Abstract
Self-assembling proteins and protein fragments encoded by the Escherichia coli genome were identified from E. coli K-12 strain MG1655. Libraries of random DNA fragments cloned into a series of λ repressor fusion vectors were subjected to selection for immunity to infection by phage λ. Survivors were identified by sequencing the ends of the inserts, and the fused protein sequence was inferred from the known genomic sequence. Four hundred sixty-three nonredundant open reading frame-encoded interacting sequence tags (ISTs) were recovered from sequencing 2,089 candidates. These ISTs, which range from 16 to 794 amino acids in length, were clustered into families of overlapping fragments, identifying potential homotypic interactions encoded by 232 E. coli genes. Repressor fusions identified ISTs from genes in every protein-based functional category, but membrane proteins were underrepresented. The IST-containing genes were enriched for regulatory proteins and for proteins that form higher-order oligomers. Forty-eight (20.7%) homotypic proteins identified by ISTs are predicted to contain coiled coils. Although most of the IST-containing genes are identifiably related to proteins in other bacterial genomes, more than half of the ISTs do not have identifiable homologs in the Protein Data Bank, suggesting that they may include many novel structures. The data are available online at http://oligomers.tamu.edu/.
For many proteins, quaternary structure is intimately coupled to function and stability. This coupling allows the regulation of many cellular processes to be controlled through specific assembly or disassembly of protein complexes as well as by conformational changes that alter how subunits contact one another.
Proteins use a wide variety of quaternary structures to assemble multisubunit complexes. Genome-wide identification of protein interactions by use of genetic (21, 36, 46, 57, 58) or biochemical (15, 19, 41) screens has provided a wealth of insight into the diversity of structures used for self-assembly. In the annotation of predicted open reading frames (ORFs), assembly interactions are an important feature that provides insights into structure and function. In addition, the involvement of a gene product in a multimeric complex suggests strategies for the generation of assembly-based inhibitors for functional studies (18). The possibility that protein interactions represent a large and largely underexploited target for drug discovery has also been discussed (6, 55)
The study of the protein interactome has focused on heterotypic interactions, as these can provide links between proteins of unknown function and proteins of known function. However, homotypic interactions, which are found in both homomultimeric proteins and as subcomplexes of heteromultimeric proteins, may be the most common way to form protein complexes in nature (31). Although by definition self-interaction does not link a protein's function to that of another protein, homotypic interactions are important in the study of protein structure, function, and evolution and should be just as useful as heterotypic interactions as potential targets for disruption in functional studies or drug development.
Homotypic interactions are poorly recovered by both two-hybrid and biochemical interaction screens. It has been shown that a modified version of a one-hybrid system based on fusion proteins to bacteriophage λ repressor can be used to identify homomultimerization domains from the Saccharomyces cerevisiae genome (36). Our general strategy is to sample genomes for self-assembling domains from libraries of genomic DNA fragments cloned downstream of the λ repressor DNA-binding domain. Clones that confer immunity to infection by phage λ identify self-assembling proteins and protein fragments. Here, we describe a more extensive study to identify and partially localize homotypic interaction domains encoded by the Escherichia coli genome.
MATERIALS AND METHODS
Strains, plasmids, and media.
The strains used in this study are derivatives of AG1688 [F′128 lacIq lacZ::Tn5/araD139 Δ(ara-leu)7697 Δ(lac)X74 galE15 galK16 rpsL(Strr) hsdR2 mcrA mcrB1] (20). The repressor fusion libraries were transformed into JH787 [AG1688 (φ80 Su-3)]. The screen for insert dependence was done with LM58 [JH787 (λLM58)] and LM59 [AG1688 (λLM58)]. λLM58 is a λimm21 specialized transducing phage that carries a pL-cat reporter. Repressor fusion libraries were constructed in pLM99 (GenBank accession no. AF308739), pLM100 (GenBank accession no. AF308740), and pLM101 (GenBank accession no. AF308741) (35-37). These vectors contain an amber mutation at codon 103 of the cI segment, between the DNA-binding domain and the DNA insert, which is used for screening for insert dependence (see below). Expression of the fusion proteins is driven by the P7107 promoter (59), a weak constitutive promoter derived from an operatorless PlacUV5. The P7107 promoter contains multiple mutations relative to PlacUV5 and has the sequences TTTATG and TACATT, respectively, at the −35 and −10 hexamers. While we do not know precisely how strong this promoter is, expression is below the basal levels observed in lacIq1 strains from multicopy expression vectors with the lacUV5 promoter lacking the lacO2 and lacO3 operators.
Luria broth (LB) and LB agar were prepared from premixed powders (Difco). 2XYT broth was prepared as described by Miller (38).
Repressor fusion library construction.
E. coli K-12 MG1655 (kindly provided by Debby Siegele) was used to prepare genomic DNA. Fifteen high-complexity libraries were generated by using either a multienzyme approach (22) or physical DNA shearing (43) to generate inserts used for the repressor fusion libraries. Enzymes were purchased from New England Biolabs (Beverly, Mass.) unless indicated otherwise.
For the multienzyme approach, a combination of restriction enzymes was used to partially digest E. coli genomic DNA and generate ends that are compatible with the cloning sites present in pLM99, pLM100, and pLM101. Ten micrograms of genomic DNA was partially digested for 1 h at the temperature recommended by the manufacturer. Equal amounts of separate CviTI (Megabase Research, Lincoln, Neb.) (8 U), BstUI (5 U), RsaI (2.5 U), and HpyCH4V (2.5 U) partial digests were pooled and cloned into the SmaI site of pLM99, pLM100, and pLM101 to generate three libraries (EB099, EB100, and EB101) in different reading frames. For the ET099, ET100, and ET101 libraries, partial digests of E. coli DNA with TaqI (0.2 U) were ligated into repressor fusion vectors digested with BstBI. The EN099, EN100, and EN101 libraries were generated from partially NlaIII (2.5 U)-digested E. coli DNA ligated into repressor fusion vectors digested with SphI. The ES099, ES100, and ES101 libraries were generated by using partially Sau3AI (2.5 U)-digested E. coli DNA ligated into repressor fusion vectors digested with BglII. In all cases, the digested vector DNA was treated with calf intestinal alkaline phosphatase (Roche) prior to use in ligations.
For mechanical shearing, the DNA was fragmented with a HydroShear apparatus (GeneMachines, San Carlos, Calif.) according to the manufacturer's instructions. Five micrograms of genomic DNA was subjected to 20 cycles at speed code 5. The average size of the resultant DNA fragments was about 2 kb. After shearing, the ends were converted to blunt ends by adding 4.5 U of T4 DNA polymerase and 21 U of Klenow fragment supplemented with 250 μM deoxynucleoside triphosphates in 1× EcoPol buffer (10 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 7.5 mM dithiothreitol), the reaction mixture was incubated for 40 min at 25°C. The blunt-ended DNA fragments were repurified with a Qiagen QIAquick PCR purification kit. The EH099, EH100, and EH101 libraries were generated by cloning sheared and DNA polymerase-treated DNA into the three repressor fusion vectors digested with SmaI and treated with alkaline phosphatase as described above.
The complexity of the libraries was estimated from the number of transformants obtained in the absence of phage selection. We estimate that each library contains on the order of 106 independent inserts. To estimate the fraction of the clones that contained inserts, primers flanking the multiple cloning site (cI-up, 5′-AGTATGCAGCCGTCACTTAG-3′, and LM3-R, 5′-GGGGTTATGCTAGTTATTGC-3′) were used to PCR amplify 60 randomly chosen clones from each library. We estimate that 95% of the clones contained a genomic insert from each of the libraries, with a typical insert size of 1,000 ± 500 bp. Amplification reactions were done by PCR with Taq DNA polymerase (Promega).
Selection and screening procedure.
Detailed procedures for selection and screening have been described previously (37). Briefly, ∼107 JH787 transformants containing unamplified fusion libraries were plated on LB-ampicillin-kanamycin plates seeded with 108 PFU of λKH54 and λKH54h80/plate. The KH54 mutation is a deletion of cI, which prevents the selection phage from forming lysogens, which would be immune to λ. λKH54 and λKH54h80 use different receptors to infect E. coli; using both phages simultaneously reduces the background of receptor mutants that would be seen with only one of the two phages. The plates were incubated at 37°C overnight, and survivors were picked into 96-well plates for further analysis. We performed insert dependence tests as described previously (35-37) for the EB099, EB100, and EB101 libraries and found that all of the survivors were insert dependent, as judged by the dependence of repressor function on an amber suppressor that allows translation of the insert. Therefore, the other libraries were not evaluated for insert dependence.
Identification of interacting fragments.
Cultures of isolated colonies were grown overnight in 1.5 ml of 2XYT+Amp broth (200 μg/ml) for plasmid preps in 2-ml-deep well plates (Whatman). Plasmid DNA was extracted from the positive clones by the Promega MagnaSil method on a BioMek 2000 laboratory automation workstation. Plasmid preps were stored at 4°C until sequencing reactions were performed.
Inserts were identified by automated dye terminator DNA sequencing from the cI-up and LM3-R primers. DNA sequencing reactions were done with the ABI Big Dye terminator kit (Applied Biosystems), and sequences were obtained at the Laboratory for Plant Genome Technologies at Texas A&M University. Sequence trace files were processed with Phred (12) or Sequencher (Gene Codes Corp., Ann Arbor, Mich.). The sequences from each end of the inserts were identified by BLAST (1) searches against the E. coli protein database (National Center for Biotechnology Information) located at ftp://ftp.ncbi.nlm.nih.gov/blast/db/ecoli.aa.Z, and the full sequences of the interacting sequence tags (ISTs) encoding self-assembling domains were inferred from the reference E. coli genome sequence. Annotations assigning gene names were from EcoGene (50). The DNA traces, FASTA files, and BLAST reports generated for the identification of ISTs were stored in the Doodle (Database of Oligomerization Domains from Lambda Experiments) database at http://oligomers.tamu.edu (L. Mariño-Ramírez, X. Tang, and J. C. Hu, unpublished data).
RESULTS
Identification of homotypic ISTs by use of repressor fusions.
The general scheme for our selection for gene fragments encoding self-assembling proteins and protein domains is shown in Fig. 1. We constructed a total of 15 libraries containing quasi-random genomic DNA fragments of the E. coli K-12 strain MG1655 as described in Materials and Methods. Each of the repressor fusion libraries was then subjected to selection for phage immunity, and the ends of the inserts from 2,089 survivor clones were sequenced. By comparing the end sequences to the MG1655 (4) reference sequence, we identified the cloned segments in each candidate, which we refer to as ISTs (21). The immune clones identified fall into two categories: ORF-encoded clones (2,005 clones) and non-ORF-encoded clones (84 clones). An ORF-encoded IST is defined as a DNA fragment from a repressor fusion that is read in the same reading frame as it is in annotated E. coli ORFs in the EcoGene database (50). The 2,005 ORF-encoded ISTs identified 463 nonredundant ORF-encoded ISTs. These ISTs were clustered into families of overlapping fragments, identifying potential homotypic interactions in 232 E. coli proteins (Table 1). Most of the non-ORF-encoded fusions were very short, typically 12 to 20 amino acids (aa) in length, similar to those observed in previous studies (23, 60). A current list of ORF-encoded ISTs is available is available in the Doodle database at http://oligomers.tamu.edu.
FIG. 1.
General scheme for selection and analysis of ISTs. Random fragments generated by partial restriction digests or mechanical shearing were cloned into a series of cloning vectors to generate fusions to the DNA-binding domain of λ cI repressor in all three reading frames. These libraries are subjected to selection for repressor function by plating on λ phage. Survivors are identified by sequencing the ends of the inserts and using BLAST to find the corresponding segment in the complete sequence of the E. coli K-12 MG1655 genome. Clones are saved as frozen cultures, and data about each clone are archived in the Doodle database at http://oligomers.tamu.edu. Details for each step are described in the text.
TABLE 1.
Genes containing ISTs identified in this study
| Gene | b no. | COG no. |
|---|---|---|
| agaR | b3131 | COG1349 |
| allR | b0506 | COG1414 |
| ansA | b1767 | COG0252 |
| araA | b0062 | COG2160 |
| argD | b3359 | COG0160 |
| argH | b3960 | COG0165 |
| argP | b2916 | COG0583 |
| argR | b3237 | COG1438 |
| asnC | b3743 | COG1522 |
| atoC | b2220 | COG2204 |
| b0298 | b0298 | COG2963 |
| b0373 | b0373 | COG2963 |
| b0540 | b0540 | COG2963 |
| b1027 | b1027 | COG2963 |
| b2088 | b2088 | COG2963 |
| betB | b0312 | COG1012 |
| caiA | b0039 | COG1960 |
| cdaR | b0162 | COG3835 |
| chbA | b1736 | COG1447 |
| citE | b0616 | COG2301 |
| clpB | b2592 | COG0542 |
| coaD | b3634 | COG0669 |
| cynR | b0338 | COG0583 |
| cynT | b0339 | COG0288 |
| cysN | b2751 | COG2895 |
| deoR | b0840 | COG1349 |
| dfp | b3639 | COG0452 |
| dnaT | b4362 | |
| dnaX | b0470 | COG2812 |
| dps | b0812 | COG0783 |
| dsdC | b2364 | COG0583 |
| ebgR | b3075 | COG1609 |
| emrE | b0543 | COG2076 |
| entA | b0596 | COG1028 |
| epd | b2927 | COG0057 |
| eutL | b2439 | |
| eutM | b2457 | |
| eutT | b2459 | |
| fabR | b3963 | COG1309 |
| fabZ | b0180 | COG0764 |
| farR | b0730 | COG2188 |
| feaB | b1385 | COG1012 |
| fepE | b0587 | COG3765 |
| fliH | b1940 | COG1317 |
| fliN | b1946 | COG1886 |
| folX | b2303 | COG1539 |
| fruR | b0080 | COG1609 |
| fsaA | b0825 | COG0176 |
| fucA | b2800 | COG0235 |
| gadB | b1493 | COG0076 |
| galS | b2151 | COG1609 |
| garL | b3126 | COG3836 |
| garR | b3125 | COG2084 |
| gcvA | b2808 | COG0583 |
| gdhA | b1761 | COG0334 |
| glcG | b2977 | COG3193 |
| gldA | b3945 | COG0371 |
| glgC | b3430 | COG0448 |
| glmM | b3176 | COG1109 |
| glnB | b2553 | COG0347 |
| glnG | b3868 | COG2204 |
| glpR | b3423 | COG1349 |
| gntR | b3438 | COG1609 |
| hdhA | b1619 | COG1028 |
| hemB | b0369 | COG0113 |
| hisI | b2026 | COG0139 |
| hofB | b0107 | COG2804 |
| iadA | b4328 | |
| ibpA | b3687 | COG0071 |
| ibpB | b3686 | COG0071 |
| ilvE | b3770 | COG0115 |
| ilvH | b0078 | COG0440 |
| ilvI | b0077 | COG0028 |
| ilvY | b3773 | COG0583 |
| ispF | b2746 | COG0245 |
| kbaZ | b3132 | |
| kch | b1250 | COG1226 |
| kdgR | b1827 | COG1414 |
| kdtA | b3633 | COG1519 |
| lldR | b3604 | COG2186 |
| lpxD | b0179 | COG1044 |
| lrp | b0889 | COG1522 |
| lsrF | b1517 | COG1830 |
| mhpC | b0349 | COG0315 |
| lsrR | b1512 | COG2390 |
| maeB | b2463 | COG0280 |
| malI | b1620 | COG1609 |
| malM | b4037 | |
| mcrB | b4346 | COG1401 |
| melA | b4119 | COG1486 |
| menG | b3929 | COG0684 |
| metB | b3939 | COG0626 |
| moaC | b0783 | COG0315 |
| mrr | b4351 | COG1715 |
| murC | b0091 | COG0773 |
| mutS | b2733 | COG0249 |
| nuoC | b2286 | COG0852 |
| paaF | b1393 | COG1024 |
| paaI | b1396 | COG2050 |
| panB | b0134 | COG0413 |
| parC | b3019 | COG0188 |
| pdxJ | b2564 | COG0854 |
| phnH | b4100 | COG3625 |
| phoH | b1020 | COG1702 |
| phoU | b3724 | COG0704 |
| pqiB | b0951 | COG3008 |
| priB | b4201 | COG2965 |
| priC | b0467 | COG3923 |
| proC | b0386 | COG0345 |
| prpC | b0333 | COG0372 |
| pspA | b1304 | COG1842 |
| pspF | b1303 | COG1221 |
| pssR | b3763 | COG0583 |
| ptsA | b3947 | COG1080 |
| purE | b0523 | COG0041 |
| radA | b4389 | COG1066 |
| rarA | b0892 | COG2256 |
| recE | b1350 | |
| recR | b0472 | COG0353 |
| recT | b1349 | COG3723 |
| relA | b2784 | COG0317 |
| rhaA | b3903 | |
| ribE | b0415 | COG0054 |
| rihC | b0030 | COG1957 |
| rssA | b1234 | COG1752 |
| rtcR | b3422 | COG1221 |
| ruvA | b1861 | COG0632 |
| ruvB | b1860 | COG2255 |
| selA | b3591 | COG1921 |
| sgcR | b4300 | COG1349 |
| sgcX | b4305 | COG1363 |
| speB | b2937 | COG0010 |
| srlR | b2707 | COG1349 |
| ssnA | b2879 | COG0402 |
| stfR | b1372 | |
| sucB | b0727 | COG0508 |
| surE | b2744 | COG0496 |
| tauD | b0368 | COG2175 |
| thiG | b3991 | COG2022 |
| tyrR | b1323 | COG3283 |
| ulaD | b4196 | COG0269 |
| uxuR | b4324 | COG2186 |
| xseB | b0422 | COG1722 |
| yafC | b0208 | COG0583 |
| yagE | b0268 | COG0329 |
| yaiN | b0357 | COG1937 |
| yajC | b0407 | COG1862 |
| ybaD | b0413 | COG1327 |
| ybaO | b0447 | COG1522 |
| ybbN | b0492 | COG3118 |
| ybcW | b0559 | |
| ybdB | b0597 | COG2050 |
| ybdG | b0577 | COG0668 |
| ybeZ | b0660 | COG1702 |
| ybgC | b0736 | COG0824 |
| ybgF | b0742 | COG1729 |
| ybjQ | b0866 | COG0393 |
| ycaC | b0897 | COG1335 |
| ycaP | b0906 | COG2323 |
| ycdL | b1011 | COG1335 |
| yceH | b1067 | COG3132 |
| yciA | b1253 | COG1607 |
| yciT | b1284 | COG1349 |
| ycjC | b1299 | COG1396 |
| ydaU | b1359 | COG3756 |
| ydfN | b1547 | |
| ydgB | b1606 | COG1028 |
| ydhT | b1669 | |
| yeaU | b1800 | COG0473 |
| yebK | b1853 | COG1737 |
| yebT | b1834 | COG3008 |
| yedX | b1970 | COG2351 |
| yeeX | b2007 | COG2926 |
| yegW | b2101 | COG2188 |
| yeiE | b2157 | COG0583 |
| yeiI | b2160 | COG2771 |
| yfaU | b2245 | COG3836 |
| yfaY | b2249 | COG1058 |
| yfbR | b2291 | COG1896 |
| yfbU | b2294 | COG3013 |
| yfdL | b2355 | |
| yfdQ | b2360 | |
| yfeC | b2398 | |
| yfeD | b2399 | |
| yfeT | b2427 | COG1737 |
| yffL | b2443 | |
| yffS | b2450 | |
| yfhH | b2561 | COG1737 |
| yfhK | b2556 | COG0642 |
| ygaE | b2664 | COG1802 |
| ygbI | b2735 | COG1349 |
| ygbL | b2738 | COG0235 |
| ygeV | b2869 | COG3829 |
| yhaJ | b3105 | COG0583 |
| yhbN | b3200 | COG1934 |
| yhcS | b3243 | COG0583 |
| yheO | b3346 | COG2964 |
| yhfS | b3376 | |
| yhjC | b3521 | COG0583 |
| yi21_1 | b0360 | COG2963 |
| yi21_2 | b1403 | COG2963 |
| yi21_3 | b1997 | COG2963 |
| yi21_4 | b2861 | COG2963 |
| yi21_5 | b3044 | COG2963 |
| yi21_6 | b4272 | COG2963 |
| yiaU | b3585 | COG0583 |
| yiaY | b3589 | COG1454 |
| yicC | b3644 | COG1561 |
| yicR | b3638 | COG2003 |
| yidW | b3695 | COG2186 |
| yihW | b3884 | COG1349 |
| yjbQ | b4056 | COG0432 |
| yjeF | b4167 | COG0063 |
| yjfQ | b4191 | COG1349 |
| yjfR | b4192 | COG2220 |
| yjgI | b4249 | COG1028 |
| yjhU | b4295 | COG2390 |
| ylbA | b0515 | COG3257 |
| ymfK | b1145 | COG1974 |
| ynaI | b1330 | COG0668 |
| yneH | b1524 | COG2066 |
| ynfL | b1595 | COG0583 |
| ynjH | b1760 | |
| ypeA | b2434 | COG0456 |
| yphG | b2549 | |
| yqeB | b2875 | COG1975 |
| yqjI | b3071 | COG1695 |
| yrbD | b3193 | COG1463 |
| yrbK | b3199 | COG3117 |
| ysgA | b3830 | COG0412 |
| ytfH | b4212 | COG1733 |
| ytfL | b4218 | COG1253 |
Annotation of proteins containing homotypic interactions.
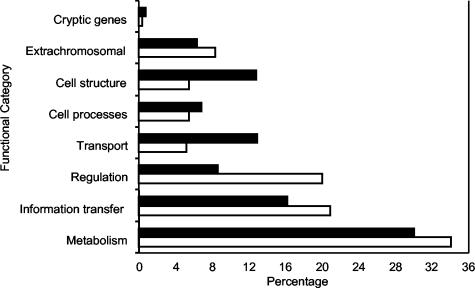
Figure 2 shows the distribution of genes with ISTs based on the functional classification in GenProtEC (48). Repressor fusions identified ISTs from genes in every protein-based functional category. Overall, the distribution of IST-containing genes is similar to that observed for the complete genome. However, relative to the genome, repressor fusions are underrepresented in the functional categories for cell structure and transport (which contains many membrane proteins).
FIG. 2.
Functional annotation of genes containing ISTs. Functional classification categories are from Blattner et al. (4) and Riley (48). Filled and open bars show the percentages of the genome and proteins containing ISTs, respectively, assigned to each functional class. Although MG1655 does not contain any plasmids, extrachromosomal genes include prophage genes that are present in the original genomic DNA used to construct the libraries.
Nevertheless, the ISTs from the 27 nonredundant proteins found in the cell structure and transport categories include 9 that are annotated as integral membrane or transmembrane proteins in SWISSPROT. In these cases, the IST could correspond to a periplasmic or cytoplasmic oligomerization domain from a membrane protein. For example, a fragment of the Kch protein corresponding to an intracellular C-terminal dimerization domain was found as an IST. This domain contains a conserved hydrophobic dimer interface also found in eukaryotic transporters (26). ISTs were also found in the C-terminal domain of YajC, a membrane-associated protein that interacts with the SecA translocation machinery (10, 42), and in EmrE, a multidrug transporter that has been shown to be oligomeric (49).
Genes from the regulation category are overrepresented among the IST-containing genes, consistent with the idea that oligomeric transcription factors are a major component of this functional category. ISTs were found in 62 proteins annotated as transcription factors. The most abundant family of transcriptional regulators in E. coli is the LysR family. Thirteen of the 46 LysR family members were identified. Nine of the 12 DeoR family members of transcriptional regulators were identified. Five of the 15 PurR family members of transcriptional regulators were also identified along with 3 of the 4 RpiR family members.
The diversity of proteins identified by ISTs is also reflected in the evolutionary families they represent. The clusters of orthologous groups of proteins (COGs) database classifies the proteins encoded by 43 sequenced genomes according to their homologous relationships (52). Of the 232 homo-oligomeric proteins identified by ISTs, 210 have a COG assignment, indicating that they are members of conserved protein families. These 210 proteins are distributed among 153 different COGs of 1,905 present in the E. coli genome (Table 1).
We are especially interested in ISTs that might identify new oligomerization domains or motifs. However, we expect that many of the ISTs will be from proteins whose structural basis for assembly has already been determined. To determine how the ISTs are distributed among known and unknown structures, we performed BLAST sequence similarity searches against the Protein Data Bank (PDB) (3) database. Using a cutoff E value of 10−6 and sequence identities of more than 70% to detect E. coli proteins or very close homologs, we found that 23 of the 232 proteins identified by ISTs have structures in the PDB. Twenty-one of the 23 structures found are annotated as homotypic oligomers in the Protein Quaternary Structure (PQS) database (17) with a variety of oligomerization states (Fig. 3a). Although repressor fusions are able to find homodimers, our selection appears to be biased towards recovering higher-order oligomers (Fig. 3b).
FIG. 3.
ISTs from proteins of known structure. (a) Fraction of E. coli ISTs that represent known or inferred structures, as described in the text; (b) distribution of oligomerization states for proteins of known or inferred structure for the complete genome (filled bars) and for E. coli ISTs (open bars).
Coiled-coil predictions for all the E. coli ORFs revealed that 495 ORFs, or 11.5%, are predicted to contain coiled coils by using the COILS2 algorithm (34). Forty-eight homo-oligomeric proteins identified here (20.7%) are predicted to form coiled coils, indicating that the homotypic interaction dataset is enriched for coiled coils. In 40 of these cases (83.3%), the IST includes the region encoding the coiled coil.
Mapping assembly domains within ORFs.
The position of an IST within a gene defines a region sufficient for forming a homotypic interaction. The sizes of the ISTs range from 16 aa for EmrE to 794 aa for YebT. Figure 4a shows the distribution of the lengths of the shortest IST found for each IST-containing gene as a fraction of the length of the complete ORF. Although a majority of the ISTs comprise >90% of the full-length gene product, in many cases the shortest ISTs suggest the presence of a distinct domain that is sufficient for oligomerization. Some genes are represented by single ISTs, whereas in other cases, several ISTs are found for the same gene. The ISTs from the multienzyme libraries generate more multiple hits to the same genes than the libraries made by random shearing. Multiple ISTs in a gene can be used to identify the minimal region or regions involved in a homotypic interaction. For example, we found eight ISTs from ParC, the A subunit of DNA topoisomerase IV; the overlap between these maps the oligomerization domain to aa 333 to 475 (Fig. 4b).
FIG. 4.
Mapping oligomerization domains within IST-containing proteins. (a) Distribution of ISTs as different fractions of the length of the complete ORF. (b) Multiple ISTs define an oligomerization domain in ParC.
In most cases the ISTs overlap, suggesting that a single region is required for oligomerization. However, ISTs only identify regions that are sufficient to self-assemble and do not rule out the possibility that more than one part of a protein can oligomerize. For MutS, the oligomerization domain in the crystal structure does not overlap with the minimal IST we identified between aa 789 and 853 (Fig. 5a). The homodimeric E. coli MutS structure was determined by using a fragment containing aa 1 to 800. The IST corresponds to an additional oligomerization domain at the carboxy terminus of the MutS protein, which allows MutS dimers to form tetramers (32).
FIG. 5.
(a) Segments of MutS in ISTs compared to MutS crystal structures. The filled bar indicates full-length MutS; open bars show ISTs found in this study. Diagonally hatched bars indicate amino acids in crystal structures of MutS from E. coli (1NG9 and 1E3M) and Thermus aquaticus (1EWQ, 1FW6, and 1EWR). (b) Segments of ClpB in ISTs compared to characterized oligomerization domains (39, 53). Filled bar, full-length ClpB; open bars, ISTs found in this study; diagonally hatched bar, N-terminal domain fragment that is monomeric in vitro; vertically hatched bar, C-terminal domain that is oligomeric in vitro.
Two ISTs, containing residues 68 to 558 and 529 to 857, were found in the ClpB protein, a heptameric ATP-dependent chaperone (28). Although the two ISTs overlap, each contains one of the two AAA motif domains identified within ClpB (Fig. 5b). The oligomerization of these two segments of ClpB that have been examined in vitro (39, 53). The C-terminal IST contains sequences shown to form hexamers. Interestingly, the N-terminal IST corresponds to a domain that behaves like a monomer in vitro, indicating that the immunity phenotype of the N-terminal IST could involve either improper folding of this fragment when fused to cI or bridging interactions with some other molecule. The N-terminal domain of ClpB has been shown to bind unfolded protein (53), raising the possibility that cI-ClpB (aa 68 to 558) self-assembles by binding to unfolded parts of the fusion protein.
In four cases, the homotypic ISTs correspond to domains of known structure (SucB, Kch, ArgR, and DnaX). The crystal structure of an oligomeric domain of SucB was determined for a fragment located between aa 173 and 405 (PDB accession no. 1E2O). The minimal IST was found between aa 191 and 405, and the amino acids not present in the IST are unstructured in the crystal structure. The oligomerization domain of the Kch protein is located between aa 241 and 393 (PDB accession no. 1ID1), and its minimal IST was found between aa 229 and 405. The hexamerization domain of the arginine repressor (ArgR) is located between aa 80 and 156 (PDB accession no. 1XXA), and its minimal IST was found between aa 48 and 156. The minimal IST for DnaX was found in between aa 247 and 455, which overlaps with the amino acids seen in the oligomerization domain of the gamma subunits in the clamp loader complex (aa 1 to 373; PDB accession no. 1JR3). Translational frameshifting within dnaX generates two gene products, tau and gamma (5, 14, 56). The domain III fragment (aa 222 to 382) of both tau and gamma has been shown to form homotetramers in vitro (7, 16).
DISCUSSION
Using large-scale functional selections with λ repressor fusions, we identified homotypic interactions for 232 proteins encoded by the E. coli genome. As with a similar study with yeast genomic fragments (37), there are several criteria to support the idea that the ISTs identified here represent bona fide oligomerization domains. First, the strong bias toward fusions from annotated ORFs and in the correct reading frame is consistent with a requirement for correct folding; at higher expression levels, peptides encoded by sequences that are not in frame with annotated ORFs are common (23, 60). Second, in several cases, structural or biochemical evidence in the literature supports the oligomerization state of specific ISTs. Third, in no case do we identify a fusion to a protein where we have been able to find evidence that the fused domain should be monomeric. Nevertheless, for many of the genes identified here, the ISTs should be viewed as strong but not definitive evidence for oligomerization. False positives, while rare, are expected in the repressor system under our conditions. For example, repressor fusions to E. coli dihydrofolate reductase, a well-characterized monomeric protein, are immune to phage infection and purify as a mixture of monomers and dimers (unpublished data).
Among the genes identified by ISTs, we find oligomerization domains that have been previously identified and many that are novel. The proteins with ISTs that have entries or close homologs in the PDB not only serve as positive controls but also give an idea of the range of different homotypic molecular architectures that can be identified by use of repressor fusions. We used annotations from the PQS database (17) to evaluate the oligomerization of ISTs that correspond to known protein structures. PQS uses an automated algorithm to guess the oligomerization state of a protein by evaluating the surface area buried by protein-protein contacts in crystal structures. While PQS annotations are not perfect, they provide a best guess in cases where biochemical data are not available. In the few cases where PQS annotations do not mark IST-containing proteins as homotypic oligomers, there is good reason to believe that they are homo-oligomeric. GlnG (NtrC) and DnaX have structures in the PDB but are not part of the homotypic PQS subset. However, it is well established that NtrC forms homotypic oligomers (30, 45), and repressor fusions have been used to study the oligomerization determinants within NtrC (13). The dnaX gene encodes two proteins, the tau and gamma subunits of DNA polymerase III holoenzyme. Translational frameshifting occurs at residue 430, which is within the DnaX ISTs. Thus, the phage immunity of these constructs could be due either to cI fusions to segments of tau, which dimerizes to hold together the two catalytic subunits in the DNA polymerase III holoenzyme (29), or to cI fusions to segments of the gamma subunit, which forms a homotetramer in vitro (7) and is part of a heteropentameric subcomplex of the clamp loader in DNA polymerase III holoenzyme in vivo (25). For FruR, a member of the PurR family of transcriptional regulators, a nuclear magnetic resonance structure is available for the N-terminal part of the protein (PDB accession no. 1UXD). However, the ISTs for FruR include C-terminal domains that are not present in the nuclear magnetic resonance structure. Although the oligomeric state of FruR is unknown, the sequences corresponding to the FruR IST form homodimers or homotetramers in other members of the PurR family.
From analysis of E. coli proteins of known structure and their close relatives, it is likely that on the order of half of the proteins encoded in the genome are involved in homotypic assemblies or subassemblies of larger heterotypic complexes (L. Mariño-Ramírez and J. C. Hu, unpublished data). The 232 proteins identified by ISTs thus represent a sampling of the possible oligomerization domains encoded in the E. coli genome rather than an exhaustive enumeration.
We find oligomeric proteins in all functional categories. The ISTs are biased toward transcription factors and against membrane proteins. The bias toward transcription factors is likely to reflect the tendency of regulators to be active oligomers at very low expression levels, comparable to those used here to avoid false positives. In addition, the low expression levels of most transcription factors may be favorable for the recovery of ISTs, as abundant dimeric proteins could interfere with the activity of repressor fusions by titrating them into inactive heteromultimeric complexes. The recovery of ISTs will also be dependent on the topology of the repressor fusions; the repressor domains must be close enough to each other to bind the operator half-sites. This may prevent fusions to integral membrane proteins from properly localizing. A recent report of the use of repressor fusion vectors specifically tailored to detect transmembrane domains identifies nine proteins that were missed here (33). That study assayed for the activity of repressor fusion proteins at expression levels above those used here.
Among the 232 proteins, we found several proteins that have been previously identified by others by use of repressor fusions: IbpB (23), PspA (11), and NtrC (13). However, there are E. coli proteins that are known to form active repressor fusions that were not found in our screen, FtsZ (8), MalK (27), PhoB (13), Fur (51), BglG (2), and YigA (23), consistent with the idea that our screen was not saturated. In addition, there are many proteins we have not recovered as ISTs that we think should be recoverable in repressor fusions. These include LacZ, LacI, CAP, TrpB, and many other well-studied stable oligomers. In several cases, we obtained ISTs for some members of a conserved family of proteins but not from others that are likely to oligomerize similarly. The LysR family of transcriptional regulators (LTTRs) is the second largest family of proteins present in the E. coli genome. It is likely that all LTTRs assemble through similar homotypic interactions into dimers or tetramers, but we recovered ISTs for only 13 of the 43 members of the LTTR family found in E. coli. Similarly, we have identified ISTs for some, but not all, members of the PurR family of transcription factors.
Despite subjecting numbers of clones that should be sufficient to provide full coverage to phage selection, the ISTs recovered from both sheared and restriction enzyme libraries are still missing many oligomeric proteins, indicating that nonrandom factors remain. Although random shearing provided a dramatic improvement in coverage compared to previous studies which used partial restriction digests only (54), several factors may skew the recovery of ISTs even from sheared DNA. First, the shearing process itself may not be perfectly random. Second, different numbers of fusion junctions may be possible for different oligomeric proteins, so that some proteins would be overrepresented even in a perfectly random library. Third, genes that are adjacent to other genes that are toxic in high copy will be underrepresented. Fourth, our ability to recover oligomerization domains depends, of course, on the ability of a fusion protein to assemble enough oligomers to provide immunity to phage λ infection. Some oligomeric proteins may simply have dissociation constants that are too high to support repressor activity at the expression levels provided by the weak constitutive promoter in our vectors (37). Based on the phenotypes of fusions to variant GCN4 leucine zippers (59, 61), we estimate that cells expressing dimeric repressor fusions with dissociation constants in the low micromolar range should be immune to λ. However, these estimates are based on many extrapolations from in vitro to in vivo conditions and may not be applicable to other proteins. Note also that steady-state expression levels are not the only factor affecting whether a clone is recovered. Freshly transformed cells must express sufficient repressor activity from the plasmid to confer immunity before encountering a phage particle seeded on the plate. We know that the plating efficiency on phage plates after transformation with plasmids carrying different repressor fusions varies over several orders of magnitude under the conditions we used to prevent the recovery of transformation siblings (data not shown). The observed bias toward recovering higher-order oligomers may reflect their improved ability to bind cooperatively to adjacent operators within OR and OL or to form looped repressor-operator complexes between OR and OL (9, 47).
While the present screen has not reached saturation, the ISTs we identified already provide us with a wealth of information about specific E. coli proteins and about oligomeric proteins in general. The identification of an IST defines oligomerization as a biochemical property of the protein containing it and often maps the oligomerization domain within the protein coding sequence. This is often the only functional annotation for hypothetical ORFs and may provide an entry for further studies of biological function. For example, repressor fusions can be used to study how the activity of specific proteins is modulated by controlling oligomerization (2, 24).
In many cases, ISTs identify an oligomerization domain in a specific region of the protein. This suggests the existence of a contiguous and independent folding unit in the protein that drives oligomerization. In other cases, the ISTs found are very close to the full length of the protein. In some cases, the entire protein may be necessary to observe the homotypic interaction. For example, we recovered multiple clones encoding FolX fusions to aa 1 to 120, suggesting that the entire protein was required for a homotypic interaction. FolX forms an octameric ring-like structure where the entire protein appears to be required for proper folding (PDB accession no. 1B9L) (44). However, we cannot conclude that subdomains are not sufficient for the oligomerization of most proteins where the IST covers the entire ORF, as we may have simply failed to sample the appropriate fragments.
More than half of the proteins identified with repressor fusions do not have an identifiable homotypic homolog in the PDB and may represent new folds. The identification of oligomeric subdomains may be useful for structure determination. Multidomain proteins are often difficult to study, and the ISTs should define independently folding domains that may be more amenable to structure determination than the full-length protein.
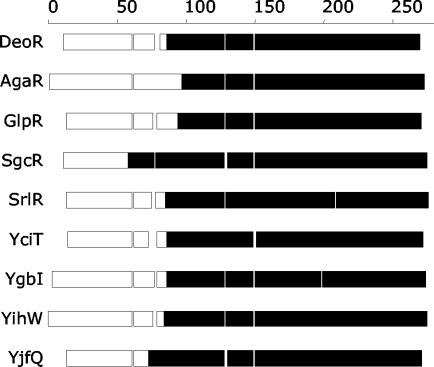
IST data can be combined with evolutionary analysis to provide better domain mapping and functional assignments. For example, multiple-sequence alignments of the DeoR family of transcription factors suggest two conserved domains separated by a nonconserved linker. Sequence analysis identified a helix-turn-helix located towards the N terminus of these proteins. The best-characterized member of this family, the DeoR repressor, appears to be an octamer in solution (40), but the location of the oligomerization domain was not previously described. Nine of the members of the DeoR family of transcriptional regulators were identified by using repressor fusions (Fig. 6). The ISTs include various amounts of the C-terminal end of the ORF, assigning oligomerization function to the conserved C-terminal domain.
FIG. 6.
Minimal ISTs found for the deoR family of transcriptional regulators with homotypic interactions in E. coli. There are 12 members of this family (COG1349) in E. coli K-12, and 9 of them have been identified with repressor fusions. Bars indicate the lengths of the full-length ORFs, aligned based on the multiple-sequence alignment in the COGS database; spaces indicate gaps in the alignment. Filled regions indicate the smallest ISTs found for each gene. The regions involved in the homotypic interaction cluster at the C terminus.
Acknowledgments
This work was supported by Public Health Service Grant R01GM63652-01 from the NIGMS to J.C.H. L.M.-R. was supported in part by a Fulbright/Colciencias/IIE predoctoral fellowship. N.R. was supported in part by the National Science Foundation REU program.
We thank Patricia Klein and Eun-Gyu No for invaluable help with library construction and DNA sequencing, respectively. Rodolfo Aramayo, John Mullet, Debby Siegele, and members of the Hu lab provided useful advice and discussions. Additional technical assistance was provided at various stages of the project by Barbara Blum, Brian Hatten, and Svenja Simon-Marshall.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amster-Choder, O., and A. Wright. 1992. Modulation of the dimerization of a transcriptional antiterminator protein by phosphorylation. Science 257:1395-1398. [DOI] [PubMed] [Google Scholar]
- 3.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 5.Blinkowa, A. L., and J. R. Walker. 1990. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 18:1725-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cochran, A. G. 2000. Antagonists of protein-protein interactions. Chem. Biol. 7:R85-94. [DOI] [PubMed] [Google Scholar]
- 7.Dallmann, H. G., and C. S. McHenry. 1995. DnaX complex of Escherichia coli DNA polymerase III holoenzyme. Physical characterization of the DnaX subunits and complexes. J. Biol. Chem. 270:29563-29569. [PubMed] [Google Scholar]
- 8.Di Lallo, G., D. Anderluzzi, P. Ghelardini, and L. Paolozzi. 1999. FtsZ dimerization in vivo. Mol. Microbiol. 32:265-274. [DOI] [PubMed] [Google Scholar]
- 9.Dodd, I. B., A. J. Perkins, D. Tsemitsidis, and J. B. Egan. 2001. Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes Dev. 15:3013-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duong, F., and W. Wickner. 1997. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 16:2756-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dworkin, J., G. Jovanovic, and P. Model. 2000. The PspA protein of Escherichia coli is a negative regulator of σ54-dependent transcription. J. Bacteriol. 182:311-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 13.Fiedler, U., and V. Weiss. 1995. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induces dimerization of the receiver modules. EMBO J. 14:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flower, A. M., and C. S. McHenry. 1990. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl. Acad. Sci. USA 87:3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 16.Glover, B. P., A. E. Pritchard, and C. S. McHenry. 2001. tau binds and organizes Escherichia coli replication proteins through distinct domains: domain III, shared by gamma and tau, oligomerizes DnaX. J. Biol. Chem. 276:35842-35846. [DOI] [PubMed] [Google Scholar]
- 17.Henrick, K., and J. M. Thornton. 1998. PQS: a protein quaternary structure file server. Trends Biochem. Sci. 23:358-361. [DOI] [PubMed] [Google Scholar]
- 18.Herskowitz, I. 1987. Functional inactivation of genes by dominant negative mutations. Nature 329:219-222. [DOI] [PubMed] [Google Scholar]
- 19.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 20.Hu, J., N. Newell, B. Tidor, and R. Sauer. 1993. Probing the roles of residues at the e and g positions of the GCN4 leucine zipper by combinatorial mutagenesis. Protein Sci. 2:1072-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jappelli, R., and S. Brenner. 1999. A genetic screen to identify sequences that mediate protein oligomerization in Escherichia coli. Biochem. Biophys. Res. Commun. 266:243-247. [DOI] [PubMed] [Google Scholar]
- 24.Jappelli, R., and S. Brenner. 1996. Interaction between cAMP-dependent protein kinase catalytic subunit and peptide inhibitors analyzed with λ repressor fusions. J. Mol. Biol. 259:575-578. [DOI] [PubMed] [Google Scholar]
- 25.Jeruzalmi, D., M. O'Donnell, and J. Kuriyan. 2001. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell 106:429-441. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, Y., A. Pico, M. Cadene, B. T. Chait, and R. MacKinnon. 2001. Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel. Neuron 29:593-601. [DOI] [PubMed] [Google Scholar]
- 27.Kennedy, K. A., and B. Traxler. 1999. MalK forms a dimer independent of its assembly into the MalFGK2 ATP-binding cassette transporter of Escherichia coli. J. Biol. Chem. 274:6259-6264. [DOI] [PubMed] [Google Scholar]
- 28.Kim, K. I., G. W. Cheong, S. C. Park, J. S. Ha, K. M. Woo, S. J. Choi, and C. H. Chung. 2000. Heptameric ring structure of the heat-shock protein ClpB, a protein-activated ATPase in Escherichia coli. J. Mol. Biol. 303:655-666. [DOI] [PubMed] [Google Scholar]
- 29.Kim, S., H. G. Dallmann, C. S. McHenry, and K. J. Marians. 1996. tau couples the leading-and lagging-strand polymerases at the Escherichia coli DNA replication fork. J. Biol. Chem. 271:21406-21412. [DOI] [PubMed] [Google Scholar]
- 30.Klose, K. E., A. K. North, K. M. Stedman, and S. Kustu. 1994. The major dimerization determinants of the nitrogen regulatory protein NTRC from enteric bacteria lie in its carboxy-terminal domain. J. Mol. Biol. 241:233-245. [DOI] [PubMed] [Google Scholar]
- 31.Klotz, I., D. Darnall, and N. Langerman. 1975. Quaternary structure of proteins, p. 293-411. In H. Neurath & R. L. Hill (ed.), The proteins, vol. 1. Academic Press, New York, N.Y.
- 32.Lamers, M. H., A. Perrakis, J. H. Enzlin, H. H. Winterwerp, N. de Wind, and T. K. Sixma. 2000. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature 407:711-717. [DOI] [PubMed] [Google Scholar]
- 33.Leeds, J. A., D. Boyd, D. R. Huber, G. K. Sonoda, H. T. Luu, D. M. Engelman, and J. Beckwith. 2001. Genetic selection for and molecular dynamic modeling of a protein transmembrane domain multimerization motif from a random Escherichia coli genomic library. J. Mol. Biol. 313:181-195. [DOI] [PubMed] [Google Scholar]
- 34.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 35.Mariño-Ramírez, L., L. Campbell, and J. C. Hu. 2003. Screening peptide/protein libraries fused to the λ repressor DNA binding domain in E. coli cells. Methods Mol. Biol. 205:235-250. [DOI] [PMC free article] [PubMed]
- 36.Mariño-Ramírez, L., and J. C. Hu. 2002. Isolation and mapping of self-assembling protein domains encoded by the Saccharomyces cerevisiae genome using lambda repressor fusions. Yeast 19:641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mariño-Ramírez, L., and J. C. Hu. 2002. Using λ repressor fusions to isolate and characterize self-assembling domains, p. 375-393. In E. Golemis and I. Serebriiskii (ed.), Protein-protein interactions: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Mogk, A., C. Schlieker, C. Strub, W. Rist, J. Weibezahn, and B. Bukau. 2003. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 278:17615-17624. [DOI] [PubMed] [Google Scholar]
- 40.Mortensen, L., G. Dandanell, and K. Hammer. 1989. Purification and characterization of the deoR repressor of Escherichia coli. EMBO J. 8:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman, J. R., and A. E. Keating. 2003. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300:2097-2101. [DOI] [PubMed] [Google Scholar]
- 42.Nouwen, N., and A. J. Driessen. 2002. SecDFyajC forms a heterotetrameric complex with YidC. Mol. Microbiol. 44:1397-1405. [DOI] [PubMed] [Google Scholar]
- 43.Oefner, P. J., S. P. Hunicke-Smith, L. Chiang, F. Dietrich, J. Mulligan, and R. W. Davis. 1996. Efficient random subcloning of DNA sheared in a recirculating point-sink flow system. Nucleic Acids Res. 24:3879-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ploom, T., C. Haussmann, P. Hof, S. Steinbacher, A. Bacher, J. Richardson, and R. Huber. 1999. Crystal structure of 7, 8-dihydroneopterin triphosphate epimerase. Struct. Fold. Des. 7:509-516. [DOI] [PubMed] [Google Scholar]
- 45.Porter, S. C., A. K. North, A. B. Wedel, and S. Kustu. 1993. Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev. 7:2258-2273. [DOI] [PubMed] [Google Scholar]
- 46.Rain, J. C., L. Selig, H. De Reuse, V. Battaglia, C. Reverdy, S. Simon, G. Lenzen, F. Petel, J. Wojcik, V. Schachter, Y. Chemama, A. Labigne, and P. Legrain. 2001. The protein-protein interaction map of Helicobacter pylori. Nature 409:211-215. [DOI] [PubMed] [Google Scholar]
- 47.Revet, B., B. von Wilcken-Bergmann, H. Bessert, A. Barker, and B. Muller-Hill. 1999. Four dimers of lambda repressor bound to two suitably spaced pairs of lambda operators form octamers and DNA loops over large distances. Curr. Biol. 9:151-154. [DOI] [PubMed] [Google Scholar]
- 48.Riley, M. 1998. Genes and proteins of Escherichia coli K-12. Nucleic Acids Res. 26:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rotem, D., N. Sal-man, and S. Schuldiner. 2001. In vitro monomer swapping in EmrE, a multidrug transporter from Escherichia coli, reveals that the oligomer is the functional unit. J. Biol. Chem. 276:48243-48249. [DOI] [PubMed] [Google Scholar]
- 50.Rudd, K. E. 2000. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 28:60-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stojiljkovic, I., and K. Hantke. 1995. Functional domains of the Escherichia coli ferric uptake regulator protein (Fur). Mol Gen Genet. 247:199-205. [DOI] [PubMed] [Google Scholar]
- 52.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 53.Tek, V., and M. Zolkiewski. 2002. Stability and interactions of the amino-terminal domain of ClpB from Escherichia coli. Protein Sci. 11:1192-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thorstenson, Y. R., S. P. Hunicke-Smith, P. J. Oefner, and R. W. Davis. 1998. An automated hydrodynamic process for controlled, unbiased DNA shearing. Genome Res. 8:848-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toogood, P. L. 2002. Inhibition of protein-protein association by small molecules: approaches and progress. J. Med. Chem. 45:1543-1558. [DOI] [PubMed] [Google Scholar]
- 56.Tsuchihashi, Z., and A. Kornberg. 1990. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. USA 87:2516-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 58.Walhout, A. J., R. Sordella, X. Lu, J. L. Hartley, G. F. Temple, M. A. Brasch, N. Thierry-Mieg, and M. Vidal. 2000. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287:116-122. [DOI] [PubMed] [Google Scholar]
- 59.Zeng, X., A. M. Herndon, and J. C. Hu. 1997. Buried asparagines determine the dimerization specificities of leucine zipper mutants. Proc. Natl. Acad. Sci. USA 94:3673-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, Z., A. Murphy, J. C. Hu, and T. Kodadek. 1999. Genetic selection of short peptides that support protein oligomerization in vivo. Curr. Biol. 9:417-420. [DOI] [PubMed] [Google Scholar]
- 61.Zhu, H., S. Celinski, J. Scholtz, and J. Hu. 2000. The contribution of buried polar groups to the conformational stability of the GCN4 coiled-coil. J. Mol. Biol. 300:1379-1389. [DOI] [PubMed] [Google Scholar]