Abstract
Naturally photoswitchable proteins offer a means of directly manipulating the formation of protein complexes that drive a diversity of cellular processes. We have developed tunable light-inducible dimerization tags (TULIPs) based on a synthetic interaction between the LOV2 domain of Avena sativa phototropin 1 (AsLOV2) and an engineered PDZ domain (ePDZ). TULIP tags can recruit proteins to diverse structures in living yeast and mammalian cells, either globally or with precise spatial control using a steerable laser. The equilibrium binding and kinetic parameters of the interaction are tunable by mutation, making TULIPs readily adaptable to signaling pathways with varying sensitivities and response times. We demonstrate the utility of TULIPs by conferring light sensitivity to functionally distinct components of the yeast mating pathway and by directing the site of cell polarization.
INTRODUCTION
Cells commonly interpret developmental cues through assemblies of structural and signaling proteins that are built up from a combination of transient protein–protein and protein–membrane interactions (PPIs and PMIs)1. PPIs and PMIs can enforce the proximity of reactant species (e.g., a kinase and its substrate) or spatially constrain molecules within the cell to create a polarized response. Designed photoactivatable proteins offer unprecedented spatial and temporal control of cellular signaling processes2. For example, a fusion of the small GTPase Rac1 and the photosensor AsLOV2 allows direct control of Rac1 activity in living tissues, and has led to ground-breaking experiments on polarity and motility3–5. Nevertheless, direct fusion does not always confer photoactivatable control on proteins of interest, even with case-by-case optimization3.
The ubiquity and modularity of PPIs and PMIs suggests that light-inducible interactions should be an especially flexible tool for triggering cellular signaling events precisely in space and time, thereby obviating the need to optimize the caging of individual proteins. Several groups have adapted light-inducible PPIs that occur naturally in Arabidopsis thaliana for use as cell-biological reagents6–8. While each of these methods has attractive features, all have drawbacks. For example, a method based on FKF1 and GIGANTEA requires a large photosensory protein (1,173 amino acids) and exhibits slow association (minutes) and dissociation (hours) kinetics6. Another, based on the large photosensory domain (908 amino acids) of phytochrome B and its interacting factor PIF6, exhibits dimerization within seconds upon illumination with 650 nm light7. However, recovery requires hours unless dissociation is accelerated by 750 nm light and precise spatial control requires simultaneous, two-wavelength illumination7. A third method, based on cryptochrome 2 and CIB1, features small domains (498 and 170 amino acids) that exhibit dimerization within ten seconds and dissociation within ten minutes8. However, it remains unclear whether the proteins can be used for spatially resolved control of cell signaling8. More broadly, the basis of all three light-mediated interactions remains poorly characterized and the ability to tune important physical parameters is limited.
An ideal light-inducible PPI for optogenetics should use small, genetically encoded interacting domains that do not require exogenous cofactors. It should exhibit switching between biologically relevant binding affinities on a range of timescales. Its components should be compatible as fusions to a variety of subcellular markers, and it should be possible to confine photoactivation to a small region of the cell. We set out to create a set of tunable light-controlled interacting protein tags (TULIPs) with these properties using small, well-characterized interacting domains. We show that the TULIPs system is a versatile and tunable optogenetic tool to localize proteins to specific regions of yeast or mammalian cells, and to trigger specific cellular signaling pathways. Remarkably, the TULIPs system is capable of regulating the activity of nucleotide exchange factors, scaffold proteins and kinases, all by recruitment to the plasma membrane.
RESULTS
Photoswitch design
As a photosensor, we chose the second LOV (Light-Oxygen-Voltage) domain of Avena sativa phototropin 1 (AsLOV2)9. LOV domains are small (~125 residue) photosensory domains based on a PAS (Per-ARNT-Sim) core that binds a flavin cofactor. Like many PAS domains, AsLOV2 features flanking N- and C-terminal α helices (the A′α and Jα helices, respectively)10,11. Upon photoexcitation with blue light (< 500 nm), the Jα helix undocks from the LOV core and unfolds11,12. This conformational change is critical to phototropin signaling and has been exploited in designed photosensors3,13,14. Numerous mutational15–17 and chemical18 methods of tuning the physical properties of AsLOV2 have been reported.
Following a broadly successful approach for making engineered photoreceptors19, we reasoned that fusion of a peptide epitope to the C-terminus of the Jα helix would allow the LOV2–Jα interaction to sterically block, or cage the epitope from binding to a cognate PDZ domain (Fig. 1a, Supplementary Fig. 1). We anticipated that successful caging would require sequence overlap, so that part of the epitope would adopt a binding-incompetent α-helical conformation in the dark, Jα-docked state14 (Supplementary Note 1). As a binding partner, we used high-affinity, high-specificity engineered variants of the Erbin PDZ domain20. These clamshell-like “ePDZ” chimeras (194 amino acids) are highly tunable; mutational variants of ePDZ and its cognate peptide vary in interaction affinity from ~ 0.5 nM to > 10 µM21.
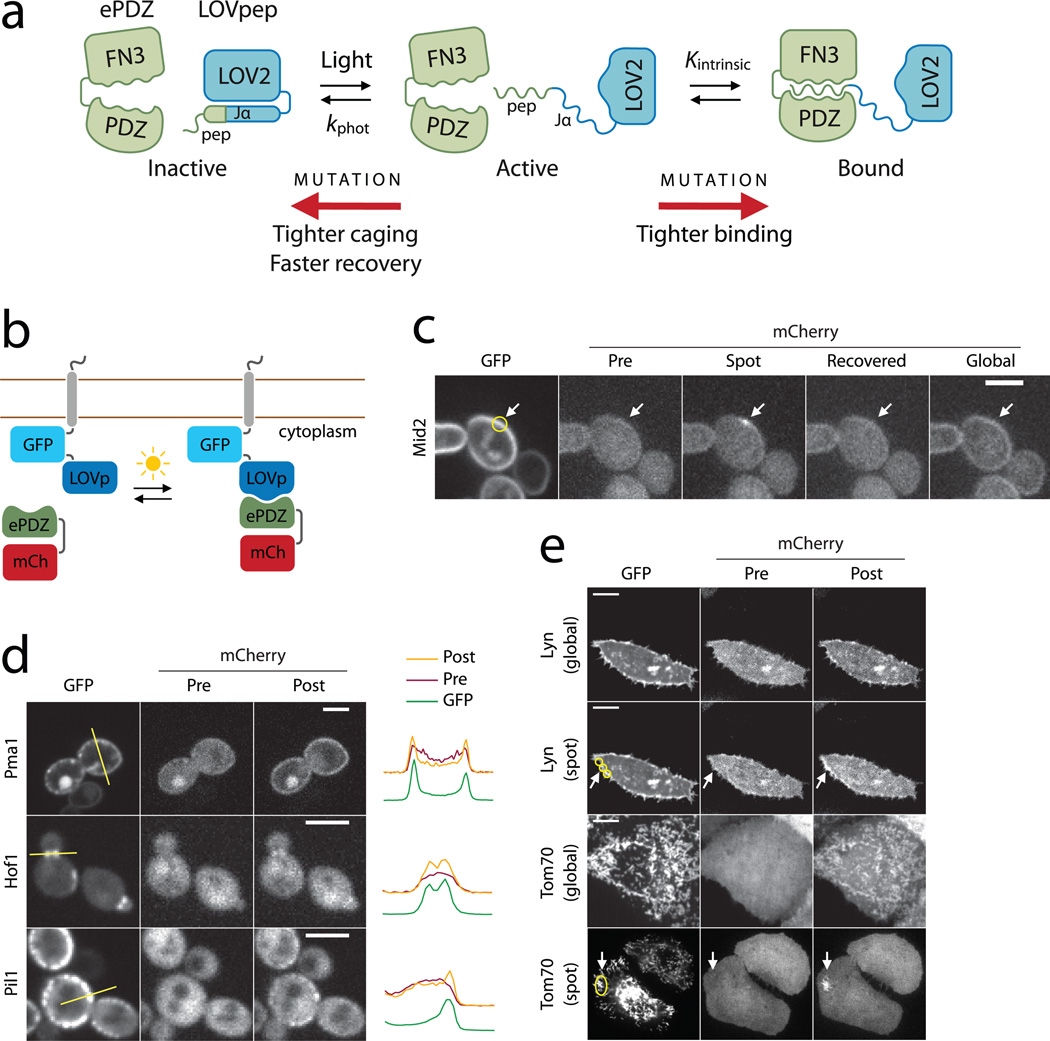
Figure 1.
Design and characterization of TULIPs. (a) Schematic design of TULIPs. In the dark, a peptide epitope is caged by docking of the Jα helix to the LOV2 core (blue). Upon photoexcitation, the Jα helix undocks and exposes the peptide epitope for binding by ePDZ (green). The caging, intrinsic ePDZ–peptide affinity (Kintrinsic) and lifetime of the photoexcited state (kphot) can all be tuned by mutations. (b) Schematic of the assay used to measure ePDZ–LOVpep binding in living yeast. (c) Recruitment of ePDZb1–mCherry to the integral plasma membrane protein Mid2 in yeast using spot (arrow) and global photoexcitation. Scale bar, 5 µm. (d) Recruitment of ePDZb–mCherry to diverse subcellular markers in yeast by global photoexcitation. Scale bars, 5 µm. The plots depict pixel intensities measured along the yellow lines indicated in the GFP images. (e) Recruitment of ePDZb1–mCherry to the plasma membrane and mitochondria of HeLa cells by global and spot (arrow) photoexcitation. Scale bars, 10 µm.
We designed a series of five AsLOV2–peptide fusions for initial screening (Supplementary Fig. 2a and Supplementary Note 1) by appending a peptide epitope (–SSADTWV–COOH) to serial truncations of the Jα helix. We fused these with GFP and the transmembrane protein Mid2 and co-expressed the constructs with mCherry-tagged ePDZ (Fig. 1b). We assayed recruitment of ePDZ–mCherry to the plasma membrane in the dark and immediately after photoexcitation with a 473 nm laser. To quantify the plasma membrane association of ePDZ–mCherry, we measured the ratio of plasma membrane and cytoplasmic fluorescence, averaged over a population of cells (<Robs>, Supplementary Fig. 3 and Online Methods). For the longest AsLOV2–peptide fusions (Registers 1–3), <Robs> was relatively high in both the dark and photoexcited states, and photoswitching was slight (Supplementary Fig. 2b). Binding was diminished for the shorter fusions (Registers 4 & 5, Supplementary Fig. 2a), probably because more of the epitope is masked in the Jα-docked conformation. Both constructs exhibited greater binding in the lit state than in the dark, indicating light-directed plasma membrane recruitment of ePDZ–mCherry.
Subcellular recruitment
We modified the Register 4 construct to make the sequence more favorable for ePDZ binding and LOV–Jα docking (LOVpep, Supplementary Fig. 2a and Supplementary Note 1). Using this construct, plasma membrane recruitment to Mid2 was reversible on minute timescales and capable of repeated cycles of photoexcitation (Supplementary Fig. 4). Using a steerable 440 nm laser to illuminate small (~ 250 nm) regions, we were able to reliably and reversibly recruit ePDZb1–mCherry (a high-affinity ePDZ variant fused to mCherry) to a cortical patch (Fig. 1c). After localized recruitment, global plasma membrane recruitment could be stimulated by global photoexcitation (Fig. 1c).
We used LOVpep fusions to recruit ePDZb–mCherry (a low- affinity ePDZ variant fused to mCherry) to a variety of subcellular compartments. We tethered GFP–LOVpep to proteins with distinctive localizations, including Hof1 (bud neck), Pil1 (eisosomes) and Pma1 (plasma membrane)22. The GFP–LOVpep fusions localized as expected (Fig. 1d). ePDZ–mCherry was predominantly cytoplasmic in the dark and colocalized with GFP–LOVpep upon global photoexcitation.
We also tested TULIP tags in cultured HeLa cells. We fused GFP–LOVpep to the plasma membrane-localization signal from Lyn kinase23, and to the mitochondrial outer membrane protein Tom7022, and co-expressed each of these with ePDZb1–mCherry. In the dark, ePDZb1–mCherry was diffuse in the cytoplasm and nucleoplasm (Fig. 1e and Supplementary Fig. 5). Upon global blue light stimulation, ePDZb1–mCherry colocalized with GFP–LOVpep (Fig. 1e). The translocation was reversible for at least three cycles of illumination and recovery (Supplementary Video 1). Using spot photoexcitation, we were able to recruit ePDZb1–mCherry to the mitochondria or plasma membrane in a confined region of the cell (Fig. 1e).
Mutational tuning of affinity and kinetics
The docking equilibrium of the LOV–Jα interaction can be changed by mutations, and thereby modulate the dynamic range of effector activity17. We tested previously described AsLOV2 mutations that either decrease (V529N12), or increase (I532A17 and T406A,T407A (J.Z, C.A, and T.R.S., unpublished data)) helix docking (Fig. 2a). In addition to ePDZb and ePDZb1, we also evaluated recruitment of PDZ–mCherry to explore lower binding affinities. For most recruited proteins, the V529N mutation increased both the lit- and dark-state values of <Robs>, indicating increased binding (Fig. 2b). In the case of PDZ–mCherry recruitment, dark-state binding was effectively undetectable for LOVpep and the V529N mutant. The T406A,T407A double mutation, which stabilizes the N-terminal A′α helix of AsLOV2 and increases Jα docking affinity, decreased <Robs> for ePDZb–mCherry recruitment, in both the lit and dark states. Unexpectedly, T406A,T407A increased <Robs> for ePDZb1–mCherry recruitment (and slightly for dark-state PDZ–mCherry recruitment), perhaps due to adventitious interactions between the mutated LOV domain and ePDZb1. When paired with ePDZb or ePDZb1, the I532A mutation decreased <Robs> relative to the T406A,T407A variant. We also tested mutations in the peptide epitope that have been shown to diminish binding to ePDZ (Supplementary Note 1 and Supplementary Fig. 6). Most previously described mutations in AsLOV2, ePDZ, and the peptide epitope exhibited predictable effects on the LOVpep–ePDZ interaction, thereby supporting our design principle (Fig. 2b and Supplementary Fig. 6). The variants provide a range of experimentally accessible dimerization affinities.
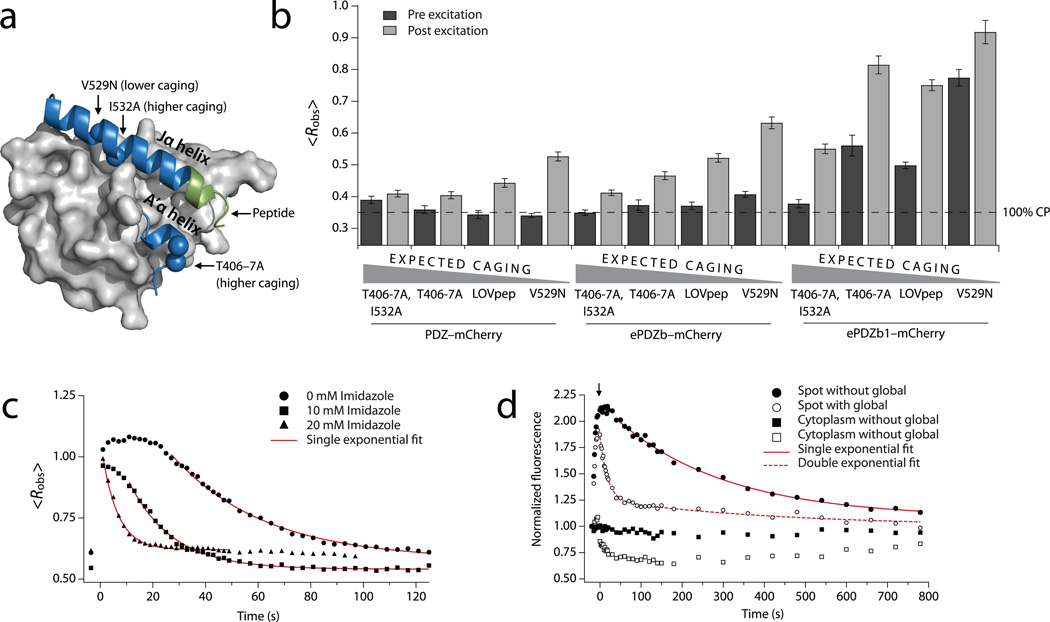
Figure 2.
Mutational and chemical control of binding. (a) AsLOV2 structure (Protein Data Bank: 2v0u) showing the location of the ePDZ epitope (green) and the caging mutations used in this study. (b) Lit- and dark-state <Robs> using LOVpep with caging mutations. Data are the means from a population (n ≥ 34) of cells; error bars show s.e.m. The dashed line represents <Robs> for ~100% cytoplasmic ePDZ–mCherry. (c) Kinetics of global recruitment and dissociation of ePDZb1–mCherry for LOVpep with wild-type dark-recovery kinetics. Imidazole is added to the media in the concentrations indicated. Data are the means from a population (n ≥ 8) of cells. Red lines are exponential fits of the dissociation phase (kobs). (d) Kinetics of spot recruitment and dissociation of ePDZb1–mCherry using slow-cycling (V416I) LOVpep. ePDZb1–mCherry is recruited to a spot as in Fig. 1c. For the filled symbols, the recruited molecules are allowed to recover without further illumination. For the open symbols, the cell is globally photoexcited at the time indicated by the arrow so as to deplete the unbound cytoplasmic pool (open squares) of ePDZb1–mCherry. Data are the means from a population (n ≥ 13) of cells. Red lines are exponential fits of the dissociation phase.
The maximum temporal resolution of experiments using ePDZ–LOVpep depends on the lifetime of the light-recruited complex. When photoexcitation ceases, light-recruited ePDZb fully dissociates from LOVpep within a few minutes in vivo (kobs = 0.041 s−1, Supplementary Table 1 and Supplementary Video 2). We investigated whether this observed dark-state dissociation rate (kobs) follows the LOVpep dark recovery rate (kphot) or the intrinsic ePDZ–peptide dissociation rate (kdiss). The rate kdiss is slower for ePDZb1 than for ePDZb when binding to model peptide substrates in vitro (~10−4 s−1 and ~10−2 s−1, respectively)20. However kobs was remarkably similar for the ePDZb–mCherry and ePDZb1–mCherry (Supplementary Table 1 and Supplementary Video 2). Using a mutated LOVpep (V416I) with approximately ten-fold slower dark recovery16, kobs was again similar for the ePDZb–mCherry and ePDZb1–mCherry, and both matched the slower kphot (Supplementary Table 1). In order to test whether kobs can be increased, we chemically accelerated kphot by adding imidazole to the media18. As with mutational tuning, changes in kobs generally tracked changes in kphot (Fig. 2c and Supplementary Table 1). Taken together, the data indicate that mutational or chemical modulation of the dark-state recovery of LOVpep also controls kobs.
These data are compatible with two models: either the ePDZ–LOVpep complex persists as long as the lit state, or the bound ePDZ rapidly exchanges with the unbound pool. To distinguish between these, we observed ePDZ–mCherry dissociation from a locally-photoexcited spot. Spot recruitment does not deplete the unbound pool; therefore the LOVpep occupancy is higher than during global recruitment (Fig. 1c). The dissociation of spot-recruited ePDZ–mCherry followed single-exponential kinetics with a rate similar to kphot when left unperturbed, but followed biphasic kinetics when the cytoplasmic pool was rapidly depleted by global photoexcitation after a few seconds (Fig. 2d, Supplementary Table 2 and Supplementary Video 3). Most of the bound ePDZ–mCherry dissociated quickly, while the remainder dissociated slowly with a rate matching kobs and kphot. Taken together, the data suggest that bound ePDZ rapidly exchanges with the unbound pool. When the unbound pool is quickly depleted by global recruitment, the amount of spot-recruited ePDZ is above the new equilibrium value. Re-equilibration occurs with the rapid dissociation of ePDZ, while the equilibrium itself changes slowly as the LOVpep reverts to the low-affinity dark state with the rate kphot. Fast dissociation kinetics and a continuously tunable photoexcitation lifetime are potentially desirable features for an optogenetic tool because together they allow precise temporal control in biological experiments.
Optical control of MAPK activation and polarity
We asked whether TULIPs can be used for light-activated cellular signaling. The yeast mating pathway normally is initiated by binding of a peptide pheromone to a G protein coupled receptor (GPCR) (Fig. 3a). Downstream, components of two conserved signaling modules are recruited to the plasma membrane by the activated G protein: one comprising a mitogen activated protein kinase (MAPK) cascade organized by the scaffold Ste5, and the other comprising the GTPase Cdc42 and its associated scaffolds, guanine nucleotide exchange factor[t1] and effectors. The MAPK module, whose activation leads to G1 arrest and transcription of mating-specific genes, has been extensively engineered in attempts to elucidate its workings24. Notably, tethering Ste5 or Ste11 to the plasma membrane robustly activates the pathway25,26. The Cdc42 module is required for polarized growth in budding yeast, and its activation is usually constrained by intrinsic or extrinsic spatial cues27. Disruption of Cdc42-mediated polarization in vegetatively growing cells prevents budding, but not isotropic growth, and leads to an enlarged terminal phenotype27.
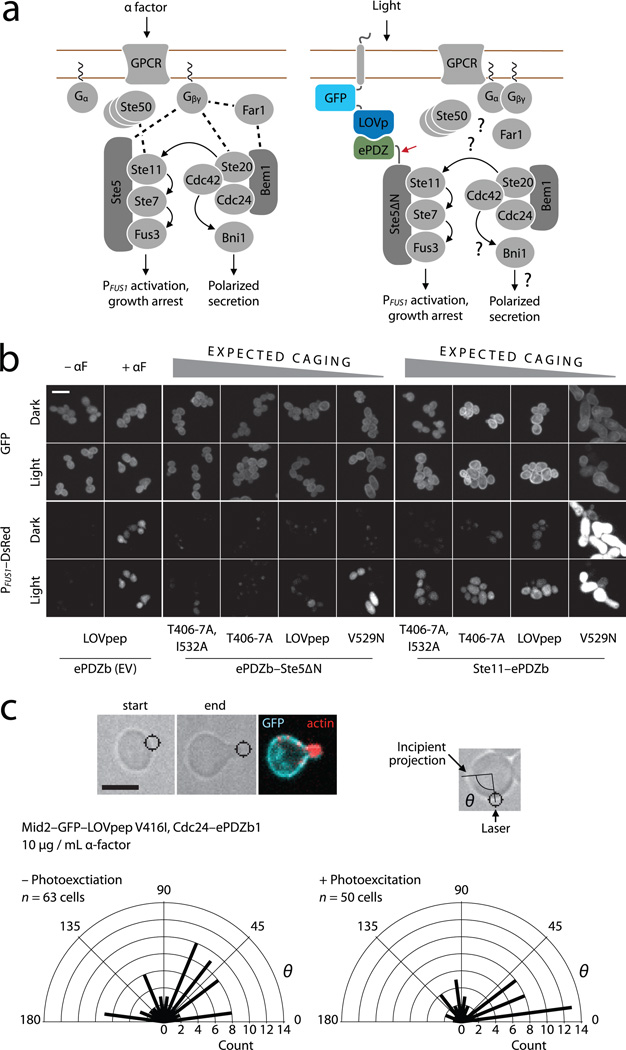
Figure 3.
Optical control of MAPK activation and polarity establishment in yeast. (a) The wild-type mating pathway in budding yeast (left), and a scheme for light-dependent plasma membrane recruitment of Ste5∆N (right). The red arrow indicates the fusion between ePDZ and Ste5∆N. Dashed lines indicate wild-type binding interactions, many of which may be absent in Ste5∆N recruitment. (b) PFUS1 promoter activation, cell cycle arrest and polarized growth in light- and dark-grown cells. ePDZb–Ste5∆N or ePDZb–Ste11 are globally recruited to plasma membrane-tethered LOVpep variants as indicated. Scale bar, 10 µm. (c) Light-directed polarized growth. Cells are exposed to mating pheromone to induce cell cycle arrest, then stimulated with spot photoexcitation to recruit Cdc24–ePDZb1 to plasma membrane-tethered LOVpep. Radial plots show quantification of light-directed polarized growth. θ is the angle between the spot of laser photoexcitation, the center of the cell and the incipient projection. Radial bars depict the number of polarization events of angle θ in each 15° sector. “– Photoexcitation” denotes a negative control experiment in which the spot photoexcitation laser was switched off. P < 0.01 for a comparison of experimental and control distributions (two-sample Kolmogorov–Smirnov). Scale bar, 5 µm.
We screened a variety of MAPK and Cdc42 associated proteins for growth defects upon plasma membrane recruitment. We performed initial experiments with a constitutively active LOVpep variant (LOVpepCA) fused to Mid2 and under control of the galactose-inducible GAL1 promoter. Several proteins, when fused to ePDZb caused growth arrest upon galactose-induced expression of LOVpepCA (Supplementary Fig. 7).
We next asked if light-dependent recruitment of ePDZb–Ste5ΔN (an allele deficient in G protein binding25) and ePDZb–Ste11 could cause MAPK pathway activation (Fig. 3a). There was little or no detectable dark state growth arrest upon galactose induction over a range of expected affinities (Supplementary Fig. 8), indicating that the ePDZb–LOVpep interaction is well caged in the dark with respect to biological activity. However, continuous illumination caused growth arrest in some strains, and the extent of arrest corresponded with the expected affinity of the LOVpep variant (Supplementary Fig. 8).
To characterize the phenotype, we illuminated cells for 4 hours in liquid culture and then examined them by microscopy and flow cytometry. As expected, αF-stimulated control cells formed mating projections (shmoos) and transcribed a PFUS1–DsRed-Max reporter gene28; these cells also did not show light-stimulated pathway activation (Fig. 3b and Supplementary Fig. 9). For ePDZb–Ste5ΔN recruitment, highly caged LOVpep variants did not measurably activate the pathway (Fig. 3b and Supplementary Fig. 9). This level of dark-state suppression is consistent with our previous observation that strong Jα docking can suppress effector activity even under full photoexcitation17. In contrast, less strongly caged LOVpep variants allowed more robust light-dependent cell-cycle arrest and polarized growth, presumably because the weaker peptide caging leads to a higher level of Ste5∆N recruitment. Conversely, more caging was required to bring ePDZb–Ste11 recruitment into a sensitive range, with the less caged variants causing constitutive activation (Fig. 3b and Supplementary Fig. 9). Intriguingly, light-stimulated cells were less polarized than αF-stimulated cells, even though both had stopped forming buds (Fig. 3b).
To determine whether TULIPs can also control GTPase signaling pathways, we examined light-directed recruitment of Cdc24–ePDZb1 to Mid2–GFP–LOVpep. Global recruitment caused a growth arrest with a terminal phenotype of large, round cells that depended on the strength of the ePDZ–peptide interaction (Supplementary Fig. 10). The ability of full length Cdc24 to block polarization upon global recruitment is surprising as earlier work indicated that the protein was autoinhibited29. This discrepancy could result from the efficiency with which the protein is recruited (Supplementary Fig. 10).
To determine whether plasma membrane-localized Cdc24 is active, we locally recruited Cdc24–ePDZb1 to specify the direction of polarized growth. Indeed, we could specify the orientation of mating projection growth in αF-arrested cells (Fig. 3c and Supplementary Video 4), suggesting that the GEF is intrinsically active. This result demonstrates that the TULIPs system can be used to control the activity of signaling molecules with high spatial precision, even in small (~ 5 µm diameter) cells.
DISCUSSION
Our approach, though conceptually similar to other strategies based on light-directed recruitment6–8, offers unique advantages. First, both components are small, facilitating genetic manipulation, and do not require an exogenous cofactor6. Second, both proteins are structurally and biophysically well characterized. The mechanism by which AsLOV2 modulates the activity of heterologous effectors is generally well-understood3,11,12,17, and our findings indicate that these principles are applicable to the ePDZ-LOVpep interaction. Numerous tuning mutations, including alterations of the intrinsic affinity, the stability of Jα helix docking, and the photocycle time, are available and can be easily incorporated into experiments (Supplementary Note 2). In principle, many activation mechanisms should be light-controllable using TULIPs, such as transcriptional regulation, enzyme–substrate enforced proximity, and protein fragment complementation.
Even in simple engineered systems, small changes in binding affinity can greatly impact function30. A failed or suboptimal implementation can be due to either design flaws or a poor choice of a parameter value; yet it is usually not obvious a priori which parameter values will be best. Just as electronics prototyping requires a ready assortment of resistors, capacitors, and transistors with different characteristics, our experiments demonstrate that biological prototyping requires protein modules that are quantifiably tunable in order to ensure robust activation of a pathway of interest.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Glick (University of Chicago), S. Koide (University of Chicago) and F. Cross (Rockefeller University) for sharing plasmids, and members of the Glotzer, Weiss, Sosnick, Munro, Kovar and Glick labs for helpful discussions. This work was supported by research (GM088668, M.G. and T.R.S.) and training grants from the US National Institutes of Health, a grant from the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust (M.G., T.R.S. and E.L.W), and by an American Cancer Society Postdoctoral Fellowship to D.S. (119248-PF-10-134-01-CCG).
Footnotes
AUTHOR CONTRIBUTIONS
D.S. and M.G. conceived of the TULIPs strategy. D.S., Y.L., E.W., E.L.W. and M.G. designed experiments. D.S., Y.L., E.W. and C.M.H performed experiments. D.S., Y.L., E.W., C.M.H., T.R.S., E.L.W. and M.G. analyzed data. J.Z., C.A. and T.R.S. provided new AsLOV2 mutations. D.S., E.L.W. and M.G. wrote the paper.
COMPETING FINANCIAL INTERESTS
A provisional patent application that includes portions of the research described in this manuscript has been filed by the University of Chicago Office of Technology and Intellectual Property.
REFERENCES
- 1.Hartman NC, Groves JT. Signaling clusters in the cell membrane. Curr. Opin. Cell Biol. 2011;23:370–376. doi: 10.1016/j.ceb.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toettcher JE, Voigt CA, Weiner OD, Lim WA. The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nat. Methods. 2011;8:35–38. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoo SK, et al. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev. Cell. 2010;18:226–236. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 2010;12:591–597. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat. Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 7.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy MJ, et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie JM, et al. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 10.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 11.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 12.Yao X, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-J alpha photoswitch. Nat. Chem. Biol. 2008;4:491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 14.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc. Natl. Acad. Sci. USA. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie JM, et al. Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochemistry. 2007;46:9310–9319. doi: 10.1021/bi700852w. [DOI] [PubMed] [Google Scholar]
- 16.Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat. Chem. Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strickland D, et al. Rationally improving LOV domain-based photoswitches. Nat. Methods. 2010;7:623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandre MT, Arents JC, van Grondelle R, Hellingwerf KJ, Kennis JT. A base-catalyzed mechanism for dark state recovery in the Avena sativa phototropin-1 LOV2 domain. Biochemistry. 2007;46:3129–3137. doi: 10.1021/bi062074e. [DOI] [PubMed] [Google Scholar]
- 19.Möglich A, Moffat K. Engineered photoreceptors as novel optogenetic tools. Photochem. Photobiol. Sci. 2010;9:1286–1300. doi: 10.1039/c0pp00167h. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Koide A, Makabe K, Koide S. Design of protein function leaps by directed domain interface evolution. Proc. Natl. Acad. Sci. USA. 2008;105:6578–6583. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Makabe K, Biancalana M, Koide A, Koide S. Structural basis for exquisite specificity of affinity clamps, synthetic binding proteins generated through directed domain-interface evolution. J. Mol. Biol. 2009;392:1221–1231. doi: 10.1016/j.jmb.2009.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 23.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pryciak PM. Designing new cellular signaling pathways. Chem. Biol. 2009;16:249–254. doi: 10.1016/j.chembiol.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pryciak PM, Huntress FA. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winters MJ, Lamson RE, Nakanishi H, Neiman AM, Pryciak PM. A membrane binding domain in the ste5 scaffold synergizes with Gβγ binding to control localization and signaling in pheromone response. Mol. Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 27.Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strack RL, et al. A noncytotoxic DsRed variant for whole-cell labeling. Nat. Methods. 2008;5:955–957. doi: 10.1038/nmeth.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimada Y, Wiget P, Gulli MP, Bi E, Peter M. The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. EMBO J. 2004;23:1051–1062. doi: 10.1038/sj.emboj.7600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301:1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.