3)pLacI, pTC120, Cmr Kmr | This study |
| AH2898 | trpC2 metC3 ΔoxdD::sp, Spr | This study |
| AH2912 | trpC2 metC3 gerE36 ΔoxdD::oxdD-gfp, Spr | This study |
| AH2913 | trpC2 metC3 gerE36 ΔcotE::cm ΔoxdD::oxdD-gfp, Spr Cmr | This study |
| AH2943 | trpC2 metC3 ΔoxdD::oxdD-gfp, Cmr | This study |
| AH2944 | trpC2 metC3 ΔsafA::sp ΔoxdD::oxdD-gfp, Cmr Spr | This study |
| AH2946 | trpC2 metC3 ΔamyE::PoxdD-oxdC ΔoxdD::sp, Spr Nmr | This study |
| AH2950 | Tuner(DE3)pLacI, pTC148, Cmr Kmr | This study |
| AH2953 | trpC2 metC3 oxdDMC, Nmr | This study |
| AH2954 | trpC2 metC3, pMK3, Nmr | This study |
BGSC, Bacillus Genetic Stock Center.
Insertional inactivation of the oxdD gene.
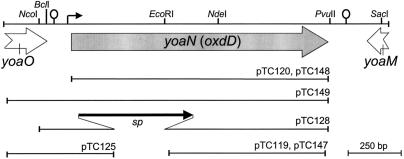
First, a 510-bp DNA fragment comprising the oxdD promoter region and the 5′ end of its coding region was PCR amplified by using primers yoaN-295D and yoaN+215R (Table 2) and doubly digested with NcoI and BglII, and the resulting 355-bp fragment was cloned between the NcoI and BamHI sites of pAH256 (17) to obtain pTC127. Next, the 3′ end of oxdD was isolated from pTC120 (see below) as a 766-bp EcoRI-XhoI fragment and cloned into the same sites of pTC127, yielding pTC128 (Fig. 1). Transformation of MB24 with ScaI-linearized pTC128 produced the spectinomycin-resistant (Spr) oxdD null mutant AH2898 (Table 1) by a double-crossover event at the oxdD locus (verified by PCR).
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′)a |
|---|---|
| yoaN-295D | GGTGATGAATTCCCGGGTGGGG |
| yoaN+215R | CCGCCGAGATCTAATTTCATGGGAGC |
| yoaN+461D | GGAACACTACTAGTTTCTGCTCG |
| yoaN-gfpR | GTTCTTCTCCTTTACTCGTACCGGGATATTTCAC |
| gfp-30D | AGTAAAGGAGAAGAACTTTTCACTGGAG |
| gfpmut2-749R | GATCCTCGAGGAATTCTTATTTGTATAGTTCATCC |
| yoaN-pETD | CGAGGTCCATGGTGTTGGAACAAC |
| yoaN-1500R | GCTGTGTAAGCTTTTACGTCTTTACG |
| gfpR | GGCGAATTCTTATTTGTATAGTTCATCCATGC |
| yoaN-fwd | GATTCATTCTCAGACGC |
| yoaN-rev-T7 | TAATACGACTCACTATAGGTTGAAAGCATTCTCCGG |
| gerE-fwd | TCGAAGCCGTCGCTAA |
| gerE-rev-T7 | TAATACGACTCACTATAGGGAGGCTCTAGCTCACCCATTC |
Restriction sites are underlined.
FIG. 1.
Genetic organization of the oxdD locus of B. subtilis. The positions, lengths, and directions of transcription of the yoaO, oxdD (yoaN), and yoaM genes are indicated below a partial restriction map of the region (27). The stem-loop structures and the bent arrow preceding the oxdD gene indicate transcription terminators and the putative oxdD promoter, respectively. The inserts present in the plasmids are also indicated. All the plasmids are described in Materials and Methods.
Construction of an oxdD-lacZ fusion.
A 510-bp fragment carrying 295 bp of DNA upstream of the oxdD start codon was PCR amplified with primers yoaN-295D and yoaN+215R (see above), doubly digested with EcoRI and BglII, and cloned between the EcoRI and BamHI sites of pSN32 (a gift from Isabel Sá-Nogueira), yielding pTC125 (Fig. 1). PstI-linearized pTC125 was used to transfer the oxdD-lacZ fusion to the amyE locus of strains MB24, AH77, and AH2721 to produce the Cmr AmyE− strains AH2886, AH2890, and AH2891, respectively (Table 1).
Construction of an oxdD-gfp fusion.
pTC119 and pTC147 (Fig. 1) were constructed in two steps. First, the oxdD 3′ region (716 bp) was PCR amplified with primers yoaN+461D and yoaN-gfpR (Table 2). Second, a 719-bp fragment comprising the coding region of the gfp gene was PCR amplified by using pEA18 (a gift from Alan Grossman) as the template and primers gfp-30D and gfpmut2-749R (Table 2). The resulting fragments were mixed and subjected to PCR with primers yoaN+461D and gfpmut2-749R (Table 2). The resulting 1,435-bp oxdD-gfp fragment was cleaved with SpeI and XhoI and cloned between the same sites of pAH256 (17) to generate pTC119 and between the same sites of pMS38 (59) to generate pTC147. Strains AH2873 (Spr) and AH2943 (Cmr) resulted from the integration of pTC119 and pTC147, respectively, into the oxdD locus of wild-type strain MB24 by a single reciprocal crossover (Campbell-type recombination) (Table 1), as verified by PCR. AH2873 was transformed with DNA from 1S105 (Table 1), yielding the Spr Cmr strain AH2883 (cotE oxdD-gfp) (Table 1). The Spr strain AH2912 (gerE oxdD-gfp) and the Spr Cmr strain AH2913 (cotE gerE oxdD-gfp) resulted from Campbell integration of pTC119 into AH94 and AH2884, respectively (Table 1). The latter strain was constructed by transforming AH94 with DNA from 1S105. The absence of congression to Ger+ was verified as described previously (47). Finally, AH2943 was transformed with chromosomal DNA from AOB68 (39) to obtain the Spr Cmr strain AH2944 (safA oxdD-gfp) (Table 1).
Construction of a B. subtilis strain bearing a multicopy allele of oxdD.
A PCR fragment (1,795 bp) comprising the entire oxdD coding region and 295 bp upstream of its transcription initiation site was generated with primers yoaN-295D and yoaN-1500R (Table 2), digested with HindIII, and cloned between the HindIII and SmaI sites of replicative plasmid pMK3 (32) to generate pTC149 (Fig. 1). Competent cells of MB24 were transformed with pTC149 and with its parental plasmid (pMK3) to obtain the neomycin-resistant (Nmr) strains AH2953 and AH2954, respectively (Table 1).
Overproduction of OxdD and OxdD-GFP.
The entire oxdD coding region (1,207 bp) was PCR amplified with primers yoaN-pETD and yoaN-1500R (Table 2), digested with NcoI and HindIII, and cloned between the same sites of pET33b(+) (Novagen) to obtain pTC120 (Fig. 1). pTC148 (Fig. 1), which can be used to overproduce OxdD-GFP, was the result of a triple ligation involving NcoI- and BamHI-digested pET28a(+) (Novagen) and the following inserts: (i) a PCR fragment obtained with primers yoaN-pETD and yoaN-1500R (see above) and digested with NcoI and EcoRI to produce a 438-bp fragment corresponding to the 5′ end of the oxdD coding region; and (ii) the 3′ oxdD coding region fused to gfp (1,458 bp), obtained by digesting with EcoRI and BamHI a 2,190-bp PCR fragment generated with primers yoaN-295D and gfpR (Table 2) and AH2873 chromosomal DNA. pTC120 and pTC148 were introduced into the E. coli host Tuner(DE3)(pLacI) (Novagen) to create strains AH2892 and AH2950 (Table 1), in which native OxdD and OxdD-GFP, respectively, could be produced under control of the T7lac promoter. Induction of OxdD production was performed as described previously (52). Following induction, the cells were harvested and lysed by passage through a French pressure cell as described previously (52).
Purification of spores and analysis of the spore coat fraction.
Spores were harvested by centrifugation of DSM cultures 24 h after the onset of sporulation. Each spore suspension was washed, and the spores were purified with a 20 to 50% Gastrografin (Schering) step gradient as described previously for Renocal-76 gradients (17, 18, 47). Coat proteins were extracted from purified spores at an optical density at 580 nm of about 2 as described previously (17, 18). Coat proteins were subjected to electrophoretic fractionation on 15% polyacrylamide gels containing sodium dodecyl sulfate (SDS). The gels were stained with Coomassie brilliant blue R-250.
Fluorescence microscopy.
Samples (0.5 ml) of DSM cultures of various strains bearing a translational oxdD-gfp fusion (see above) were collected about 8, 10, and 24 h after the initiation of sporulation and resuspended in 0.2 ml of phosphate-buffered saline. Aliquots were applied to agarose-coated microscope slides, and images were acquired with a Leica fluorescence microscope (DMRA2) by using phase-contrast optics and a standard filter for visualization of the GFP. All samples were observed with a ×63 objective lens. Images were acquired with a Cool Snap HQ camera (Roper Scientific, Tucson, Ariz.), recorded, and processed by using Adobe Photoshop.
Enzyme assays.
The activity of β-galactosidase was determined with the substrate o-nitrophenyl-β-d-galactopyranoside, as previously described (18, 47). The specific activity of oxalate decarboxylase was determined by spectrophotometry at 37°C by using a coupled reaction assay based on the method described by Magro et al. (31, 52). One unit of enzyme activity was defined as the amount of enzyme required to reduce 1 μmol of NAD per min. All specific activities are the mean values of three assays. The protein concentration was determined with a Bio-Rad assay kit (Bio-Rad Laboratories, Hercules, Calif.) used as described by the manufacturer.
RNA isolation and Northern blot analysis of gerE and oxdD.
Samples were collected from sporulating cultures of a wild-type strain (JH642) and a congenic ΔsigK::erm mutant (MO1027) (Table 1). RNA was isolated by mechanical disruption of the liquid nitrogen-frozen cell pellets in a Teflon vessel by using a Micro-Dismembrator (B. Braun Biotec International, Melsungen, Germany) as described by Petersohn et al. (41). Total RNA (5 μg per lane) was separated in a 1.5% agarose gel containing 6% (vol/vol) formaldehyde and vacuum blotted onto nylon membranes (Biodyne Plus; Pall). Digoxigenin (DIG)-labeled antisense RNA probes were generated by using T7 polymerase and gene-specific PCR products as templates. Two primer pairs (yoaN-fwd plus yoaN-rev-T7 and gerE-fwd plus gerE-rev-T7) were used in PCR with JH642 chromosomal DNA. In each of the PCRs one of the DNA primers carried the sequence of the T7 promoter. PCR fragments were subsequently used for in vitro RNA synthesis with a MAXIScript kit (Ambion, Inc., Austin, Tex.) and DIG-labeled UTP (Roche, Basel, Switzerland), which yielded hybridization probes for oxdD (444 nucleotides) and gerE (186 nucleotides). Hybridization and signal detection were performed as previously described (46).
Protein identification by peptide mass fingerprinting.
Protein spots were excised from Coomassie brilliant blue R-250-stained gels, destained, and digested with trypsin (Promega Corporation, Madison, Wis.); peptides were then extracted (38). Peptide mixtures were desalted on Poros R2/R3 tips and directly eluted onto a sample template of a matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) mass spectrometer with an elution solution containing 70% (vol/vol) acetonitrile, 0.1% (vol/vol) trifluoroacetic acid, and saturating amounts of α-cyano-3-hydroxycinnamic acid. Peptide masses were determined in the positive ion reflector mode with a Voyager Elite MALDI-TOF mass spectrometer (Applied Biosystems, Foster City, Calif.) with internal calibration. The mass accuracy was better than 30 ppm. Peptide mass fingerprints were compared to databases by using the MASCOT program (http://www.matrixscience.com/cgi/index.pl?page=../home.html). The searches considered oxidation of methionine and pyroglutamic acid formation at the N-terminal glutamine. Proteins were considered identified when the database search revealed a significant MASCOT score with a probability (P) of <0.05 that the observed match was a random event.
RESULTS
Identification of the OxdD protein.
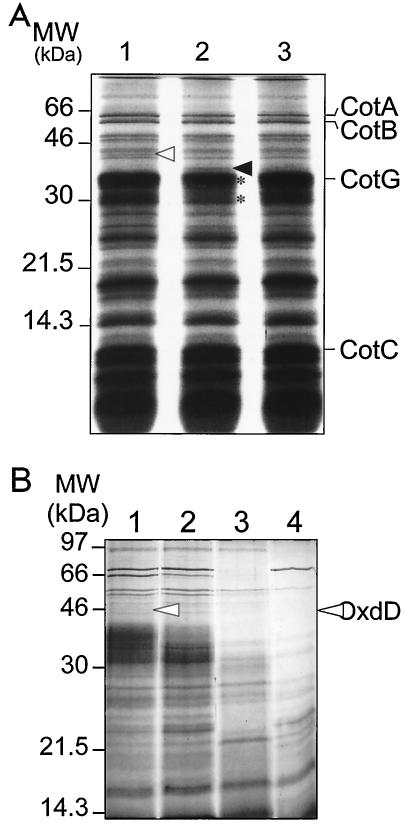
To learn more about the polypeptide composition of the spore coat layers, we used MALDI-TOF mass spectrometry to identify polypeptides extracted from highly purified spore preparations. Our strategy was to subject spores collected 24 h after the onset of sporulation in DSM to extensive washes with water and then to further purify the enriched spore preparation by centrifugation through 20 to 50% metrizoic acid step gradients (see Materials and Methods) to eliminate remnants of sporulating cells or cell debris. The purified spores were then subjected to an extraction regimen known to preferentially solubilize a fraction (about 70%) of the total spore coat-associated proteins (19), which were resolved on one-dimensional SDS—15% polyacrylamide gel electrophoresis (PAGE) gels. As a further criterion for specific association with the coat integuments, bands that were present in the wild type but were present at reduced levels in the coats of a cotE mutant were processed for MALDI-TOF analysis. By using this approach, the 43-kDa product of the yoaN locus (27) (Fig. 1) was identified as a protein associated with the spore coat layers (Fig. 2). YoaN was recently found to have oxalate decarboxylase activity when it was expressed in E. coli cells and was accordingly renamed OxdD (52). In two other recent studies the workers employed mass spectrometry techniques to identify proteins associated with the spore or spore coat (28, 29). In one of these studies, OxdD was found to be a spore-associated protein (28). The results reported here suggest that the OxdD protein specifically associates with the coat layers of B. subtilis spores.
FIG. 2.
Spore coat polypeptides extracted from spores of several strains. Spores were purified, and the coat proteins were extracted as described in Materials and Methods and electrophoretically resolved on SDS—15% PAGE gels. (A) Spore coat protein extracts of the following strains: MB24 (wild type) (lane 1), AH2898 (oxdD) (lane 2), and AH2873 (oxdD-gfp) (lane 3). (B) Profile of coat proteins extracted from spores of the following strains: MB24 (wild type) (lane 1), AH2898 (oxdD) (lane 2), AH2835 (cotE) (lane 3), and AH94 (gerE) (lane 4). The open and solid arrowheads indicate the positions of the OxdD and Hag proteins, respectively. The asterisks in panel A indicates the position of proteins that appear to be less abundant in the oxdD mutant. The positions of molecular mass markers (MW) are indicated on the left.
The 43-kDa OxdD polypeptide is absent from spores produced by an oxdD insertional mutant.
To confirm the association of OxdD with the coat and to examine its role, we constructed an oxdD insertional mutant, AH2898 (Table 1). oxdD is flanked by the yoaO gene upstream and by the yoaM gene downstream (Fig. 1); the latter genes encode proteins with unknown functions. yoaM is convergently oriented relative to oxdD (27) (Fig. 1). Moreover, results described below indicate that oxdD is a monocistronic unit transcribed from a promoter just upstream of the gene's coding sequence. Thus, the mutation is unlikely to cause a polar effect. We purified spores from MB24 and AH2898 and analyzed the collection of proteins that were extracted from their coat layers on SDS—15% PAGE gels (Fig. 2A). In spores formed by AH2898 (oxdD::sp), the 43-kDa band identified as the OxdD protein was missing (Fig. 2). The set of proteins extracted from AH2898 spores differed further from the proteins extracted from wild-type spores. First, an additional band present in the AH2898 coat extract was identified by MALDI-TOF analysis as flagellin (Hag), a major component of the flagellum (20, 30) (Fig. 2). Several preparations of AH2898 spores were examined, and in all cases Hag was found in the coat fraction (data not shown). Since the hag gene is required for motility of vegetative cells (30), it seems plausible that residual Hag protein present in the sporulation medium can associate with the spore after its release from the mother cell. Hag is tightly associated with spores of the oxdD mutant, as washes with 1 M KCl, a treatment known to release proteins loosely associated with the coat (47), did not reduce its level or preclude its presence in the collection of extractable coat polypeptides. Perhaps disruption of oxdD induces a subtle change in the properties of the coat that causes the tight adherence of Hag to the coat. Second, two proteins appeared to be less abundant in AH2898 coat extracts. One of these proteins is the normally prominent 36-kDa CotG protein (45), and the other produced a wide diffuse band in the 30-kDa region of the gel, which we were not able to identify by either MALDI-TOF or N-terminal sequence analysis (Fig. 2A). In contrast, most of the other proteins, including CotA, CotB, and CotC, remained unchanged compared to the wild-type proteins (Fig. 2A). Finally, we noted that proteins in the 43-kDa region of the gel, presumably including OxdD, were absent from the coats of cotE (AH2835) (Table 1) or gerE (AH94) (Table 1) mutant spores, or the amounts were greatly reduced (Fig. 2B). Together, these results are consistent with localization of OxdD to the spore coat (see below). Disruption of the oxdD locus had no detectable effect on spore resistance to heat or lysozyme (Table 3) or on the capacity to germinate in response to l-alanine or l-asparagine (data not shown). Oxalate is known to increase the permeability of the spore coat to certain electron microscopy dyes (25). Moreover, spores become sensitive to lysozyme after incubation for 5 min in the presence of oxalate at 80°C but not when they are incubated for 30 min at 30°C (25). To determine if OxdD contributed to protection of spores against oxalic acid under these conditions, we exposed wild-type or oxdD spores to oxalic acid (0.025, 0.25, 2.5, 5, or 25 mM) at 30°C (30 min) or 80°C (5 min). Treatment with 25 mM oxalic acid at 80°C caused similar reductions in spore viability (from about 108 to 105 spores per ml) or lysozyme resistance (from 108 to 104 spores per ml) for wild-type and mutant spores. Also, no effect on viability or resistance of wild-type and mutant spores was observed when the other concentrations of oxalic acid were used, at either 80 or 30°C.
TABLE 3.
Heat resistance and lysozyme resistance of various strains
| Strain | Relevant genotype | Sporulation (CFU/ml)a
|
||
|---|---|---|---|---|
| Viable cells | Heat-resistant cells | Lysozyme-resistant cells | ||
| MB24 | Wild type | 8.9 × 108 | 5.2 × 108 | 5.5 × 108 |
| AH2898 | ΔoxdD::sp | 6.5 × 108 | 6.2 × 108 | 8.9 × 108 |
| AH2953b | oxdDMC, pTC149 | 1.2 × 108 | 2.7 × 107 | 1.5 × 106 |
| AH2954c | Wild type, pMK3 | 4.8 × 108 | 4.8 × 108 | 2.5 × 108 |
| AH2873 | ΔoxdD::oxdD-gfp | 5.6 × 108 | 4.9 × 108 | 5.4 × 108 |
| AOB68 | ΔsafA::sp | 4.0 × 108 | 5.4 × 108 | 2.4 × 107 |
| AH2944 | ΔsafA::sp ΔoxdD::oxdD-gfp | 2.4 × 108 | 7.1 × 108 | 1.3 × 107 |
The total (viable), heat-resistant, or lysozyme-resistant cell count was determined 24 hs after the onset of sporulation in liquid medium (DSM), as described in Materials and Methods.
Strain AH2953 carries a full-length copy of oxdD in replicative plasmid pTC149 and hence a multicopy allele of oxdD (oxdDMC).
Strain AH2954 carries replicative vector pMK3 (32), which is the parental plasmid of pTC149.
Spore-associated OxdD can exhibit oxalate decarboxylase activity.
Recently, the spore coat protein CotA was shown to be a laccase, which retains enzymatic activity when it is embedded within the endospore coat (21, 32). To determine whether coat-associated OxdD also retained enzymatic activity, we assayed purified wild-type (MB24) and oxdD mutant spores (AH2898) for oxalate decarboxylase activity (see Materials and Methods). However, we were unable to detect enzymatic activity in wild-type spores. We repeated the assay with various amounts of whole-cell extracts of sporulating cells or intact spores in the presence of 25 μM MnCl2 at different pH values and substrate concentrations, after treatment of spore suspensions at 80°C for 10 min to facilitate access of the substrate (32) and after induction of spore germination. In all cases no enzymatic activity was detected. We then generated an oxdD multicopy allele (oxdDMC) by inserting the oxdD gene (including its promoter [see Materials and Methods]) into the pMK3 replicative plasmid (32). Suspensions of spores purified from the oxdDMC strain (AH2953) (Table 1) and from a strain bearing the pMK3 vector (AH2954) (Table 1) were tested for the ability to degrade oxalate. We found enzymatic activity in spores of oxdDMC strain AH2953 (about 15 mU/optical density unit of a spore suspension) but not in spores produced by strain AH2954 harboring the parental pMK3 vector. We also looked for enzymatic activity in whole-cell extracts prepared from cultures of AH2953 and AH2954 after 6 and 8 h of sporulation. Under the assay conditions used no activity was found in whole-cell extracts of AH2953 or AH2954. Therefore, enzyme activity can be detected even in the oxdDMC strain only when the enzyme accumulates at the spore surface.
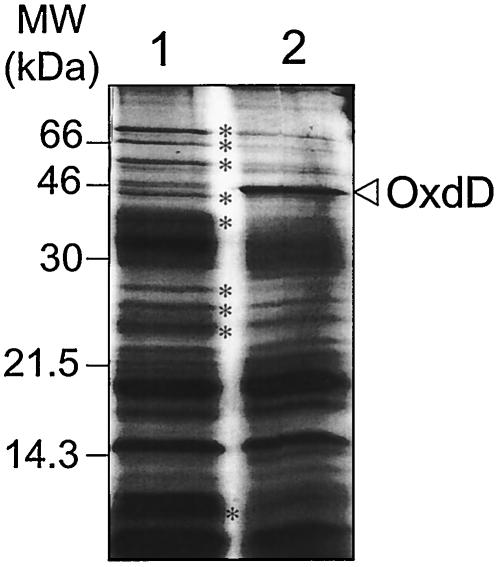
SDS-PAGE analysis of the coat polypeptides indicated that increased amounts of OxdD were present in AH2953 spores (Fig. 3, lane 2). However, the representation of several other coat polypeptides was greatly altered in AH2953 spores compared to that in spores produced by AH2954 (Fig. 3), and the resulting spores exhibited reduced resistance to heat and lysozyme treatments (Table 3), indicating that abnormal (increased) levels of OxdD perturb both the spore coat composition and spore resistance. Presumably, the coat structure of AH2953 spores is also altered. However, these spores have not been examined by electron microscopy. In any case, the results indicate that spore-associated OxdD has enzymatic activity.
FIG. 3.
Analysis of a multicopy allele of oxdD. Spore coat proteins were extracted from purified spores as described in Materials and Methods and electrophoretically resolved on SDS—15% PAGE gels. Lane 1, AH2954 containing pMK3 (control plasmid); lane 2, AH2953 containing pTC149 (oxdDMC). The open arrowhead indicates the position of OxdD. The asterisks indicate the positions of polypeptides that are absent in strain AH2953 expressing a multicopy allele of oxdD or whose amounts are reduced. The positions of molecular mass markers (MW) are indicated on the left.
oxdD gene is transcribed during sporulation under σK control.
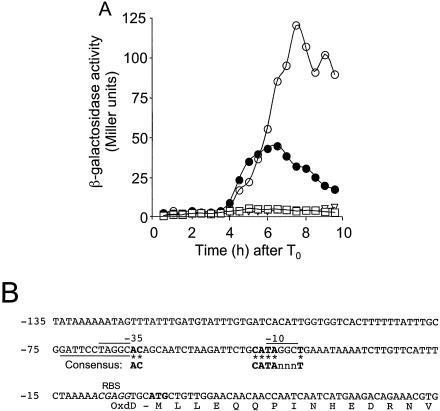
With one possible exception (47), all the genes shown to be involved in spore coat assembly are transcribed in the mother cell compartment of the sporulating cell (10, 19). In addition, expression of a significant number of the genes known to encode coat structural components relies upon σK and is either positively regulated or repressed by GerE (3, 7, 17, 45, 49, 54, 57). We used an oxdD-lacZ fusion inserted at the amyE locus to study the regulation of the oxdD gene. Expression of oxdD-lacZ was monitored throughout growth and sporulation in DSM in an otherwise wild-type strain (AH2886) and congenic sigK (AH2890) and gerE (AH2891) mutants (Table 1). We found that in AH2886, expression of oxdD-lacZ was induced around hour 4 of sporulation (Fig. 4), a temporal profile shared by several other σK-controlled genes (10, 15, 19, 57). No β-galactosidase production was detected in the sigK mutant AH2890 (Fig. 4). In contrast, expression of oxdD-lacZ increased about threefold in the gerE mutant AH2891 (Fig. 4A). Consistent with involvement of GerE in the regulation of oxdD expression, three possible GerE-binding sites were recognized in the oxdD promoter region (Fig. 4B) (22). Identical results were obtained when the oxdD-lacZ fusion was inserted at the oxdD locus (data not shown). We inferred that expression of oxdD during sporulation occurs from a promoter present in the 295 bp upstream of the gene's start codon (see Materials and Methods), which is utilized by σK and repressed by GerE.
FIG. 4.
Regulation of oxdD-lacZ expression. (A) An oxdD-lacZ fusion was inserted at the amyE locus of various strains, and samples were taken at different times after the initiation of sporulation in DSM (T0) to assay for β-galactosidase accumulation. The following strains were used: AH2886 (amyE::oxdD-lacZ) (•), AH2890 (sigK::erm amyE::oxdD-lacZ) (□), and AH2891 (gerE36 amyE::oxdD-lacZ) (○). The endogenous levels of β-galactosidase production were determined in wild-type strain MB24 (▿). (B) Sequence of the putative oxdD promoter and the −10 and −35 sequences aligned with the consensus for σK-dependent promoters (16). Bases identical to the bases in the consensus sequence are indicated by boldface type and asterisks. n represents any base. The lines above the DNA sequence indicate bases in the putative −35 and −10 regions that match the bases in the core of the GerE binding site consensus sequence (TRGGY); the line below the sequence indicates a region in the complementary strand that matches the larger consensus region for GerE binding (RWWTRGGYnnY) (22). The ribosome binding site (RBS) is indicated by italics, and the start codon is indicated by boldface type just downstream of the ribosome binding site.
oxdD is monocistronic.
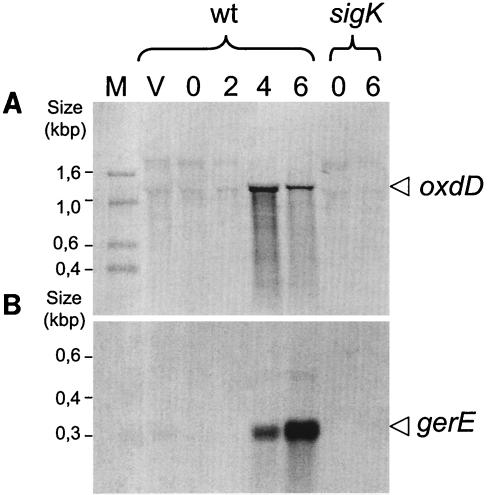
To ensure that no other upstream promoter contributed significantly to the expression of oxdD (as implied by the analysis of an oxdD-lacZ fusion at the oxdD locus), we performed a Northern blot analysis. RNA samples were prepared from a wild-type strain and from a sigK mutant at various times during sporulation and were analyzed with probes specific for oxdD or for gerE. In wild-type cells, a transcript was detected with the oxdD-specific probe at hours 4 and 6 of sporulation (Fig. 5A). The size of this transcript (about 1,200 nucleotides) is consistent with the size of the oxdD coding region (1,176 bp). No other signal appeared to be present in the wild type at any time tested, and no signal was found in the sigK mutant strain (Fig. 5A). Similarly, the gerE transcript (300 nucleotides) was detected at hours 4 and 6 during development in the wild type but not in a sigK mutant (Fig. 5B). The results indicate that transcription of oxdD during sporulation occurs mainly if not exclusively in a monocistronic mode and that it temporally coincides with the transcription of a gene (gerE) known to be under the control of σK (6, 57). We tried to map the 5′ end of the oxdD transcript by primer extension, but several attempts in which different primers were used were unsuccessful. Of note, however, was the presence of a possible canonical σK-dependent promoter (16) just upstream of oxdD (Fig. 4B).
FIG. 5.
oxdD is monocistronic. A wild-type strain (wt) and a congenic sigK mutant were grown in DSM. Samples were taken during the exponential growth phase (vegetative growth [lane V]), at the onset of sporulation (time zero [lane 0]), and at various times throughout sporulation (times [in hours] are indicated after time zero [lanes 2, 4, and 6]). Total RNA was prepared as described in Materials and Methods. RNA samples were electrophoretically resolved on denaturing agarose-formaldehyde gels and transferred to nylon membranes. The RNA blots were hybridized with DIG-labeled probes complementary to the mRNA of oxdD (A) and gerE (B). Transcript sizes were determined based on the position of the DIG-labeled marker (lane M).
OxdD-GFP is a functional oxalate decarboxylase.
The genetic requirements for assembly of OxdD were studied by using a GFP fusion. Since the oxdD mutant did not exhibit any spore resistance or germination phenotype, we wanted to determine whether the fusion protein retained oxalate decarboxylase activity. The product of the oxdD gene exhibits oxalate decarboxylase activity when it is produced in E. coli (52). We used the same activity assay to test the functionality of the OxdD-GFP fusion protein after overproduction in E. coli. The oxdD-gfp fusion was cloned under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7lac promoter, and the resulting plasmid was introduced into an appropriate E. coli host for overproduction (strain AH2950) (Table 1). Induction was performed as previously described (52) (see Materials and Methods). In parallel, we overproduced the native (unfused) OxdD protein as a positive control for activity (AH2892) (Table 1). Oxalate decarboxylase activity was then assayed in whole-cell extracts prepared from induced cultures of AH2950 and AH2892. Activity was detected in whole-cell extracts of both induced AH2950 and induced AH2892 cultures but not in extracts prepared from uninduced cultures of the same strains (data not shown). The level of overproduction of OxdD was similar to that of OxdD-GFP (about 10% of the total protein) (data not shown), and the levels of enzyme activity were comparable (2.6 U/mg of protein for OxdD and 1.6 U/mg for OxdD-GFP). Moreover, the levels of activity are comparable to those reported by Tanner et al. (52) for the activity of OxdD in E. coli crude lysates. We inferred that the overall fold of the fusion protein was not altered by fusion to GFP and that assembly of OxdD-GFP into the coat is likely to reflect assembly of the native OxdD coat protein.
Assembly of OxdD into the coat of wild-type spores.
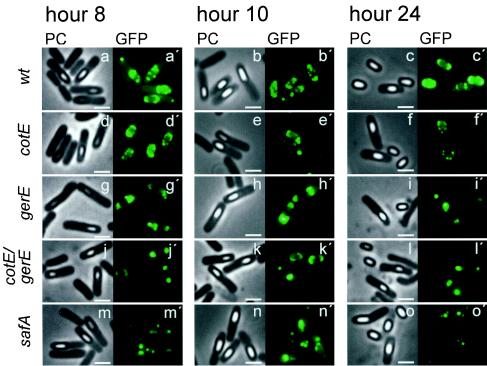
Next, we used OxdD-GFP to investigate the assembly of OxdD into the coat. Strain AH2873 (OxdD-GFP fusion in MB24) (Table 1) produced spores with wild-type resistance (Table 3) and germination properties (data not shown). The 43-kDa OxdD band was absent from the collection of AH2873 spore coat polypeptides, but no additional band corresponding to the size of OxdD-GFP (73 kDa) was detected on the Coomassie brilliant blue-stained gels (Fig. 2A, lane 3). This failure to detect OxdD-GFP could have been caused by comigration with other proteins, to reduced extractability of the fusion protein, or to both of these factors. In any case, immunoblot analysis with an anti-GFP antibody confirmed the association of OxdD-GFP with the coat of AH2873 spores (data not shown). Moreover, the results of fluorescence microscopy experiments (described below) confirmed the presence of OxdD-GFP in the coat of wild-type spores. No other changes were seen in the pattern of coat proteins, indicating that expression of OxdD-GFP does not interfere in any significant way with the assembly process (Fig. 2A). To monitor the assembly of OxdD-GFP, samples were harvested from DSM cultures 8, 10, and 24 h after the onset of sporulation, and the live AH2873 cells were mounted on agarose slides for examination by fluorescence microscopy (Fig. 6). We also used phase-contrast optics to mark the position of the whole cell and to visualize the developing spore (Fig. 6). The number of partially refractile (phase-grey), fully refractile (phase-bright), or free spores and the pattern of GFP decoration of sporulating cells or spores were recorded for each time point (Table 4). Control experiments showed that, as expected, no fluorescence was detected at any time tested for a strain having a mutation in the sigK gene (data not shown). Moreover, under the conditions used, no fluorescence signal was found to be associated with cells or spores of a wild-type strain (MB24) bearing no gfp gene or fusion for any time tested (data not shown). Fluorescence was detected for AH2873 (OxdD-GFP) at hours 4 and 6 of sporulation, when the expression of oxdD commenced and reached a maximum, respectively (Fig. 4 and 5), in less than 1% of the specimens observed. The frequency of decoration of the developing spore by OxdD-GFP was greatly enhanced around hour 8 of sporulation (Fig. 6a and a′). The fusion protein was detected as caps at both poles of the developing spore in about 6% of the specimens examined; these caps were phase grey. However, the majority of the specimens showed fluorescence around the entire developing spore (Fig. 6a and a′; Table 4). In most specimens (76%), the spore was partially refractile, whereas in some specimens (18%) the spore was phase bright (Table 4). The polar cap pattern has been observed for other coat proteins (e.g., for CotE by using either GFP fusions [53] or immunofluorescence [4, 43]) and is also consistent with observations made by using immunoelectron microscopy (11). The fact that the polar cap pattern preferentially associates with partially refractile spores suggests that it represents an early stage in the assembly process. By hour 10 of sporulation, 87% of the sporulating cells had spores fully encircled by OxdD-GFP (Fig. 6b and b′), but the frequency of decoration of phase-bright spores relative to the partially refractile spores had increased to 54% (Table 4). The overall representation of the polar cap pattern associated with phase-grey spores was maintained (4%), but this pattern was also observed for cells having phase-bright spores (4%). Finally, 24 h after the initiation of the developmental process, OxdD-GFP fully encircled the spore in 98% of the specimens examined; 84% of these specimens were free phase-bright spores, and about 14% were cells containing bright spores (Fig. 6c and c′; Table 4). We noted that the prespore surface often had a punctuated pattern of decoration rather than being uniformly covered (see Discussion). No fluorescence was detected throughout the mother cell cytoplasm in the wild-type strain, but occasionally spots of fluorescence were observed close to the cell pole opposite where the spore was formed (Fig. 6a′ and b′). Together, the results suggest that following synthesis most of the fusion protein is targeted to the poles of the developing spore and then, as the developing spore attains full refractility, gains access to the entire surface.
FIG. 6.
Assembly of OxdD-GFP into the spore coat. A functional OxdD-GFP fusion was introduced into a wild-type strain (wt) and into strains bearing mutations in loci known to be involved in assembly of the spore coat. The strains were grown in DSM, and samples were taken 8, 10, and 24 h after the onset of sporulation. Sporulating cells were observed by phase-contrast microscopy (PC) (a to o) and by fluorescence microscopy (a′ to o′) to detect OxdD-GFP (GFP). The following strains were used: AH2873 (oxdD-gfp), AH2883 (cotE oxdD-gfp), AH2912 (gerE oxdD-gfp), AH2913 (cotE gerE oxdD-gfp), and AH2944 (safA oxdD-gfp). Representative specimens are shown in each case. Quantification of the decoration patterns is shown in Table 4. Scale bars = 2 μm.
TABLE 4.
Quantification of OxdD-GFP localization patterns
| Strain (genotype) | Time (h)a | No. countedb | Spore morphologyc | No. of cells or spores with the following decoration patterns
|
||||
|---|---|---|---|---|---|---|---|---|
| Polar caps | Entire prespore | Mother cell spots | Entire spore | None | ||||
| AH2873 (wild type) | 8 | 122 | Phase grey | 7 | 93 | 0 | —d | — |
| Phase bright | 0 | 22 | 0 | — | — | |||
| Free | — | — | — | 0 | 0 | |||
| 10 | 121 | Phase grey | 5 | 40 | 0 | — | — | |
| Phase bright | 5 | 65 | 0 | — | — | |||
| Free | — | — | — | 6 | 0 | |||
| 24 | 146 | Phase grey | 0 | 2 | 0 | — | — | |
| Phase bright | 2 | 20 | 0 | — | — | |||
| Free | — | — | — | 122 | 0 | |||
| AH2883 (cotE) | 8 | 134 | Phase grey | 25 | 85 | 0 | — | — |
| Phase bright | 11 | 12 | 1 | — | — | |||
| Free | — | — | — | 0 | 0 | |||
| 10 | 137 | Phase grey | 11 | 41 | 5 | — | — | |
| Phase bright | 14 | 31 | 29 | — | — | |||
| Free | — | — | — | 0 | 6 | |||
| 24 | 127 | Phase grey | 1 | 3 | 0 | — | — | |
| Phase bright | 0 | 8 | 36 | — | — | |||
| Free | — | — | — | 3 | 76 | |||
| AH2912 (gerE) | 8 | 47 | Phase grey | 35 | 1 | 1 | — | — |
| Phase bright | 10 | 0 | 0 | — | — | |||
| Free | — | — | — | 0 | 0 | |||
| 10 | 72 | Phase grey | 42 | 0 | 0 | — | — | |
| Phase bright | 29 | 0 | 0 | — | — | |||
| Free | — | — | — | 0 | 1 | |||
| 24 | 88 | Phase grey | 3 | 0 | 0 | — | — | |
| Phase bright | 17 | 0 | 0 | — | — | |||
| Free | — | — | — | 0 | 68 | |||
| AH2913 (cotE gerE) | 8 | 118 | Phase grey | 96 | 0 | 0 | — | — |
| Phase bright | 22 | 0 | 0 | — | — | |||
| Free | — | — | — | 0 | 0 | |||
| 10 | 110 | Phase grey | 33 | 0 | 0 | — | — | |
| Phase bright | 76 | 0 | 0 | — | — | |||
| Free | — | — | — | 0 | 1 | |||
| 24 | 110 | Phase grey | 4 | 0 | 0 | — | — | |
| Phase bright | 49 | 0 | 1 | — | — | |||
| Free | — | — | — | 0 | 56 | |||
| AH2944 (safA) | 8 | 121 | Phase grey | 1 | 0 | 103 | — | — |
| Phase bright | 0 | 0 | 17 | — | — | |||
| Free | — | — | — | 0 | 0 | |||
| 10 | 130 | Phase grey | 0 | 0 | 30 | — | — | |
| Phase bright | 1 | 0 | 91 | — | — | |||
| Free | — | — | — | 1 | 7 | |||
| 24 | 121 | Phase grey | 0 | 0 | 5 | — | — | |
| Phase bright | 0 | 0 | 41 | — | — | |||
| Free | — | — | — | 1 | 74 | |||
Hours after the onset of sporulation.
Number of cells or free spores.
Morphology as determined by phase-contrast microscopy.
—, the pattern does not apply to the spore morphology.
cotE and gerE are required for stable association of OxdD with the coat.
Native OxdD absent from the coats of cotE or gerE mutant spores (Fig. 2B) or the amount was greatly reduced, but it was still possible that in the mutants OxdD was targeted to the developing spore but was not retained upon spore release. We therefore investigated the localization of OxdD-GFP in cells with a cotE mutation, which failed to assemble the outer coat (56), in cells with a gerE mutation, which lacked several of the inner and outer coat proteins (2, 3, 7, 10, 19, 33, 34, 45, 49), or in cells with both mutations. The cotE gerE double mutant formed spores that were missing both coat layers (10). In strain AH2883 (cotE), at hour 8 of sporulation, we observed decoration of the polar cap region of the developing spores in both phase-grey spores (19%) and phase-bright spores (8%) (Fig. 6d and d′; Table 4). Since representation of the polar cap pattern increased in the mutant (to 27% of the specimens observed, compared to 6% in the wild type) and this pattern was also found in association with phase-bright spores, it seems that complete encircling of the spore by OxdD-GFP was slowed down in the mutant and uncoupled from spore maturation. Accordingly, the proportions of phase-grey spores (63%) and phase-bright spores (9%) in cells of the cotE mutant at hour 8 which were fully encircled by OxdD-GFP decreased to a total of about 72% (Table 4). As in the wild type, the prespore surface often displayed a punctuated pattern of decoration (see above and Discussion). By hour 10, fluorescence was also observed as spots or patches in the mother cell in 25% of the cotE mutant cells carrying phase-grey or phase-bright spores (Fig. 6e and e′; Table 4), a pattern that persisted at hour 24 (Table 4). Decoration of the entire surface of the developing spore was greatly reduced by hour 24 (2% of the specimens scored) (Table 4). Moreover, the fluorescence signal was found in only 4% of the free spores (Fig. 6f and f′). The results suggest that OxdD-GFP is initially targeted to the surface of the developing spore following the polar cap pattern, as in the wild type, and then fully encircles the spore, albeit more slowly. However, the accumulation of fluorescence in the mother cell cytoplasm from hour 10 onward and the lack of decoration of free spores at hour 24 suggest that OxdD-GFP does not stably associate with the forming coat and does not persist in spores released by the cotE mutant cells.
In contrast to the cotE mutant, decoration of the entire surface of the developing spore was never observed for the gerE or cotE gerE mutants, even at a late time in assembly (Fig. 6g to l; Table 4). Indeed, sporulating cells of both of these mutants exhibited only polar cap pattern of spore decoration, and no fluorescence signal was ever found to be associated with free spores (Table 4). These observations indicate that the gerE36 mutation is epistatic over the cotE null allele. Consistent with the observation that GerE represses expression of oxdD (see above), increased fluorescence was observed in cells bearing the gerE36 allele. It seems that the initial targeting of OxdD-GFP to the polar regions of the developing spore does not require gerE but that migration of the fusion protein to completely surround the spore is a gerE-dependent event. The lack of fluorescence associated with free spores produced by AH2883, AH2912, or AH2913 is consistent with the observation that native OxdD is absent from the coats of purified cotE or gerE spores or the amount is greatly reduced (Fig. 2B).
SafA directs OxdD to the developing spore coat.
The cotE-independent targeting of OxdD-GFP to the surface of the developing spore suggested that the fusion protein could reside in the inner layers of the coat and that its assembly could depend on the action of morphogenetic proteins whose assembly is itself CotE independent. Localization of SpoIVA, SpoVID, and SafA is CotE independent (11, 40). We found that targeting of OxdD-GFP to the spore surface was prevented by mutations in spoIVA or spoVID (data not shown). Since spoIVA governs the assembly of SpoVID and since SpoVID recruits SafA, we also analyzed the localization of OxdD-GFP in sporulating cells of a safA mutant (AH2944). In the safA cells at hour 8 of sporulation, OxdD-GFP fluorescence tended to accumulate at the mother cell-prespore border as spots or large patches (Fig. 6m and m′), a pattern that became more accentuated as sporulation proceeded (Fig. 6n and n′) (for simplicity this pattern is recorded as mother cell spots in Table 4). Essentially no fluorescence was associated with free safA spores (Fig. 6o and o′) (Table 4). Importantly, expression of oxdD-gfp did not aggravate the lysozyme sensitivity of safA mutant spores (Table 3). The results suggest that the initial targeting of OxdD-GFP to the spore surface is safA dependent and support the view that OxdD resides in the inner layers of the coat.
DISCUSSION
Our results indicate that the OxdD protein of B. subtilis is a component of the coat layers. Several lines of evidence support this claim. First, the OxdD protein was identified by MALDI-TOF analysis among the collection of polypeptides that can be extracted from the coats of wild-type spores but not from cotE or gerE spores, which have abnormal coats (10, 19, 33, 56). Second, disruption of oxdD consistently led to the absence of the OxdD protein from coat extracts prepared from spores of the mutant. Moreover, oxdD was found to be expressed in the mother cell from hour 4 of sporulation onward under the control of σK, which coincided temporally and spatially with the expression of most genes encoding coat structural components (10, 19). Finally, studies in which a functional OxdD-GFP fusion was used revealed assembly of the fusion protein around the developing spore in a manner that was influenced by loci known to play key roles in coat biogenesis.
OxdD shows a high degree of sequence similarity to another B. subtilis protein, OxdC (formerly YvrK), which was recently characterized as an acid-inducible oxalate decarboxylase, and both proteins are very similar to well-characterized fungal enzymes (12, 51). OxdD is likely to have the overall fold and structural features of OxdC and was able to convert oxalate to formate and CO2 in an Mn-dependent manner when it was overproduced in E. coli cells (52). Both OxdD and OxdC were identified in a previous study in which liquid chromatography coupled to tandem mass spectrometry was used to analyze proteins extracted from whole spores (28). However, no expression data were reported for either oxdD or oxdC, and none of the products were assigned to a specific spore layer or structure (28). While our results indicate that OxdD is a component of the inner coat layers, we did not observe expression of an oxdC-gfp fusion during sporulation and did not see decoration of mature spores by the fusion protein (data not shown). It is possible that in contrast to OxdD (this study), minute levels OxdC are associated with the spore, presumably with the spore core. No OxdD activity was reported in B. subtilis, nor were the conditions that induced expression of the oxdD gene in B. subtilis reported. Using a combination of Northern blot analysis and a fusion of the oxdD promoter region to the lacZ gene, we were able to show that a single monocistronic transcript was produced during sporulation under the control of σK and that expression of oxdD was normally repressed by the GerE ancillary regulator. At this time, σK and GerE are the only known regulators of oxdD expression in B. subtilis.
The role of OxdD was examined by analyzing the coat polypeptide composition and the resistance and germination properties of spores produced by an oxdD insertional mutant. As observed for mutations in several other coat structural components, disruption of oxdD did not interfere significantly with the assembly or organization of the coat layers. Spores of the mutant did not differ from the wild-type spores in the ability to resist lysozyme exposure or the ability to germinate in response to l-alanine or l-asparagine. Also, they were not more sensitive to oxalate treatment or to oxalate treatment followed by lysozyme treatment. However, we did not detect oxalate decarboxylase activity in wild-type spores of B. subtilis, as assayed previously for E. coli cells producing OxdC or OxdD and for OxdC in B. subtilis extracts (51, 52). We do not think that the lack of activity is caused by poor access of oxalate to the enzyme, since the spore coat is permeable to oxalate up to the inner coat-outer coat junction and even further following treatment at 80°C (25;this study). The activity assay is based on the reduction of NAD upon conversion of formate to CO2 by formate dehydrogenase (31). Again, it seems unlikely that the smaller formate molecule does not come in contact with formate dehydrogenase. It may be that upon assembly, OxdD is in a microenvironment that does not support enzyme activity. For example, it is possible that another coat protein inhibits the enzyme. The observation that expression of a multicopy allele of oxdD results in spores that can convert oxalate into formate indicates that the protein that is synthesized during spore development and that becomes associated with the spore coat is a functional oxalate decarboxylase. We suggest that the activity observed results in part from the disorganization of the coat structure caused by the increased representation of OxdD (as shown by the altered coat protein profile and lysozyme sensitivity), which could free the enzyme from its putative inhibitory microenvironment, and in part from increased levels of enzyme. Some fungi produce oxalic acid, which can chelate manganese and stimulate the activity of an extracellular Mn peroxidase involved in lignin degradation (12). In this context, oxalate decarboxylases appear to confer protection against excess oxalate (12, 51). Since spore germination in the soil can occur in association with the growth and development of fungal hyphae (35), it is tempting to speculate that OxdD could also protect the spore (or the germinating spore) from the harmful effects of oxalic acid. It is also possible that OxdD plays only a structural role in coat assembly. In any case, the biological significance of the association of OxdD with the B. subtilis spore coat is unclear at present. Database searches suggest that the genome of at least one other spore-forming microorganism, Bacillus cereus, encodes an oxalate decarboxylase (24), but this protein has not been characterized.
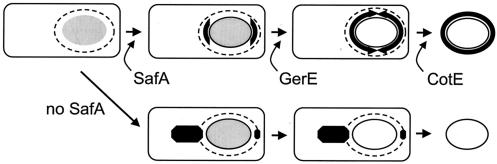
A functional OxdD-GFP fusion protein seems to be assembled in two steps. In the first step, OxdD-GFP localizes as caps at both poles of the engulfed prespore. In the second step, the fusion protein fully encircles the developing spore. The punctuated pattern of fluorescence often seen in sporulating cells or spores suggests that the distribution of OxdD-GFP is not uniform in the prespore surface but rather is patchy. While this pattern could be an artifact caused by the presence of the GFP moiety, we suspect that it may reflect the possible multimeric nature of OxdD, as suggested by its similarity to the hexameric OxdC molecule (1). Our results indicate that SafA is necessary for the initial targeting of OxdD to the cap regions of the prespore (Fig. 7). In the absence of SafA, fluorescence from OxdD-GFP is observed as patches or spots in the mother cell cytoplasm (Fig. 7). As observed for SafA (39), it is possible that OxdD localizes to the inner coat layers. This notion is consistent with the observation that the amounts of both OxdD and OxdD-GFP are greatly reduced or these molecules are missing from the coats of cotE or gerE spores, and it is further supported by the drastic effect that the oxdDMC allele has on the structure and properties of the coat. We note that a multicopy allele of cotA results in lysozyme-resistant spores with a normal complement of coat proteins except for CotA (32). Since CotA is an outer coat protein (56), one interpretation is that the effects of the oxdDMC allele result from the more internal localization of OxdD, perhaps in conjunction with its larger size (see above). We found that the initial targeting of OxdD-GFP to the prespore polar regions was gerE and cotE independent. However, OxdD-GFP fails to fully encircle the prespore in a gerE mutant, implying that at least one GerE-dependent protein is necessary for the second stage of OxdD localization (Fig. 7). In contrast, OxdD-GFP was capable of encircling the prespore in cotE mutant cells but was not retained in the mature released spores (Fig. 7). Presumably, OxdD is lost from mature spores lacking complete inner and outer coat layers. Expression of gerE is required for the development of the morphological features normally associated with the inner coat, even though a mutation also interferes with assembly of the outer coat (10, 19, 33). Mutations in cotE appear to have a much more specific effect on the assembly of the outer coat (11, 56). OxdD could be assembled at the border between the inner and outer coats, which could explain the fact that the protein is retained in association with the inner coat found in cotE spores. For example, CotH is synthesized under the control of σK, and since its assembly is both cotE and gerE dependent, it has been proposed that CotH is close to CotE, at the inner coat-outer coat border (34, 58). Another coat protein, CotS, produced under the joint control of σK and GerE, was found by immunoelectron microscopy in the inner coat and pericortex, yet it was not detected in cotE spores (49). We speculate that like OxdD, CotS cannot be retained in association with the inner coat in spores lacking the outer coat. This pattern of assembly may have a broader distribution, suggesting that care should be taken in assignment of a protein to the outer coat on the basis of its absence from cotE spores. OxdD is presently the only protein whose targeting to the developing spore specifically requires expression of safA. It will be interesting to determine whether the targeting of OxdD involves a direct interaction with SafA.
FIG. 7.
Model for the assembly of OxdD. The model predicts that OxdD is initially targeted to the polar cap regions of the developing spore in a safA-dependent manner. Localization of OxdD-GFP becomes apparent when the spore develops refractility (initially the spore is phase grey, as shown). In the absence of SafA, OxdD-GFP accumulates at the mother cell-prespore border as spots or patches, which persist until late times in development, but not in association with the released spore. Complete encircling of the developing spore by OxdD-GFP requires expression of gerE but not expression of cotE and occurs as the spore becomes phase bright (open ellipse). The requirement for safA and gerE but not for cotE suggests that OxdD associates with the inner coat layers. However, OxdD does not persist in stable association with the coat layers in the absence of the outer coat assembly in a cotE mutant. The dashed line indicates the position of the CotE ring, which marks the site of assembly of the outer coat. The CotE ring forms before the spore shows any signs of refractility or decoration by OxdD-GFP (initial cell).
Acknowledgments
We thank C. P. Moran, Jr., for critically reading the manuscript.
This work was supported by grants from the Max Planck Institute for Terrestrial Microbiology (Marburg, Germany) and the Bundesministerium für Bildung und Forschung to U.V. and by internal grants from the Instituto de Tecnologia Química e Biológica to A.O.H. T.C. holds a Ph.D. fellowship (PRAXIS XXI/BD/1167/00) from Fundação para a Ciência e a Tecnologia.
REFERENCES
- 1.Anand, R., P. C. Dorrestein, C. Kinsland, T. P. Begley, and S. E. Ealick. 2002. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution. Biochemistry 41:7659-7669. [DOI] [PubMed] [Google Scholar]
- 2.Aronson, A. I., L. Ekanayake, and P. C. Fitz-James. 1992. Protein filaments may initiate the assembly of the Bacillus subtilis spore coat. Biochimie 74:661-667. [DOI] [PubMed] [Google Scholar]
- 3.Aronson, A. I., H. Y. Song, and N. Bourne. 1989. Gene structure and precursor processing of a novel Bacillus subtilis spore coat protein. Mol. Microbiol. 3:437-444. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, T., S. Little, A. G. Stover, and A. Driks. 1999. Functional regions of the Bacillus subtilis spore coat morphogenetic protein CotE. J. Bacteriol. 181:7043-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beall, B., A. Driks, R. Losick, and C. P. Moran, Jr. 1993. Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J. Bacteriol. 175:1705-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cutting, S., S. Panzer, and R. Losick. 1989. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J. Mol. Biol. 207:393-404. [DOI] [PubMed] [Google Scholar]
- 7.Cutting, S., L. B. Zheng, and R. Losick. 1991. Gene encoding two alkali-soluble components of the spore coat from Bacillus subtilis. J. Bacteriol. 173:2915-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutting, S. M., and P. B. V. Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 9.Donovan, W., L. B. Zheng, K. Sandman, and R. Losick. 1987. Genes encoding spore coat polypeptides from Bacillus subtilis. J. Mol. Biol. 196:1-10. [DOI] [PubMed] [Google Scholar]
- 10.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driks, A., S. Roels, B. Beall, C. P. Moran, Jr., and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8:234-244. [DOI] [PubMed] [Google Scholar]
- 12.Dunwell, J. M., S. Khuri, and P. J. Gane. 2000. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64:153-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 14.Enguita, F. J., L. O. Martins, A. O. Henriques, and M. A. Carrondo. 2003. Crystal structure of a bacterial endospore coat component. A laccase with enhanced thermostability properties. J. Biol. Chem. 278:19416-19425. [DOI] [PubMed] [Google Scholar]
- 15.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 17.Henriques, A. O., B. W. Beall, and C. P. Moran, Jr. 1997. CotM of Bacillus subtilis, a member of the alpha-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J. Bacteriol. 179:1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henriques, A. O., B. W. Beall, K. Roland, and C. P. Moran, Jr. 1995. Characterization of cotJ, a sigma E-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J. Bacteriol. 177:3394-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95-110. [DOI] [PubMed] [Google Scholar]
- 20.Hirose, I., K. Sano, I. Shioda, M. Kumano, K. Nakamura, and K. Yamane. 2000. Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology 146:65-75. [DOI] [PubMed] [Google Scholar]
- 21.Hullo, M. F., I. Moszer, A. Danchin, and I. Martin-Verstraete. 2001. CotA of Bacillus subtilis is a copper-dependent laccase. J. Bacteriol. 183:5426-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichikawa, H., R. Halberg, and L. Kroos. 1999. Negative regulation by the Bacillus subtilis GerE protein. J. Biol. Chem. 274:8322-8327. [DOI] [PubMed] [Google Scholar]
- 23.Ichikawa, H., and L. Kroos. 2000. Combined action of two transcription factors regulates genes encoding spore coat proteins of Bacillus subtilis. J. Biol. Chem. 275:13849-13855. [DOI] [PubMed] [Google Scholar]
- 24.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 25.Kozuka, S., and K. Tochikubo. 1991. Permeability of dormant spores of Bacillus subtilis to malachite green and crystal violet. J. Gen. Microbiol. 137:607-613. [DOI] [PubMed] [Google Scholar]
- 26.Kroos, L., and Y. T. Yu. 2000. Regulation of sigma factor activity during Bacillus subtilis development. Curr. Opin. Microbiol. 3:553-560. [DOI] [PubMed] [Google Scholar]
- 27.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 28.Kuwana, R., Y. Kasahara, M. Fujibayashi, H. Takamatsu, N. Ogasawara, and K. Watabe. 2002. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology 148:3971-3982. [DOI] [PubMed] [Google Scholar]
- 29.Lai, E. M., N. D. Phadke, M. T. Kachman, R. Giorno, S. Vazquez, J. A. Vazquez, J. R. Maddock, and A. Driks. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 185:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaVallie, E. R., and M. L. Stahl. 1989. Cloning of the flagellin gene from Bacillus subtilis and complementation studies of an in vitro-derived deletion mutation. J. Bacteriol. 171:3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magro, P., P. Marciano, and P. Di Lenna. 1988. Enzymatic oxalate decarboxylation in isolates of Sclerotinia sclerotiorum. FEMS Microbiol. Lett. 49:49-52. [Google Scholar]
- 32.Martins, L. O., C. M. Soares, M. M. Pereira, M. Teixeira, T. Costa, G. H. Jones, and A. O. Henriques. 2002. Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J. Biol. Chem. 277:18849-18859. [DOI] [PubMed] [Google Scholar]
- 33.Moir, A. 1981. Germination properties of a spore coat-defective mutant of Bacillus subtilis. J. Bacteriol. 146:1106-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naclerio, G., L. Baccigalupi, R. Zilhão, M. De Felice, and E. Ricca. 1996. Bacillus subtilis spore coat assembly requires cotH gene expression. J. Bacteriol. 178:4375-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholson, W. L. 2002. Roles of Bacillus endospores in the environment. Cell Mol. Life Sci. 59:410-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biology methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 38.Otto, A., B. Thiede, E. C. Muller, C. Scheler, B. Wittmann-Liebold, and P. Jungblut. 1996. Identification of human myocardial proteins separated by two-dimensional electrophoresis using an effective sample preparation for mass spectrometry. Electrophoresis 17:1643-1650. [DOI] [PubMed] [Google Scholar]
- 39.Ozin, A. J., A. O. Henriques, H. Yi, and C. P. Moran, Jr. 2000. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 182:1828-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozin, A. J., C. S. Samford, A. O. Henriques, and C. P. Moran, Jr. 2001. SpoVID guides SafA to the spore coat in Bacillus subtilis. J. Bacteriol. 183:3041-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 43.Pogliano, K., E. Harry, and R. Losick. 1995. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol. 18:459-470. [DOI] [PubMed] [Google Scholar]
- 44.Roels, S., A. Driks, and R. Losick. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sacco, M., E. Ricca, R. Losick, and S. Cutting. 1995. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J. Bacteriol. 177:372-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serrano, M., S. Hovel, C. P. Moran, Jr., A. O. Henriques, and U. Völker. 2001. Forespore-specific transcription of the lonB gene during sporulation in Bacillus subtilis. J. Bacteriol. 183:2995-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serrano, M., R. Zilhão, E. Ricca, A. J. Ozin, C. P. Moran, Jr., and A. O. Henriques. 1999. A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J. Bacteriol. 181:3632-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevens, C. M., R. Daniel, N. Illing, and J. Errington. 1992. Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takamatsu, H., Y. Chikahiro, T. Kodama, H. Koide, S. Kozuka, K. Tochikubo, and K. Watabe. 1998. A spore coat protein, CotS, of Bacillus subtilis is synthesized under the regulation of σK and GerE during development and is located in the inner coat layer of spores. J. Bacteriol. 180:2968-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takamatsu, H., T. Kodama, T. Nakayama, and K. Watabe. 1999. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J. Bacteriol. 181:4986-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanner, A., and S. Bornemann. 2000. Bacillus subtilis YvrK is an acid-induced oxalate decarboxylase. J. Bacteriol. 182:5271-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanner, A., L. Bowater, S. A. Fairhurst, and S. Bornemann. 2001. Oxalate decarboxylase requires manganese and dioxygen for activity. Overexpression and characterization of Bacillus subtilis YvrK and YoaN. J. Biol. Chem. 276:43627-43634. [DOI] [PubMed] [Google Scholar]
- 53.Webb, C. D., A. Decatur, A. Teleman, and R. Losick. 1995. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J. Bacteriol. 177:5906-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, J., H. Ichikawa, R. Halberg, L. Kroos, and A. I. Aronson. 1994. Regulation of the transcription of a cluster of Bacillus subtilis spore coat genes. J. Mol. Biol. 240:405-415. [DOI] [PubMed] [Google Scholar]
- 55.Zheng, L., R. Halberg, S. Roels, H. Ichikawa, L. Kroos, and R. Losick. 1992. Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J. Mol. Biol. 226:1037-1050. [DOI] [PubMed] [Google Scholar]
- 56.Zheng, L. B., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 2:1047-1054. [DOI] [PubMed] [Google Scholar]
- 57.Zheng, L. B., and R. Losick. 1990. Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 212:645-660. [DOI] [PubMed] [Google Scholar]
- 58.Zilhão, R., G. Naclerio, A. O. Henriques, L. Baccigalupi, C. P. Moran, Jr., and E. Ricca. 1999. Assembly requirements and role of CotH during spore coat formation in Bacillus subtilis. J. Bacteriol. 181:2631-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zilhão, R., M. Serrano, R. Isticato, E. Ricca, C. P. Moran, Jr., and A. O. Henriques. 2004. Interactions among CotB, CotG, and CotH during assembly of the spore coat in Bacillus subtilis. J. Bacteriol. 186:1110-1119. [DOI] [PMC free article] [PubMed]