Abstract
Bacillus subtilis produces hemicellulases capable of releasing arabinosyl oligomers and arabinose from plant cell walls. In this work, we characterize the transcriptional regulation of three genes encoding arabinan-degrading enzymes that are clustered with genes encoding enzymes that further catabolize arabinose. The abfA gene comprised in the metabolic operon araABDLMNPQ-abfA and the xsa gene located 23 kb downstream most probably encode α-l-arabinofuranosidases (EC 3.2.1.55). Here, we show that the abnA gene, positioned immediately upstream from the metabolic operon, encodes an endo-α-1,5-arabinanase (EC 3.2.1.99). Furthermore, by in vivo RNA studies, we inferred that abnA and xsa are monocistronic and are transcribed from σA-like promoters. Transcriptional fusion analysis revealed that the expression of the three arabinases is induced by arabinose and arabinan and is repressed by glucose. The levels of induction by arabinose and arabinan are higher during early postexponential growth, suggesting a temporal regulation. Moreover, the induction mechanism of these genes is mediated through negative control by the key regulator of arabinose metabolism, AraR. Thus, we analyzed AraR-DNA interactions by in vitro quantitative DNase I footprinting and in vivo analysis of single-base-pair substitutions within the promoter regions of xsa and abnA. The results indicate that transcriptional repression of the abfA and xsa genes is achieved by a tightly controlled mechanism but that the regulation of abnA is more flexible. We suggest that the expression of genes encoding extracellular degrading enzymes of arabinose-containing polysaccharides, transport systems, and intracellular enzymes involved in further catabolism is regulated by a coordinate mechanism triggered by arabinose via AraR.
Hemicellulose is the second-most abundant renewable biomass polymer, next to cellulose. This fraction of plant cell walls comprises a complex mixture of polysaccharides that includes xylans, arabinans, galactans, mannans, and glucans. Enzymes responsible for degrading plant cell wall polysaccharides have many agroindustrial applications, such as biobleaching of pulps in the pulp and paper industry, improving digestibility of animal feedstock, processing of flour in the baking industry, and clarifying juices (references 6, 26, and 27, and references therein). Although many hemicellulases have been purified and characterized from both fungi and bacteria, including mesophilic and thermophilic Bacillus spp., knowledge concerning regulation at the molecular level of hemicellulolytic genes is scarce (reference 34 and references therein).
The saprophytic endospore-forming gram-positive bacterium Bacillus subtilis participates in enzymatic dissolution of plant cell walls in its natural reservoir, the soil. l-Arabinose is distributed in hemicelluloses and is present at high concentrations in arabinoxylans, arabinogalactans, and arabinan. The latter is composed of α-1,5-linked l-arabinofuranosyl units, some of which are replaced with α-1,3- and α-1,2-linked chains of l-arabinofuranosyl residues (2). The two major enzymes that hydrolyze arabinan are α-l-arabinofuranosidases (AFs) (EC 3.2.1.55) and endo-α-1,5-arabinanases (ABNs) (EC 3.2.1.99). AFs remove arabinose side chains, allowing ABNs to attack the glycosidic bonds of the arabinan backbone and releasing a mixture of arabinooligosaccharides and l-arabinose (9). B. subtilis synthesizes at least three enzymes, an ABN and two AFs, capable of releasing arabinosyl oligomers and l-arabinose from plant cell walls (12, 13, 28, 39).
Previous work by our group characterized the genes involved in the utilization of l-arabinose that belong to the araABDLMNPQ-abfA operon (32) and the divergently arranged araE and araR genes (31, 33), located in distinct regions of the B. subtilis chromosome. The first three genes of the l-arabinose metabolic operon, araA, araB, and araD, encode the enzymes required for the intracellular conversion of l-arabinose into d-xylulose 5-phosphate, which is further catabolized through the pentose phosphate pathway (30). The product of the araE gene is a permease, the main transporter of l-arabinose into the cell (33). The araR gene encodes the regulatory protein of l-arabinose metabolism in B. subtilis, negatively controlling the expression from the l-arabinose-inducible promoters of the ara genes (22, 23). Additionally, the ara regulon is subjected to carbon catabolite repression by glucose and glycerol (11). The last gene of the l-arabinose metabolic operon, abfA, and the xsa gene located 23 kb downstream from the operon (32, 40) (Fig. 1) most probably encode AFs belonging to the glycosyl hydrolase (GH) family G51 (see the Carbohydrate-Active Enzymes website [http://afmb.cnrs-mrs.fr/∼cazy/CAZY]). The gene abnA, located immediately upstream from the metabolic operon (32, 40) (Fig. 1), most likely encodes an ABN grouped in the GH43 family (http://afmb.cnrs-mrs.fr/∼cazy/CAZY).
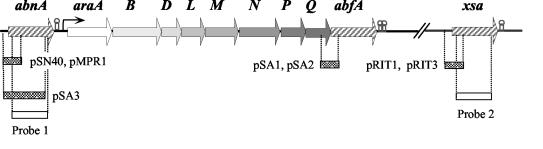
FIG. 1.
Localization of the abnA, abfA, and xsa genes on the B. subtilis chromosome. The three genes are represented by striped arrows pointing in the direction of transcription. abfA belongs to the araABDLMNPQ-abfA metabolic operon, abnA is located immediately upstream, and the xsa gene is positioned 23 kb downstream of the metabolic operon. Hairpin structures indicate potential terminators. The dotted boxes below the physical map represent the extension of the inserts fused to the lacZ gene in the indicated plasmids, and the open boxes represent the fragments used as probes for Northern analysis of the abnA (probe 1) and xsa (probe 2) transcripts. Plasmids pMPR1, pSA1, and pRIT3 were integrated into the host chromosome by means of a single-crossover (Campbell-type) recombinational event that occurred in the region of homology of the resulting strains (Table 1). Linearized DNA from plasmids pSN40, pSA3, pSA2, and pRIT1 was used to transform B. subtilis strains (Table 1), and the fusions were integrated into the chromosome via double recombination with the back and front sequences of the amyE gene.
Our work focuses on the regulation of expression of the abfA, xsa, and abnA genes. Additionally, functional analysis of abnA revealed that this gene encodes an ABN. In vivo RNA studies demonstrated the monocistronic nature of xsa and abnA and allowed us to characterize their promoter regions. We show that the expression of the abfA, xsa, and abnA genes is positively controlled at the transcriptional level by arabinose and arabinan, repressed by glucose, and most likely subjected to temporal regulation. Moreover, in vivo and in vitro studies indicate that the transcription factor AraR plays a major role in the control of the expression of the arabinan-degrading genes. It is hypothesized that coordinate expression of genes involved in the degradation of arabinose-containing polysaccharides is triggered by arabinose and mediated by AraR.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. subtilis strains used in this study are listed in Table 1. Escherichia coli DH5α (Gibco BRL) was used for routine molecular cloning work and was grown on Luria-Bertani (LB) medium (20). Ampicillin (75 μg ml−1), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg ml−1), or IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) was added as appropriate. The B. subtilis strains were grown on LB medium (20) or C minimal medium (24), and chloramphenicol (5 μg ml−1) or kanamycin (10 μg ml−1) was added as appropriate. Solid medium was made with LB or C minimal medium containing 1.6% (wt/vol) Bacto Agar (Difco). The abnA phenotype was tested in C minimal medium (24) plates supplemented with 0.4% (wt/vol) debranched arabinan (Megazyme). The amyE phenotype was tested by plating strains on tryptose blood agar base medium (Difco) containing 1% (wt/vol) potato starch, and after overnight incubation, the plates were flooded with a solution of 0.5% (wt/vol) I2 and 5.0% (wt/vol) KI for the detection of starch hydrolysis. For the β-galactosidase assays and RNA preparation, the B. subtilis strains were grown in liquid C minimal medium supplemented with 1% (wt/vol) casein hydrolysate. When necessary, 0.4% (wt/vol) l-arabinose, 0.4% (wt/vol) arabinan (sugar beet; Megazyme), and 0.4% (wt/vol) glucose were added to the cultures. The transformation of E. coli and B. subtilis strains was performed as previously described (23).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype or description | Source or referencea |
|---|---|---|
| 168T+ | Prototroph | F. E. Young |
| IQB215 | ΔaraR::Km | 31 |
| IQB405 | amyE::[xsa′-lacZ cat] | pRIT1 → 168T+b |
| IQB406 | amyE::[xsa′-lacZ cat] ΔaraR::Km | pRIT1 → IQB215b |
| IQB407 | xsa::pRIT3[xsa′-lacZ cat] | pRIT3 → 168T+ |
| IQB410 | amyE::[abnA′-lacZ cat] | pSN40 → 168T+b |
| IQB411 | amyE::[abnA′-lacZ cat] ΔaraR::Km | pSN40 → IQB215b |
| IQB412 | abnA::pMPR1[abnA′-lacZ cat] | pMPR1 → 168T+ |
| IQB413 | ΔabnA::Km | pMPR4 → 168T+b |
| IQB448 | amyE::[abnA′-lacZ cat] | pSA3 → 168T+b |
| IQB449 | amyE::[abnA′-lacZ cat] ΔaraR::Km | pSA3 → IQB215b |
| IQB450 | abfA::pSA1[abfA′-lacZ cat] | pSA1 → 168T+ |
| IQB451 | amyE::[abfA′-lacZ cat] | pSA2 → 168T+b |
| IQB453 | abfA::pSA1[abfA′-lacZ cat] ΔaraR::Km | pSA1 → IQB215 |
| IQB464 | amyE::[abnA′(−38 G → T)-lacZ cat] | pZI15 → 168T+b |
| IQB465 | amyE::[xsa′(−27 G → T)-lacZ cat] | pZI19 → 168T+b |
The arrows indicate transformation and point from donor DNA to the recipient strain.
Transformation was carried out with linearized plasmid DNA.
DNA manipulations and sequencing.
DNA manipulations were carried out as described by Sambrook et al. (29). Restriction enzymes were purchased from MBI Fermentas and New England Biolabs and used according to the manufacturer's instructions. DNA was eluted from agarose gels with a GENECLEANII kit (Bio101). DNA sequencing was performed with a Sequenase version 2.0 kit (USB) or an ABI PRIS BigDye terminator ready reaction cycle sequencing kit (Applied Biosystems). PCR amplifications were done by using high-fidelity native Pfu DNA polymerase (Stratagene), and the products were purified by using a QIAquick PCR purification kit (QIAGEN).
Construction of plasmids and strains.
The xsa and abnA promoter regions were amplified by PCR of chromosomal DNA of wild-type strain B. subtilis 168T+ with oligonucleotides ARA87 and ARA88 and oligonucleotides ARA85 and ARA86 (Table 2), respectively. Plasmid pRIT1 was obtained by cloning a 281-bp EcoRI-EcoRV DNA fragment, obtained from the PCR product bearing the xsa promoter region, into pSN32 (22) digested with EcoRI and SmaI. An insertion of the same fragment into pJM783 (25) restricted with EcoRI and SmaI yielded plasmid pRIT3. To construct plasmid pSN40, the PCR product bearing the abnA promoter region was digested with EcoRI and XmnI, and the resulting 292-bp product was inserted into pSN32 restricted with EcoRI and SmaI. Plasmid pMPR1 was obtained by ligation of a 300-bp EcoRI-BamHI DNA fragment from pSN40, containing the abnA promoter region, to the pJM783 EcoRI-BamHI sites. A PCR product bearing the abnA promoter region, amplified from chromosomal DNA of wild-type strain B. subtilis 168T+ with oligonucleotides ARA85 and ARA89 (Tables 1 and 2) and digested with EcoRI-SacI, was inserted into the pBluescript II SK(+) (Stratagene) EcoRI-SacI sites, to yield pMPR2. To construct plasmid pSA1, a 687-bp DNA fragment from the araABDLMNPQ-abfA operon carrying the 3′ end of the araQ gene and the 5′ end of the abfA gene (Fig. 1), obtained by EcoRI-HincII digestion of plasmid pTN13 (32), was ligated to the pJM783 (25) EcoRI-SmaI sites. The same DNA fragment inserted into pSN32 restricted with EcoRI and SmaI yielded pSA2. Plasmid pSA3 was constructed by insertion of a 1,024-bp PCR product, amplified from chromosomal DNA of wild-type strain B. subtilis 168T+ with oligonucleotides ARA85 and ARA92 (Tables 1 and 2) and digested with EcoRI-BglII, into the pSN32 EcoRI-BamHI sites. The sequences of all the inserts obtained by PCR were confirmed by DNA sequencing.
TABLE 2.
B. subtilis oligonucleotides and sequences used in this study
| Primer | Sequences (5′ → 3′)a | Complementary sequence |
|---|---|---|
| ARA1 | −39TAAGGGTAACTATTGCCG−22 | pSN32 |
| ARA32 | +124GAATTCAGGATCCTTTGTCTGAAGC+100 | araA |
| ARA72 | +35AGTGTATCAACAAGCTGG+17 | pSN32 |
| ARA85 | −171GTGATGATTATGAATTCGCGG−157 | abnA |
| ARA86 | +160CACTCGAAAAGTGTAAGAAGCG+139 | abnA |
| ARA87 | −207AAAATAGCGGATTACGGCATCG−186 | xsa |
| ARA88 | +189CCTAAATGCTCAGCAAAATGACC+167 | xsa |
| ARA89 | +936ATCCAAAATGGTGCCTCCG+918 | abnA |
| ARA90 | +124GAATCACTGCTTGATGTTCAGACATGCC+97 | xsa |
| ARA91 | +1509GTATTGTCTGCAGGATTCCGG+1489 | xsa |
| ARA92 | +879GCCTGTAATGCTTTTAGATCTTCC+856 | abnA |
| ARA113 | −44GAAAATGTCGTTGACATTTACGAACATATATAATATGG−7 | xsa |
| ARA114 | −7CCATATTATATATGTTCGTAAATGTCAACGACATTTTC−44 | xsa |
| ARA129 | −52GATTCTATTTTTTTTTCTGTACAAATTACAGC−21 | abnA |
| ARA130 | −21GCTGTAATTTGTACAGAAAAAAAAATAGAATC−52 | abnA |
The numbers in the primers sequences refer to the positions of the sequence in abnA or xsa relative to the transcription start point of each gene or to the positions in pSN32 relative to the EcoRI site (+1) in the multiple cloning site. The following restriction sites are underlined in the oligonucleotides sequences: EcoRI, GAATTC; PstI, CTGCAG; BglII, AGATCT; BamHI, GGATCC.
To construct pZI15, which carries a single-base-pair substitution in the AraR binding site ORB1 (−38 G→T), plasmid pZI13, a pBluescript II SK(+) (Stratagene) derivative containing a 302-bp EcoRI-BamHI insert from pSN40, was used as the target DNA for site-directed mutagenesis with the QuikChange kit (Stratagene) and the overlapping oligonucleotides ARA129 and ARA130 (Table 2). An EcoRI-BamHI DNA fragment from the resulting plasmid, pZI14, was subcloned into those sites of pSN32 to yield pZI15. To perform a single-nucleotide substitution in the AraR binding site ORX2 (−27 G→T), plasmid pZI17, a pBluescript II SK(+) (Stratagene) derivative containing a 291-bp EcoRI-BamHI insert from pRIT1, was used as target DNA for site-directed mutagenesis and for the overlapping primers ARA113 and ARA114 (Table 2), as described above. An EcoRI-BamHI DNA fragment from the resulting plasmid, pZI18, was ligated to the pSN32 EcoRI-BamHI sites to obtain pZI19. The single point mutations were confirmed by DNA sequencing.
Linearized plasmid DNA from pSN40, pRIT1, pSA2, pSA3, pZI15, and pZI19 (Fig. 1), carrying the different promoter-lacZ transcriptional fusions, was used to transform B. subtilis strains (Table 1), and the fusions were integrated into the chromosome via double recombination with the back and front sequences of the amyE gene. This event led to the disruption of the amyE locus and was confirmed as described above. Plasmids pRIT3, pMPR1, and pSA1 (Fig. 1) were integrated into the host chromosome by means of a single-crossover (Campbell-type) recombinational event that occurred in the region of homology (Table 1).
A PCR product containing the entire abnA gene, amplified from chromosomal DNA of wild-type strain B. subtilis 168T+ with oligonucleotides ARA32 and ARA85 (Tables 1 and 2), was digested with DdeI-BamHI, and this fragment bearing the 3′ end of abnA was inserted in the pSN32 SmaI-BamHI sites to yield pSN41. Plasmid pSN42 was the result of the insertion of a 535-bp EcoRI-BamHI DNA fragment from pSN41 into the pBluescript II SK(+) (Stratagene) EcoRI-BamHI sites. To construct plasmid pMPR3, a 1.5-kb SphI-SmaI DNA fragment from pAH248 (31) containing a kanamycin resistance (Kmr) gene was inserted into the pSN42 SphI-SmaI site. By subcloning a 1,626-bp SalI DNA fragment from pSN40 (see above) at the unique SalI site of pMPR3, pMPR4 was obtained. This plasmid was used, after linearization, to delete the abnA gene from the wild-type B. subtilis 168T+ chromosome.
β-Galactosidase assays.
Strains of B. subtilis harboring the transcriptional lacZ fusions were grown as described above. Samples of cell culture were collected 2 h (exponential growth phase) and 4 h (late exponential growth phase) after induction, and the level of β-galactosidase activity was determined as previously described (32). The ratio of β-galactosidase activity from cultures grown in the presence or absence of an inducer (arabinose or arabinan) was taken as a measure of AraR repression in each strain analyzed (regulation factor). The ratio of β-galactosidase activity from cultures grown in the presence or absence of glucose was taken as a measure of glucose repression (glucose repression factor).
RNA preparation, Northern blot analysis, and primer extension analysis.
B. subtilis strains were grown as described above, and cells were harvested 2 h after induction. Total RNA was prepared by using an RNeasy kit (QIAGEN) according to the manufacturer's instructions. For Northern blot analysis, 10 μg of total RNA was run in a 1.2% (wt/vol) agarose formaldehyde denaturing gel and transferred to positively charged Hybond-N+ (Amersham) nylon membranes according to standard procedures (29). A size determination was done by using an RNA ladder (9 to 0.5 kb [New England Biolabs] or 6 to 0.2 kb [MBI Fermentas]). A DNA fragment of 763 bp used as an abnA probe was obtained by PCR amplification of chromosomal DNA with primers ARA85 and ARA89 followed by PstI digestion. PCR amplification with chromosomal DNA as a template and primers ARA87 and ARA91 (Table 2) yielded a DNA fragment that, after digestion with BclI, resulted in a 1.4-kb xsa DNA probe. The DNA probes were labeled with a Megaprime DNA labeling system (Amersham) and [α-32P]dCTP (3,000 Ci/mmol; Amersham).
Primer extension analysis was performed essentially as described by Sambrook et al. (29). Primer ARA86, complementary to the abnA sequence (Table 2), and primer ARA90, complementary to the xsa sequence (Table 2), were end labeled with [γ-32P]ATP (3,000 Ci/mmol) by using T4 polynucleotide kinase (NEB). A total of 2.5 ρmol of each labeled primer was mixed with 50 to 100 μg of RNA in separate experiments, denatured by heating to 85°C for 10 min, and annealed by incubation at 45°C overnight. The extension reaction was conducted for 2 h at 37°C by using 50 U of avian Moloney murine leukemia virus reverse transcriptase (RevertAid; MBI Fermentas). Analysis of the extended products was carried out on 6% (wt/vol) polyacrylamide urea gels.
DNase I footprinting.
The target DNA fragments from the xsa and abnA promoters were obtained by PCR amplification with oligonucleotides ARA1 and ARA72 (Table 2) using pRIT1 and pSN40 (see above) as templates and yielding 339- and 350-bp DNA fragments, respectively. The labeling of the fragments and the DNase I footprinting experiments were performed by using purified native AraR as previously described by Mota et al. (22). The apparent dissociation constant (Kapp) for the different operators was determined as the total concentration of AraR required for half-maximal site protection.
RESULTS
The abfA and xsa genes and functional analysis of the abnA gene
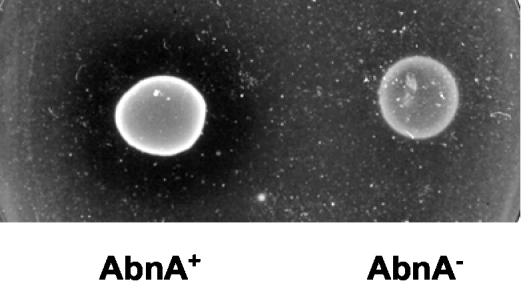
The abfA and the xsa genes most probably encode AFs (EC 3.2.1.55). The amino acid sequence of AbfA displays a high level of identity (71%) to characterized AbfA from Geobacillus stearothermophilus T-6 (8), and AF activity was reported for the B. subtilis abfA gene product (40). Xsa is highly homologous to characterized AFs from Thermobacillus xylanilyticus AbfD3 (64% identity) (3), Clostridium cellulovorans ArfA (60% identity) (15), and Clostridium stercorarium ArfB (56% identity) (42). Based on primary amino acid sequence analysis, the abnA gene most likely encodes an ABN (EC 3.2.1.99), with 52% identity to a characterized thermostable ABN from Bacillus thermodinitrificans (36) and 38% identity to ArbA from Cellvibrio japonicus (19). Previously, Sakamoto et al. (28) reported the cloning of the gene ppc from B. subtilis strain IFO 3134 that encodes an ABN displaying 94% identity to the product of the abnA gene from B. subtilis 168T+. However, it is unclear whether the arabinan-degrading activity measured in that strain reflects the expression of the ppc gene (28). To characterize the function of the abnA gene, we constructed an insertion-deletion mutation in the abnA region (see Materials and Methods). The abnA-null mutant strain (IQB413) (Table 1) was able to grow on minimal medium plates supplemented with debranched arabinan (see Materials and Methods), although more slowly than the wild-type strain. However, the clear halo of hydrolysis observed for the wild-type strain was absent in the mutant (Fig. 2), indicating that the product of the abnA gene is an ABN.
FIG. 2.
Functional analysis of the abnA gene. The B. subtilis wild-type strain 168T+ (AbnA+) and the abnA-null mutant strain IQB413 (AbnA−) were grown on C minimal medium plates (see Materials and Methods) supplemented with 0.4% (wt/vol) debranched arabinan for 48 h at 37°C.
abnA and xsa transcript analysis
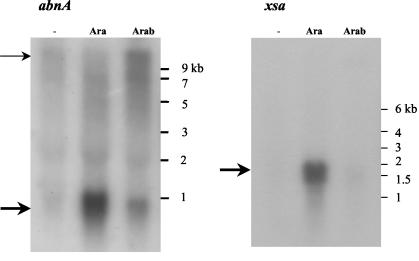
Previous work showed that the abfA gene encodes a 500-amino-acid polypeptide and belongs to the araABDLMNPQ-abfA operon, a polycistronic transcriptional unit responsive to arabinose (32) (Fig. 1). The abnA and xsa genes encode 323- and 495-amino-acid polypeptides, respectively, and both potential open reading frame terminators were found downstream (32, 40) (Fig. 1). To study transcription of the abnA and xsa genes, total RNA isolated from the wild-type strain grown for 2 h in the absence or presence of arabinose and arabinan (potential inducers) was annealed separately to DNA probes for abnA and xsa. Arabinose-inducible abnA- and xsa-specific transcripts of about 0.9 and 1.6 kb, respectively, were detected (Fig. 3). Weaker hybridization signals of the same size were also visible with RNA from cells grown in the presence of arabinan. No hybridization signals were detected in the absence of sugars, suggesting that both arabinose and arabinan might function as inducers. The extent of both abnA and xsa mRNA signals closely matched the expected sizes (1 and 1.6 kb, respectively) and confirm their monocistronic nature. However, when arabinan was used as an inducer, a weak high-molecular-weight RNA signal was visible with the abnA DNA probe. One possibility for this finding is that this weak message corresponds to cotranscription with upstream genes and/or the downstream araABDLMNPQ-abfA operon.
FIG. 3.
Northern blot analysis of the abnA- and xsa-specific transcripts. Ten micrograms of total RNA extracted from the wild-type strain grown in the absence of sugar (−), in the presence of arabinose (Ara), or in the presence of arabinan (Arab) was run in a 1.2% (wt/vol) agarose formaldehyde denaturing gel (see Materials and Methods). The RNA ladder used as molecular size markers is indicated to the right of each gel. The abnA-specific (left) and xsa-specific (right) transcripts detected with DNA probe fragments abnA (767 bp) and xsa (1,420 bp) are indicated by heavy arrows. The additional weak high-molecular-weight RNA signal visible with the abnA DNA probe (left) is indicated by a light arrow.
Expression of abnA and xsa is driven from σA-like promoters
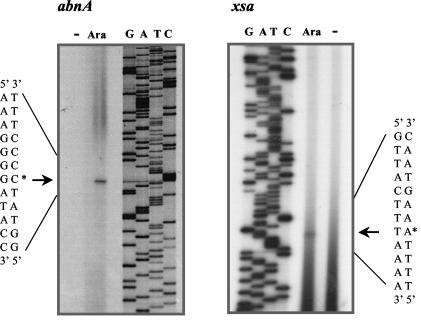
Primer extension analysis of total RNA isolated from cells grown in the presence of arabinose showed that the 5′ end of the abnA message corresponds to a G residue 117 bp upstream from the initiation TTG codon (Fig. 4 and 5A). Centered at −35 and −10 bp upstream from the abnA transcription start site are two sequences, TGTACA and TACAAT, respectively (Fig. 5A), that are similar to the consensus sequences for recognition by B. subtilis σA-containing RNA polymerase (TTGACA-17bp-TATAAT) (10, 21). By using the same technique, the apparent transcriptional start point of xsa was assigned to a T nucleotide 100 bp upstream from the initiation ATG codon (Fig. 4 and 5B). The potential −35 and −10 regions (TTGACA-17bp-TATGGT) (Fig. 5B) closely match the σA consensus (see above). No abnA- or xsa-specific extension products were seen with RNA extracted from cells grown in the absence of sugar, results parallel to those observed by Northern blot analysis.
FIG. 4.
Mapping of the transcriptional start site of the abnA and xsa genes. Radiolabeled oligonucleotides ARA86 and ARA90 (Table 2), complementary to the abnA and xsa sequences, respectively, were hybridized and used to direct cDNA synthesis from total B. subtilis 168T+ RNA isolated from exponentially growing cells in the absence (−) or presence (Ara) of arabinose (see Materials and Methods). After extension, the products were analyzed by gel electrophoresis together with a set of dideoxynucleotide chain termination sequencing reactions by using the same primers and plasmids pMPR2 and pRIT3, respectively, as templates. Arrows and asterisks indicate the positions of the abnA- and xsa-specific primer extension products and the deduced start site of transcription, a G residue in the abnA sequence (left) and a T residue in the xsa sequence (right).
FIG. 5.
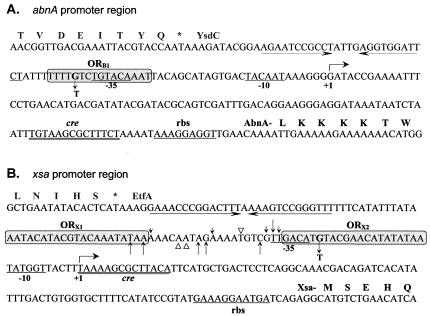
Promoter regions of the abnA and xsa genes. The nucleotide sequences of the abnA (A) and xsa (B) nontranscribed strands are shown in the 5′-to-3′ direction. The transcription start site (+1) defined by primer extension analysis and the −35 and −10 regions of each promoter are indicated below the nucleotide sequence. The putative ribosome binding sites (rbs) are represented, and the potential catabolic repression-associated sequences (CRE) are double underlined. (A) The predicted primary structure of AbnA and the polypeptide encoded by ysdC (the upstream gene) is given in single-letter code above the nucleotide sequence. A putative terminator sequence of ysdC is represented by convergent arrows. The AraR binding site, ORB1, deduced by similarity and confirmed by site-directed mutagenesis, is represent by a grey box. A single nucleotide change introduced in ORB1 at position −38 (G→T) is indicated. (B) The predicted primary structure of Xsa and the polypeptide encoded by etfA (the upstream gene) is given in single-letter code above the nucleotide sequence. A putative terminator sequence of etfA is represented by convergent arrows. AraR binding regions detected in DNase I footprinting experiments, ORX1 and ORX2, are shown in grey boxes. The sites of enhanced (black arrows) and diminished (open triangles) DNase I cleavage outside of the protected regions detected in the noncoding strand are shown above the sequence and in the coding strand below the sequence. The size of the arrow reflects the intensity of enhanced cleavage by DNase I. A single-base-pair substitution introduced in ORX2 at position −27 (G→T) is indicated.
abnA, xsa, and abfA transcription is responsive to arabinose and arabinan and is repressed by glucose.
To study the functionality of the abnA and xsa promoters, they were fused to the lacZ gene of E. coli and integrated at the amyE locus of the B. subtilis wild-type chromosome (strains IQB410 and IQB405, respectively) (Fig. 1 and Table 1). Since transcription of the abfA gene is driven from a σA-like promoter located upstream from the araA gene of the metabolic operon, expression of abfA was analyzed by the construction of a transcriptional lacZ fusion at the abfA locus (strain IQB450) (Fig. 1 and Table 1). The same DNA fragment, harboring the 3′ end of the araQ gene and the 5′ end of the abfA gene, was also fused to lacZ in a different vector and integrated at the amyE locus of the B. subtilis wild-type chromosome (strain IQB451) (Fig. 1 and Table 1). As expected, this strain did not show promoter activity in the experiments described below (data not shown). The levels of accumulated β-galactosidase activity of the strains bearing the various transcriptional fusions were examined in the absence of sugars and in the presence of arabinose, arabinan, and arabinose plus glucose. Samples were collected 2 and 4 h after induction (t2 and t4, respectively), which corresponds to the exponential growth phase and early postexponential phase, respectively. In the presence of arabinose, expression from the abfA′-lacZ, xsa′-lacZ, and abnA′-lacZ fusions (strains IQB450, IQB405, and IQB410) (Table 3) increased during exponential growth about 96-, 24-, and 3-fold, respectively, and a small increment in expression was observed at early postexponential phase (t4) (Table 3). The presence of arabinan also stimulated expression from the same abfA′-lacZ, xsa′-lacZ, and abnA′-lacZ fusions, about five-, three-, and fivefold, respectively (t2) (Table 3). However, these lower-level responses, compared to those observed in the presence of arabinose, increased more dramatically at the end of the exponential growth phase (t4) (Table 3). These results confirm the Northern blot analysis indicating that both arabinose and arabinan function as inducers of the abnA, xsa, and abfA genes. The possibility that other regulatory regions upstream from the DNA fragments used to construct the xsa′-lacZ and abnA′-lacZ fusions integrated at the amyE locus might influence expression from xsa and abnA was examined. We constructed transcriptional lacZ fusions at the xsa and abnA loci (strains IQB407 and IQB412) (Table 1), and the regulation factor calculated for these strains was similar to that observed for strains IQB405 and IQB410, respectively (data not shown).
TABLE 3.
Expression from abfA′-lacZ, xsa′-lacZ, and abnA′-lacZ fusions in a wild-type and araR-null mutant backgrounda
| Promoter fusion and strainb | Time | β-Galactosidase activity (Miller units)c
|
Regulation/repression factord
|
|||||
|---|---|---|---|---|---|---|---|---|
| −Ara | +Ara | +Arab | +Ara +Glc | Ara | Arab | Glc | ||
| abfA′-lacZ | ||||||||
| IQB450 (WT) | t2 | 3.2 ± 0.7 | 303.9 ± 28.9 | 16.8 ± 2.6 | 13.0 ± 2.3 | 96.4 | 5.3 | 21.8 |
| t4 | 3.2 ± 0.4 | 490.4 ± 10.1 | 88.6 ± 6.3 | 27.8 ± 4.5 | 153.0 | 27.6 | 17.6 | |
| IQB453 (AraR−) | t2 | 1,363.5 ± 17.4 | 750.5 ± 125.1 | 660.1 ± 31.2 | NDe | 0.6 | 0.5 | ND |
| t4 | 1,957.6 ± 111.0 | 485.3 ± 62.6 | 766.1 ± 55.0 | ND | 0.3 | 0.4 | ND | |
| xsa′-lacZ | ||||||||
| IQB405 (WT) | t2 | 7.5 ± 0.2 | 177.3 ± 3.9 | 21.8 ± 0.7 | 9.2 ± 0.5 | 23.7 | 2.9 | 19.3 |
| t4 | 9.0 ± 0.8 | 287.4 ± 38.6 | 245.8 ± 23.2 | 17.2 ± 1.6 | 31.9 | 27.2 | 16.7 | |
| IQB406 (AraR−) | t2 | 564.0 ± 57.6 | 503.8 ± 13.4 | 397.4 ± 4.9 | ND | 0.8 | 0.7 | ND |
| t4 | 561.7 ± 74.4 | 296.2 ± 39.3 | 679.1 ± 46.7 | ND | 0.5 | 1.2 | ND | |
| abnA′-lacZ | ||||||||
| IQB410 (WT) | t2 | 3.2 ± 0.7 | 10.0 ± 0.8 | 15.2 ± 2.1 | 1.6 ± 0.3 | 3.1 | 4.7 | 6.5 |
| t4 | 3.0 ± 0.4 | 11.0 ± 0.4 | 25.7 ± 5.2 | 1.7 ± 0.0 | 3.7 | 8.6 | 7.5 | |
| IQB411 (AraR−) | t2 | 31.3 ± 2.0 | 35.2 ± 0.4 | 25.9 ± 0.1 | ND | 1.1 | 0.8 | ND |
| t4 | 39.6 ± 4.2 | 25.8 ± 2.4 | 42.8 ± 1.7 | ND | 0.7 | 1.1 | ND | |
| IQB448 (WT) | t2 | 2.4 ± 0.6 | 13.2 ± 0.9 | 17.3 ± 2.7 | 1.8 ± 0.2 | 5.6 | 7.2 | 7.2 |
| t4 | 1.8 ± 0.2 | 13.6 ± 1.9 | 31.5 ± 5.2 | 2.0 ± 0.3 | 7.4 | 17.1 | 6.8 | |
| IQB449 (AraR−) | t2 | 58.8 ± 7.5 | 53.7 ± 4.1 | 36.8 ± 4.7 | ND | 0.9 | 0.6 | ND |
| t4 | 59.1 ± 3.1 | 67.5 ± 16.5 | 46.7 ± 7.6 | ND | 1.1 | 0.8 | ND | |
The strains containing different promoter-lacZ fusions were grown on C minimal medium supplemented with casein hydrolysate in the absence of sugar (−Ara), in the presence of arabinose (+Ara), in the presence of arabinan (+Arab), and in the presence of arabinose plus glucose (+Ara +Glc). Samples were analyzed 2 h (t2) and 4 h (t4) after the addition of sugars.
WT, wild type.
The levels of accumulated β-galactosidase activity represent the average of results from three independent experiments with wild-type strains and two independent experiments with AraR− strains.
The regulation factor, was calculated as the ratio of the level of expression (in Miller units) obtained in the presence of arabinose (Ara) or arabinan (Arab) to the value determined in the absence of sugar (−Ara), was taken as a measure of AraR repression. Glucose repression (Glc) was calculated as the ratio of the level of expression (in Miller units) obtained in the presence of arabinose to the value determined in the presence of glucose (+Ara +Glc).
ND, not determined.
The addition of glucose caused a 21.8-fold repression of the abfA′-lacZ fusion expression, a 19.3-fold repression of xsa′-lacZ expression, and a 6.5-fold repression of abnA′-lacZ expression (t2) (Table 3). No significant differences in the levels of glucose repression were observed during early postexponential phase (t4) (Table 3). Previously, it has been shown that glucose repression of the araABDLMNPQ-abfA metabolic operon is mainly regulated by CcpA via binding to two catabolite responsive elements (CREs), one located between the promoter region of the operon and the araA gene and one located 2 kb downstream within the araB gene (11). Mutagenesis studies of B. subtilis revealed the CRE consensus sequence TGWAARCGYTWNCW (W = A or T, R = A or G, Y = C or T, N = any base) (14, 18, 38, 41). Based on sequence homology studies, potential CRE sequences were detected in the promoter region of the abnA and xsa genes. The CRE for abnA is positioned between the promoter and the TTG initiation codon (+79TGTAAGCGCTTTCT+92) (Fig. 5A), and the CRE for xsa overlaps the transcription start site of the gene (+1TAAAAGCGCTTACA+14) (Fig. 5B).
AraR plays a major role in transcriptional control of abnA, xsa, and abfA.
In previous work, a search of the B. subtilis database was done (SubtiList) for sequences similar to the AraR consensus operator (ATTTGTACGTACAAAT) (22), the key regulator of arabinose utilization. Among the potential AraR binding sites detected, two were located in the promoter region of the xsa and abnA genes (22). Thus, we investigated the expression from the abfA′-lacZ, xsA′-lacZ, and abnA′-lacZ fusions in an araR-null mutant background. The levels of accumulated β-galactosidase activity of the resulting strains (IQB453, IQB406, and IQB411, respectively) were examined as described above, and the results are shown in Table 3. The degree of AraR repression (regulation factor) was determined indirectly by the ratio of the values obtained in induced and noninduced cultures. Disruption of the araR gene led to a total derepression of the expression from the abfA′-lacZ, xsA′-lacZ, and abnA′-lacZ fusions (strains IQB453, IQB406, and IQB411, respectively) in comparison to that from the wild type. These results suggest that AraR plays a major role in the transcriptional control of the abnA, xsa, and abfA genes. Interestingly, we found another sequence within the abnA coding region (AAACAGTACGTACAAAA, at a position +831 relative to the transcription start site) similar to that of the AraR consensus operator (see above). We constructed an abnA′-lacZ fusion bearing this element to determine its involvement in the regulation of abnA expression (Fig. 1). The transcriptional fusion was integrated in a single copy at the amyE locus of the wild-type and araR-null mutant backgrounds, and the resulting strains IQB448 and IQB449 (Table 1), respectively, were analyzed as described above. In the presence of arabinose and arabinan, strain IQB448 showed a twofold increase in the level of regulation by AraR relative to that of strain IQB410 (Table 3), indicating that this putative operator might contribute to the regulation of abnA expression at the transcriptional level.
Binding of AraR to the promoter region of xsa and abnA genes.
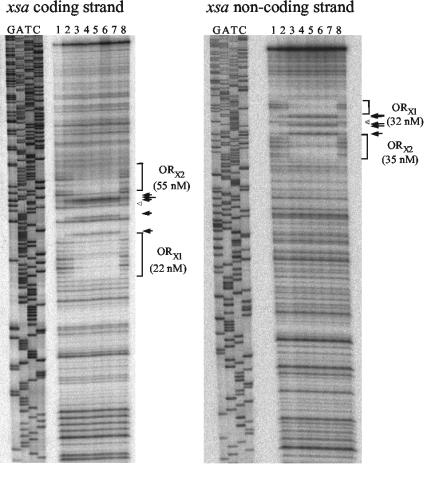
The ability of AraR to bind to the promoter region of xsa and abnA genes was determined by quantitative DNase I footprinting with DNA fragments from the plasmids harboring the transcriptional fusions as targets (see Materials and Methods). Two AraR binding sites were detected by DNase I footprinting in the xsa promoter region (Fig. 6). In the xsa coding strand, AraR protects the regions between positions −72 and −52 (ORX1) and between positions −32 and −11 (ORX2); similar sequences are protected in the noncoding strand (Fig. 6). A pattern of DNase I-enhanced and -diminished cleavage was observed between ORX1 and ORX2 (Fig. 5 and 6) that resembled the DNase I footprintings of the araABDLMNPQ-abfA operon and araE (22). AraR binding to the two in-phase operators of the xsa metabolic operon and of the transport gene promoter regions thus seems to occur in similar ways, producing in both cases a distortion of the DNA helix. Binding of AraR to ORX1 and ORX2 was also inhibited by the presence of arabinose (Fig. 6), indicating that arabinose is the effector which modulates AraR binding to DNA. The Kapp for each individual binding site was determined as the repressor concentration at which half-maximal site occupancy was observed (Fig. 6). Although these values were calculated for a single experiment, the relative affinity of AraR to ORX1 and ORX2 is comparable to that observed for the AraR binding sites in the promoter regions of the metabolic operon and the araE gene (22). Binding of AraR to the putative ORB1 operator identified by sequence analysis within the abnA promoter was not detected in the same range of protein concentrations used for the xsa promoter (data not shown).
FIG. 6.
DNase I protection experiments of the xsa promoter by the AraR protein. Each strand of a 339-bp DNA fragment carrying the xsa promoter region was end labeled with γ-32P in separate experiments. AraR concentrations were calculated considering a pure dimeric protein. Lane 1, no protein; lane 2, 25 nM AraR; lane 3, 50 nM AraR; lane 4, 100 nM AraR; lane 5, 150 nM AraR; lane 6, 200 nM AraR; lane 7, 250 nM AraR; lane 8, 250 nM AraR plus 0.02% (wt/vol) l-arabinose. Protected regions, termed ORX1 and ORX2, are indicated in the autoradiograms by brackets. The sites of enhanced (black arrows) and diminished (open triangles) DNase I cleavage outside of the protected regions are both indicated in the autoradiograms. The size of the arrow reflects the intensity of enhanced cleavage by DNase I. Total repressor concentration at which half-maximal site occupancy is achieved (a value that represents Kapp, the apparent affinity of AraR to each site) is indicated within parentheses for each operator and was calculated from a single experiment.
To assess the functionality of the AraR operators in vivo, we introduced the same single-base-pair substitution in both ORX2 and the putative ORB1 (Fig. 5). This mutation in a highly conserved position of the AraR target sequence (22) was designed to prevent the binding of AraR to ORX2 in the xsa promoter and to ORB1 in the abnA promoter. The two mutant promoters were fused to the lacZ gene of E. coli and were analyzed in a B. subtilis wild-type background as described above. Both mutations resulted in a loss of regulation by AraR (Table 4). These results indicate that just one base pair change in one of the two operators (ORX1 and ORX2) is sufficient to abolish repression and suggest cooperative binding to the two in-phase operators in the xsa promoter region. Furthermore, ORB1 is an active cis-acting element for the regulation of the abnA promoter.
TABLE 4.
Site-directed mutagenesis of the xsa and abnA promoters
| Promoter fusion and strain (base substitution) | β-Galactosidase activity (Miller units)a
|
Repression factorb
|
|||
|---|---|---|---|---|---|
| −Ara | +Ara | +Arab | Ara | Arab | |
| xsa′-lacZ | |||||
| IQB405 (wild type) | 7.5 ± 0.2 | 177.3 ± 3.9 | 21.8 ± 0.7 | 23.7 | 2.9 |
| IQB465 (ORX2-27 G→T) | 728.6 ± 67.6 | 580.3 ± 49.5 | 740.1 ± 102.1 | 0.8 | 1.0 |
| abnA′-lacZ | |||||
| IQB410 (wild type) | 3.2 ± 0.7 | 10.0 ± 0.8 | 15.2 ± 2.1 | 3.1 | 4.7 |
| IQB464 (ORB1-38 G→T) | 63.7 ± 5.3 | 27.1 ± 3.3 | 73.2 ± 3.6 | 0.4 | 1.1 |
The strains containing different promoter-lacZ fusions were grown on C minimal medium supplemented with casein hydrolysate in the absence of sugar (−Ara), in the presence of arabinose (+Ara), or in the presence of arabinan (+Arab). Samples were analyzed 2 h (t2) after the addition of sugars. The levels of accumulated β-galactosidase activity represent the average of results from three independent experiments.
AraR repression was calculated as the ratio of the level of expression (Miller units) obtained in the presence of arabinose (+Ara) or arabinan (+Arab) to the value determined in the absence of sugar (−Ara).
DISCUSSION
In B. subtilis, the genes encoding hemicellulolytic enzymes are clustered with genes encoding enzymes that further catabolize these carbon sources (reference 35 and references therein). In this study, we analyzed the mechanisms that regulate the expression of three arabinan-degrading genes, abfA, xsa, and abnA, which are assembled with genes involved in arabinose catabolism (Fig. 1). The abfA and xsa genes most probably encode AFs (EC 3.2.1.55) belonging to the GH51 family (http://afmb.cnrs-mrs.fr/∼cazy/CAZY). Although the exact subcellular localization of Xsa and AbfA is unknown, these enzymes are believed to be intracellular (1, 37). Nonetheless, AFs from G. stearothermophilus and B. subtilis were purified from supernatants of cultures at the end of the stationary phase (8, 13, 39). Based on primary amino acid sequence analysis, the abnA gene most likely encodes an ABN (EC 3.2.1.99) grouped in the GH43 family (http://afmb.cnrs-mrs.fr/∼cazy/CAZY), which was shown to be extracellular (1). Here, we determined the function of the abnA gene by showing that an insertion-deletion mutation in this gene led to a loss of arabinanase activity (Fig. 2). However, the abnA-null mutant is still able to grow on minimal medium with arabinan as the sole carbon source. This result might be due to the activity of the yxiA gene product, a hypothetical arabinanase displaying 27% identity to AbnA (17).
During exponential growth, the expression from abfA′-lacZ, xsA′-lacZ, and abnA′-lacZ transcriptional fusions revealed that abfA and xsa are strongly induced by arabinose, whereas the induction of abnA is weak (Table 3). The levels of induction in response to arabinan are very similar for the three genes. However, the level of arabinan-mediated induction of abfA and xsa is considerably lower than that observed with arabinose, while the induction levels of abnA are similar in both cases. This result is most probably due to different kinetics of induction by arabinose among the promoters (see below). A disruption of the araR gene abolished the regulation of the three arabinan-degrading genes, suggesting that AraR plays a major role in the transcriptional control of these genes. Previously, it has been reported that the addition of arabinose to the medium causes an immediate cessation of growth in an araR-null mutant background, which is probably due to an intracellular increase of arabinose and consequently an increase of the metabolic sugar phosphate intermediates that are toxic to the cell (31). This effect is not observed when arabinan is added to the cultures, indicating that the reduced levels of expression observed in the presence of this carbon source might be due to lower levels of intracellular arabinose during the exponential growth phase. Since the arabinan-mediated induction of the three promoters is strictly dependent on AraR, and arabinan itself is not the molecular inducer (data not shown), the observed arabinan induction should be the result of low levels of intracellular arabinose. As mentioned above, the arabinose-mediated induction of xsa and abfA promoters is higher than that of the abnA promoter, while the arabinan induction levels are identical in the three cases. Therefore, a likely explanation for these results is that a lower level of arabinose is enough to rapidly and fully induce the abnA promoter but not the abfA and xsa promoters. Taken together, these observations indicate that the AraR protein exerts a tight control of arabinose- and arabinan-inducible transcription of the abfA and xsa genes but that repression of the abnA gene is more flexible.
One of the mechanisms involved in the synthesis of many degradative enzymes in B. subtilis is mediated by transition phase regulation (7). Accordingly, when the transcriptional fusions are analyzed at early postexponential phase, the levels of expression of abfA, xsa, and abnA in response to arabinose and arabinan are higher than those observed during exponential growth (Table 3). This finding suggests that the arabinan-degrading genes are subject to temporal regulation. In agreement with our observations, by extracellular proteome analysis, Antelmann et al. (1) detected AbnA at higher levels during stationary phase. Interestingly, the temporal differences among the induced levels of expression that we noticed are more striking in the case of arabinan than for arabinose. This finding suggests that arabinan, or one of its degradation products, may play an important role in this process. However, it may also be due simply to a higher amount of intracellular free arabinose as a result of a higher level of AbnA during the stationary phase. The mechanisms underlying the temporal regulation of the arabinan-degrading genes are unknown, but studies are currently in progress to address this question. Nonetheless, the data presented here indicate that the arabinose repressor, AraR, also plays a crucial role in the control of abfA, xsa, and abnA expression during early postexponential phase. Additionally, glucose repression previously characterized for the araABDLMNPQ-abfA operon (11) seems to be mediated by a similar mechanism in the case of the abnA and xsa genes.
The DNase I footprinting analysis of the xsa promoter suggests that this gene should be regulated by AraR by a mechanism similar to that proposed for the araABDLMNPQ-abfA operon and the araE transport gene (22, 23). As in these two cases, the AraR binding sites are separated by approximately four turns of the DNA helix (41 bp), and a pattern of DNase I hypersensibility was observed in the interoperator region (Fig. 5 and 6). Furthermore, a single-base-pair change in one of the two operators (ORX1 and ORX2) is sufficient to abolish AraR repression in vivo. By analogy, this finding suggests that the binding of AraR to the operators in the xsa promoter is cooperative, resulting in a distortion of the DNA helix that may be in the form of a small DNA loop. In contrast, noncooperative binding of AraR to one operator in the promoter region of the abnA gene, and possibly to a second operator located downstream within the abnA coding region (Tables 3 and 4), is less effective, as observed in the case of autoregulation of araR expression (22, 23). The fact that we could not detect in vitro binding of AraR to the promoter region of abnA might indicate a low affinity of the regulator to its operator site. However, one cannot exclude the possibility of additional trans-acting factors involved in the regulation of abnA expression which may contribute to AraR binding or which may directly control abnA expression. Together, these observations might explain the different mode of response to arabinose and arabinan of abnA expression compared to those of xsa and abfA during exponential growth (Table 3), which may reflect distinct physiological requirements. A tight control of the xsa and abfA genes ensures the expression of these intracellular enzymes solely when the arabinose inducer is present. On the other hand, a weak control of abnA allows for a low level of basal transcription of this extracellular enzyme.
Bacilli secrete a vast number of polysaccharide backbone-degrading enzymes, which produce relatively large oligosaccharide products. These units, disaccharides, trisaccharides, and oligosaccharides, enter the cell by specific transport systems and are further broken down by intracellular enzymes (4, 35). We have shown that in B. subtilis, arabinan is degraded by at least one extracellular hemicellulase, AbnA. The resulting products, arabinose, arabinobiose, arabinotriose, and arabinooligosaccharides, are transported by different systems. Arabinose enters the cell mainly through the AraE permease (33), and the uptake of arabinose oligomers most likely occurs via AraNPQ, an ABC-type transporter (32). These latter products might be further digested intracellularly by AbfA and Xsa. Interestingly, the AraE permease is also responsible for the transport of xylose and galactose into the cell (16). These three structurally different sugars, arabinose, xylose, and galactose, are frequently found associated in hemicelluloses. Furthermore, xylan- and xylose-utilizing genes are controlled by the XylR repressor, and no regulatory protein specifically controlling galactose utilization has been found (reference 35 and references therein). These observations suggest a coordinated expression, triggered by arabinose and mediated by AraR, of genes encoding enzymes responsible for extracellular degradation of arabinose-containing polysaccharides and transport systems and intracellular catabolism of arabinose, xylose, and galactose. Concerted regulation of the production of all pectin side-chain-cleaving enzymes in response to arabinose seems likely to occur in Aspergillus spp. (5). Thus, it will be interesting to know how this regulatory circuitry in response to arabinose is disseminated among hemicellulase-producing microorganisms.
Acknowledgments
We thank Rita Teodoro and Susana S. Silva for constructing some plasmids and strains.
This work was supported by grant no. POCTI/AGR/36212/00 from Fundação para a Ciência e Tecnologia and FEDER.
REFERENCES
- 1.Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484-1502. [DOI] [PubMed] [Google Scholar]
- 2.Beldman, G., H. A. Schols, S. M. Piston, M. J. F. Searl-van Leewen, and A. G. J. Vorangen. 1997. Arabinans and arabinan degrading enzymes. Adv. Macromol. Carbohydr. Res. 1:1-64. [Google Scholar]
- 3.Debeche, T., N. Cummings, I. Connerton, P. Debeire, and M. J. O'Donohue. 2000. Genetic and biochemical characterization of a highly thermostable α-l-arabinofuranosidase from Thermobacillus xylanilyticus. Appl. Environ. Microbiol. 66:1734-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deutscher, J., A. Galinier, and I. Martin-Verstraete. 2002. Carbohydrate uptake and metabolism, p. 129-150. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 5.de Vries, R. P. 2003. Regulation of Aspergillus genes encoding plant cell wall polysaccharide-degrading enzymes; relevance for industrial production. Appl. Microbiol. Biotechnol. 61:10-20. [DOI] [PubMed] [Google Scholar]
- 6.de Vries, R. P., and J. Visser. 2001. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 65:497-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari, E., A. S. Jarnagin, and B. F. Schmidt. 1993. Commercial production of extracellular enzymes, p. 917-937. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 8.Gilead, S., and Y. Shoham. 1995. Purification and characterization of an α-l-arabinofuranosidase from Bacillus stearothermophilus T-6. Appl. Environ. Microbiol. 61:170-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazlewood, G. P., and H. J. Gilbert. 1998. Structure and function analysis of Pseudomonas plant cell wall hydrolases. Prog. Nucleic Acid Res. Mol. Biol. 61:211-241. [DOI] [PubMed] [Google Scholar]
- 10.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 11.Inácio, J. M., C. Costa, and I. de Sá-Nogueira. 2003. Distinct molecular mechanisms involved in carbon catabolite repression of the arabinose regulon in Bacillus subtilis. Microbiology 149:2345-2355. [DOI] [PubMed] [Google Scholar]
- 12.Kaji, A., and T. Saheki. 1975. Endo-arabanase from Bacillus subtilis F-11. Biochim. Biophys. Acta 410:354-360. [DOI] [PubMed] [Google Scholar]
- 13.Kaneko, S., M. Sano, and I. Kusakabe. 1994. Purification and some properties of α-l-arabinofuranosidase from Bacillus subtilis 3-6. Appl. Environ. Microbiol. 60:3425-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, J. H., and G. H. Chambliss. 1997. Contacts between Bacillus subtilis catabolite regulatory protein CcpA and amyO target site. Nucleic Acids Res. 25:3490-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosugi, A., K. Murashima, and R. H. Doi. 2002. Characterization of two noncellulosomal subunits, ArfA and BgaA, from Clostridium cellulovorans that cooperate with the cellulosome in plant cell wall degradation. J. Bacteriol. 184:6859-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krispin, O., and R. Allmansberger. 1998. The Bacillus subtilis AraE protein displays a broad substrate specificity for several different sugars. J. Bacteriol. 180:3250-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Verstraete, I., J. Stülke, A. Klier, and G. Rapoport. 1995. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J. Bacteriol. 177:6919-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKie, V. A., G. W. Black, S. J. Millward-Sadler, G. P. Hazlewood, J. I. Laurie, and H. J. Gilbert. 1997. Arabinanase A from Pseudomonas fluorescens subsp. cellulosa exhibits both an endo- and an exo- mode of action. Biochem. J. 323:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Moran, C. P., Jr., N. Lang, S. F. J. LeGrice, G. Lee, M. Stephens, A. L. Sonenshein, J. Pero, and R. Losick. 1982. Nucleotide sequences that signal the initiation of transcription in Bacillus subtilis. Mol. Gen. Genet. 186:339-346. [DOI] [PubMed] [Google Scholar]
- 22.Mota, L. J., P. Tavares, and I. Sá-Nogueira. 1999. Mode of action of AraR, the key regulator of l-arabinose metabolism in Bacillus subtilis. Mol. Microbiol. 33:476-489. [DOI] [PubMed] [Google Scholar]
- 23.Mota, L. J., L. M. Sarmento, and I. de Sá-Nogueira. 2001. Control of the arabinose regulon in Bacillus subtilis by AraR in vivo: crucial roles of operators, cooperativity, and DNA looping. J. Bacteriol. 183:4190-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pascal, M., F. Kunst, J. A. Lepesant, and R. Dedonder. 1971. Characterization of two sucrase activities in Bacillus subtilis Marburg. Biochimie 53:1059-1066. [DOI] [PubMed] [Google Scholar]
- 25.Perego, M. 1993. Integrational vectors for genetic manipulation in Bacillus subtilis, p. 615-624. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 26.Saha, B. C. 2000. α-l-Arabinofuranosidases: biochemistry, molecular biology and application in biotechnology. Biotechnol. Adv. 18:403-423. [DOI] [PubMed] [Google Scholar]
- 27.Saha, B. C. 2003. Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 5:279-291. [DOI] [PubMed] [Google Scholar]
- 28.Sakamoto, T., M. Yamada, H. Kawasaki, and T. Sakai. 1997. Molecular cloning and nucleotide sequence of an endo-1,5-α-l-arabinase gene from Bacillus subtilis. Eur. J. Biochem. 245:708-714. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Sá-Nogueira, I., and H. de Lencastre. 1989. Cloning and characterization of araA, araB, and araD, the structural genes for l-arabinose utilization in Bacillus subtilis. J. Bacteriol. 171:4088-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sá-Nogueira, I., and L. J. Mota. 1997. Negative regulation of l-arabinose metabolism in Bacillus subtilis: characterization of the araR (araC) gene. J. Bacteriol. 179:1598-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sá-Nogueira, I., T. V. Nogueira, S. Soares, and H. Lencastre. 1997. The l-arabinose (ara) operon of Bacillus subtilis: nucleotide sequence, genetic organization and expression. Microbiology 143:957-969. [DOI] [PubMed] [Google Scholar]
- 33.Sá-Nogueira, I., and S. S. Ramos. 1997. Cloning, functional analysis, and transcriptional regulation of the Bacillus subtilis araE gene involved in l-arabinose utilization. J. Bacteriol. 179:7705-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shallom, D., and Y. Shoham. 2003. Microbial hemicellulases. Curr. Opin. Microbiol. 3:219-228. [DOI] [PubMed] [Google Scholar]
- 35.Stülke, J., and W. Hillen. 2000. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54:849-880. [DOI] [PubMed] [Google Scholar]
- 36.Takao, M., A. Yamaguchi, K. Yoshikawa, T. Terashita, and T. Sakai. 2002. Molecular cloning of the gene encoding thermostable endo-1,5-alpha-l-arabinase of Bacillus thermodenitrificans TS-3 and its expression in Bacillus subtilis. Biosci. Biotechnol. Biochem. 66:430-433. [DOI] [PubMed] [Google Scholar]
- 37.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weickert, M. J., and G. H. Chambliss. 1990. Site-directed mutagenesis of a catabolic repression operator sequence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:6238-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinstein, L., and P. Albersheim. 1979. Structure of plant cell walls. IX. Purification and partial purification of a wall-degrading endoarabanase and an arabinosidase from Bacillus subtilis. Plant Physiol. 63:425-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wipat, A., N. Carter, S. C. Brignell, B. J. Guy, K. Piper, J. Sanders, P. T. Emmerson, and C. R. Harwood. 1996. The dnaB-pheA (256 degrees-240 degrees) region of the Bacillus subtilis chromosome containing genes responsible for stress responses, the utilization of plant cell walls and primary metabolism. Microbiology 142:3067-3078. [DOI] [PubMed] [Google Scholar]
- 41.Zalieckas, J. M., L. V. Wray, Jr., and S. H. Fisher. 1998. Expression of the Bacillus subtilis acsA gene: position and sequence context affect cre-mediated carbon catabolite repression. J. Bacteriol. 180:6649-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zverlov, V. V., W. Liebl, M. Bachleitner, and W. H. Schwarz. 1998. Nucleotide sequence of arfB of Clostridium stercorarium, and prediction of catalytic residues of α-l-arabinofuranosidases based on local similarity with several families of glycosyl hydrolases. FEMS Microbiol. Lett. 164:337-343. [DOI] [PubMed] [Google Scholar]