Abstract
Background
Optimal care of persons infected with human immunodeficiency virus type 2 (HIV-2) requires an accurate assessment of HIV-2 plasma viral load (VL), but no clinically-approved quantitative HIV-2 RNA VL assay exists.
Objectives
To validate a novel quantitative HIV-2 RNA assay for clinical and research use.
Study Design
The Abbott m2000sp/rt platform was adapted for quantification of HIV-2 RNA in plasma. Amplification targeted a region of the long terminal repeat conserved in Group A and B HIV-2. Electron microscopy-counted-HIV-2 standards, the WHO/NIBSC HIV-2 International Standard and clinical specimens (N=162) were used to determine the precision, sensitivity, specificity, linear range, accuracy, and clinical performance of the assay.
Results
The quantitative linear range of the HIV-2 RNA assay was 10–1,000,000 copies/mL (R2 >0.99), with a limit of detection of 8 copies/mL (95% CI, 5–18 copies/ml). The assay did not cross-react with HIV-1, and quantification of HIV-2 RNA was not affected by the presence of >5 log10 HIV-1 RNA copies/mL. The total standard deviation (SD) and intra- and inter- run SD were 0.095, 0.093 and 0.162, respectively, at nominal inputs of 3.7, 1.7 and 1.0 log10 HIV-2 RNA copies/mL. The HIV-2 WHO/NIBSC International Standard (1000 IU) was shown to contain 152 RNA copies/mL (95% CI 141–163). Overall, HIV-2 RNA was quantified at ≥ 10 copies/mL from 86 (53%) clinical specimens (median, 2.24 log10copies/mL; range 10–16870), and nine specimens (6%) had HIV-2 RNA detected at <10 copies/mL.
Conclusions
We developed and validated a highly-sensitive HIV-2 VL assay that is suitable for clinical and research use.
Keywords: HIV-2, PCR, Real-time, diagnostic, plasma viral loads
1. Background
HIV-2 is endemic in West Africa and has spread globally with an estimated 1–2 million infections worldwide.1–4 Compared to HIV-1, patients infected with HIV-2 often have a longer asymptomatic stage, slower decline in CD4cells and decreased AIDS-associated mortality: this is thought to primarily be due to the very low HIV-2 plasma viral loads found in the majority of untreated patients, with 25–35% behaving as “elite-controllers”.5–8 In areas where HIV-2 and HIV-1 co-circulate, some patients are dually infected with both HIV types.9–11 Eight different HIV-2 Groups (A-H) have been reported, however, only Groups A and B are found to commonly infect humans.12,13
As is the case for HIV-1, sensitive and accurate measurements of HIV-2 viral load (VL) are critical for clinical management and monitoring of antiretroviral therapy. 14,15 United States FDA-approved and commercial HIV VL assays available internationally do not accurately detect or quantitate HIV-2 RNA in plasma16,17 and assays specific for HIV-2 RNA are limited to “in-house” methods designed for research use. 18,19 Thus, there is a global need to develop an independently validated VL assay for clinical care of HIV-2-infected patients that can be used in international settings.
2. Objectives
Our objectives were to develop a quantitative HIV-2 plasma RNA assay that uses the Abbott m2000 platform and to validate the assay following Clinical and Laboratory Standards Institute (CLSI) guidelines for clinical use20,21 and guidelines recommended by the pSMILE project (Patient Safety Monitoring in International Laboratories) (www.psmile.org).
3. Study design
3.1 HIV-2 and internal control (IC) multiplex RT-PCR
The following HIV-2 LTR sequence-specific primers and probe were used: forward primer (5′-GCGGAGAGGCTGGCAGAT-3′); reverse primer (5′-GAACACCCAGGCTCTACCTGCTA-3′); and probe (5′-6FAM-AGAGAACCTCCCAGG-NFQMGB-3′), as previously described.22 A non-competitive IC was ~1000-nt-long RNA synthesized in vitro (E. Ramos, MSc Thesis, UW, 2009).23 The forward and reverse primers for the amplification of IC RNA were 5’-GCAAATGTTAGCTAGTGCATCCA-3’ and 5’- TGTCACTTCCCCTTGGTTCTCTC-3’, respectively; the probe sequence was 5’-VIC-ATTGTAGTTGGTAGGAC-NFQMGB-3’. Reactions contained 1× RT-PCR buffer and 1x RT-PCR Enzyme Mix (AgPath™ One-Step RT-PCR Reagents Kits, Invitrogen Co, Carlsbad, CA), 600 nM of HIV-2 primers, 200 nM HIV-2 probe, 100 nM IC primers and 50 nM IC probe in a total volume of 100 µL. The following real-time PCR cycling parameters were used: 45°C for 10’ and 95°C for 10’ followed by 45 cycles at 95°C for 20“ and 60°C for 1’.
3.2 HIV-2 standards
We used the electron microscopy viral particle counted HIV-2 strain NIH-Z (Group A) as a standard (7.2 × 1010 viral particles/ml; Advanced Biotechnologies, Inc., Columbia, MD). Since two RNA genomes are packed in each viral particle, we calculated nominal RNA copies/mL by multiplying viral particles/mL by two. Serial dilutions of HIV-2 standards and positive controls were prepared in HIV-seronegative defibrinated human plasma (Basematrix 53, SeraCare Life Sciences, Inc., Milford MA). Following establishment of the linear range, for each assay run, four HIV-2 calibrators were analyzed at concentrations of 100000, 5000, 500 and 100 copies/mL. An HIV-2 Group B EHO viral stock (obtained from Jan McClure, UW) was also used to verify the assay linear range.24
3.3 Clinical samples
Plasma controls from HIV-uninfected individuals were confirmed using the Genetic Systems HIV1/2-Plus-O-EIA (Bio-Rad, Redmond, WA). HIV-1 RNA levels in all HIV-1-seropositive plasma samples were quantified using the Abbott HIV-1 RealTime assay (Abbott Molecular, Des Plaines, IL). HBV and HCV clinical specimens were provided by the UW Molecular Diagnostic Laboratory. United States (US) HIV-2– infected clinical samples originated from California, New York, Maryland, Massachusetts, Texas and Washington states. All clinical specimens were de-identified as required by the UW Humans Subjects Division.
Senegalese HIV-2 patient plasma samples were collected as part of ongoing studies of immune correlates of HIV-2 infection in antiretroviral (ARV)-naïve subjects (IE)25 and antiretroviral therapy for HIV-2 (H2A).22,26 HIV-2–infected individuals with clinical AIDS, CD4 counts <200/mm3, or CD4 counts <350/mm3 with clinical HIV-related symptoms were treated with antiretrovirals as part of the Senegalese Government Antiretroviral program (ISAARV) at the Clinique Des Maladies Infectieuses Ibrahima DIOP MAR, Dakar, Senegal. All study subjects provided written informed consent, and specimens were collected with human subjects approval from the UW, the University of Dakar, and the Senegalese Ministry of Health Institutional Review Boards.
3.4 HIV-2 Assay procedure
The Abbott m2000sp/rt platform was used to process patient plasma samples and amplify extracted RNA according to a standard operating procedure developed by UW CFAR Clinical Retrovirology Laboratory. The Abbott m2000sp instrument was operated by a customer-specific protocol that processed 1-mL aliquots of EDTA plasma and generated a 65-µL RNA eluate. A Master Mix Addition protocol was used to load 60 µL of RT-PCR reagent mix and transfer 40 µL of RNA eluate to the PCR plate. Approximately 600 copies of IC RNA per sample were co-extracted and co-amplified along with the HIV-2 RNA, and the amplified products were detected using the Abbott m2000 instrument. HIV-2 RNA levels were calculated using the m2000rt LDA software. The threshold setting for HIV-2 6FAM fluorescence was set at 0.02.
3.5 Potential interfering substances
The OptiChallenge Inhibition Panel (Acrometrix Life Technologies, Benicia, CA) was used to determine the extent to which each of seven different factors interfered with HIV-2 RNA quantification. The Certificate of Analysis for this panel listed tested concentrations of hemoglobin, heparin, bilirubin and triglycerides. HIV-2 standard was added to diluted aliquots, in HIV-negative plasma, of each panel member to achieve a final HIV-2 RNA concentration of 2.48 log10 copies/mL.
3.6 Statistical analyses
To determine the linearity of the assay, serial dilutions of HIV-2 standards were prepared in HIV-seronegative EDTA plasma at concentrations of 10, 50, 100, 1000, 5000, 100,000, 500,000 and 1,000,000 RNA copies/ml, and the samples were subjected to VL testing. Data used to determine the linear range of the assay were evaluated by linear and polynomial regression analyses using the trendline/regression tool of Excel 2010 (Microsoft Corp, Redmond WA). The FDIST function of Excel was used to calculate the correlation coefficients produced by linear, second- and third-order polynomial equations. The lower limit of detection was determined by probit regression analysis (SPSS version 13, Chicago, IL). For assay precision, the statistical method designed for the one-per-day run described in CLSI document EP5-A2 27 was used to calculate total standard deviation (SD), within-run SD, between-run SD and total coefficient of variation (CV). Data from experiments with the Optichallenge Inhibition Panel were tested for statistically significant differences using generalized linear models with Bonferroni’s corrections (PROC GLM, SAS version 9.3, SAS Inc., Cary NC).
4. RESULTS
4.1 HIV-2 Assay Linear range and lower limit of detection
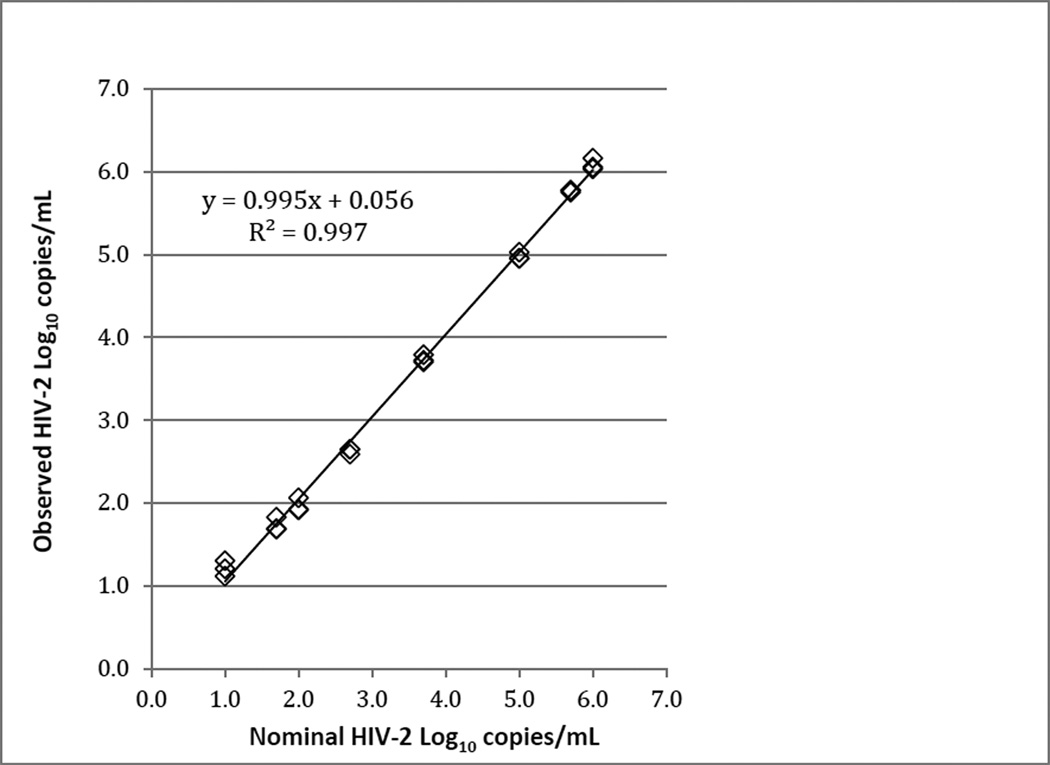
The linear range of the assay was determined using serial dilutions of EM-counted HIV-2 NIH-Z stocks (Group A) prepared in HIV-seronegative EDTA plasma. We consistently observed a linear response across the entire range of NIH-Z RNA concentrations tested (nominal 10–1,000,000 copies/ml)(Figures 1 and S1). Fitting of the data to second- or third-order polynomial equations did not significantly change the regression coefficient, indicating that the relationship between observed and nominal RNA values was well-described by the linear model (data not shown). Similar results were obtained using plasma samples spiked with 10-fold serial dilutions of HIV-2 EHO (Group B) through a linear range of 1.0 to 6.5 log10 RNA copies/ml (Figure S2).
Figure 1.
Representative data demonstrating the linear range of the HIV-2 VL assay. Viral RNA levels were measured in HIV-seronegative plasma samples that were spiked with EM-counted HIV-2 NIH-Z at final concentrations of 10, 50, 100, 500, 5000, 100,000, 500,000 and 1,000,000 RNA copies/ml. Slope and Y-intercept values for the resultant trend line were 0.995 and 0.056, respectively, as determined by linear regression (R2 = 0.997). Each sample was assayed in triplicate.
To determine the lower limit of detection (LLOD), HIV-2 NIH-Z standards were prepared at 20, 10, 5, 2, 1 and 0.5 RNA copies/mL. A total of 18 replicates were tested for each RNA concentration on three different days. The sensitivity of detection was 100% for RNA concentrations ≥10 copies/mL (Table 1). Based on probit regression analysis, the mean LLOD for the assay was 8 HIV-2 RNA copies/mL (95% CI, 5–18 copies/mL).
Table 1.
Lower limit of detection using HIV-2 NIH-Z virion standard.
| Concentration HIV-2 RNA copies/mL |
Log10 (conc.) | Number tested | Number Detected | Percent detected |
|---|---|---|---|---|
| 20 | 1.30 | 18 | 18 | 100% |
| 10 | 1.00 | 18 | 18 | 100% |
| 5 | 0.70 | 18 | 15 | 83% |
| 2 | 0.30 | 18 | 7 | 39% |
| 1 | 0.00 | 18 | 6 | 33% |
| 0.5 | −0.30 | 18 | 1 | 6% |
4.2 HIV-2 Assay Precision and Accuracy
Assay precision was evaluated by preparing standards at nominal concentrations of 5000, 50 and 10 RNA copies/mL and quantifying the level of HIV-2 RNA in each sample daily in duplicate for 21 days (Table 2). The observed mean of the nominal 3.7 log10 copies/mL was 3.67 log10 copies/mL (95% CI 3.47–3.87 log10 copies/mL). However, the nominal 1.0 log10 copies/mL control had a higher than expected observed mean of 1.48 log10 copies/mL (95% CI 1.17–1.79 log10 copies/mL). The average slope of the observed calibration curve (−3.40) was less than the expected value (−3.32), which resulted in an inflated quantification of the low-copy number samples. Additionally, HIV-2 standards at 1.0 log10 copies/mL were found to have a lower than expected cycle number (CN) (Figures 1, S1, S2).
Table 2.
Precision of HIV-2 RNA quantification at nominal 5000, 50 and 10 copies/mL.
| Nominal Standards | |||
| High Control | Mid Control | Low Control | |
| Copies/mL | 5000 | 50 | 10 |
| Log10 copies/mL | 3.70 | 1.70 | 1.00 |
| Assay (mean log10copies/mL) | 3.67 | 1.91 | 1.48 |
| Repeatability (within-run) SD a | 0.050 | 0.079 | 0.129 |
| Reproducibility (between-day) SD a | 0.080 | 0.048 | 0.098 |
| Total SD a | 0.095 | 0.093 | 0.162 |
| Total CV(%)b | 22 | 25 | 34 |
SD on the log10 scale.
calculated by dividing untransformed total SD copies/mL with mean copies/mL.
The National Institute for Biological Standardization and Control (NIBSC)/WHO recently developed an HIV-2 international reference standard for calibration of HIV-2 assays (lyophilized HIV-2 CAM2 Group A, set at 1000 International Units (IU)). 28 We determined the relationship between our HIV-2 assay copy number and IU. Four 1-mL aliquots of reconstituted viral stock were tested in two different runs and we found a mean of 152 copies HIV-2 RNA/mL per 1000 IU (95% CI: 141–163). Our conversion is within the reported range, 1.96–4.20 log10 copies per 1000 IU, as tested by nine international laboratories using in-house quantitative assays.28
4.3 HIV-2 Assay Specificity
To investigate the potential for false-positive or falsely-inflated results due to off-target amplification (i.e. cross-reactivity), we tested twenty US and twenty Senegal HIV-seronegative plasma samples, all tested negative in the HIV-2 assay. In addition, 30 HIV-1–seropositive plasma samples were tested the HIV-2 VL assay. The level of HIV-1 RNA in these samples, as quantified by the Abbott HIV-1 RealTime Assay, ranged from undetectable to 5.6 log10 RNA copies/mL. All 30 HIV-1 samples tested negative for HIV-2 (data not shown). To determine whether HIV-1 might affect quantitation of HIV-2, eight HIV-1 plasma samples were spiked with the HIV-2 viral standard. The efficiency of detecting HIV-2 and quantification of the HIV-2 was not significantly changed by the presence of HIV-1 RNA ranging from 3.5 to 5.6 log10 copies/mL (P=0.9) (Table 3). Four, HIV negative, Hepatitis B virus (VL range: 3.25–7.24 log10 IU/mL) and four Hepatitis C virus (VL range: 2.82–6.57 log10 IU/mL) containing clinical samples tested negative for HIV-2 in the assay. Similarly, RNA extracted from an estimated 200 Plasmodium falciparum cultured in human erythrocytes tested negative for HIV-2 in the assay.
Table 3.
Interference with HIV-1 RNA
| Spike of HIV-2 viral stock | ||||
|---|---|---|---|---|
| Sample IDa | Estimated HIV-1 Log10 copies/mLb |
HIV-2 quantity (Log10 copies/mL) |
HIV-2 CNc | ICd CN |
| v265 | 5.61 | 1.46 | 37.48 | 32.13 |
| v650 | 5.40 | 1.62 | 36.91 | 32.06 |
| v564 | 5.34 | 1.48 | 37.43 | 32.08 |
| v608 | 4.44 | 1.51 | 37.32 | 32.32 |
| v168 | 3.73 | 1.54 | 37.16 | 31.73 |
| v349 | 3.61 | 1.54 | 37.16 | 31.93 |
| v734 | 3.49 | 1.66 | 36.76 | 32.12 |
| Negative plasma | Not Detected | 1.49 | 37.34 | 32.36 |
The last three digits of container identification number (9-digit number) are listed.
Quantified by Abbott HIV-1 RealTime assay.
CN named by Abbott LDA and is equivalent to the quantification cycle defined by MIQE.34
Internal competitive RNA control
4.4 HIV-2 Assay extraction efficiency and effects of Interfering substances commonly found in clinical blood samples
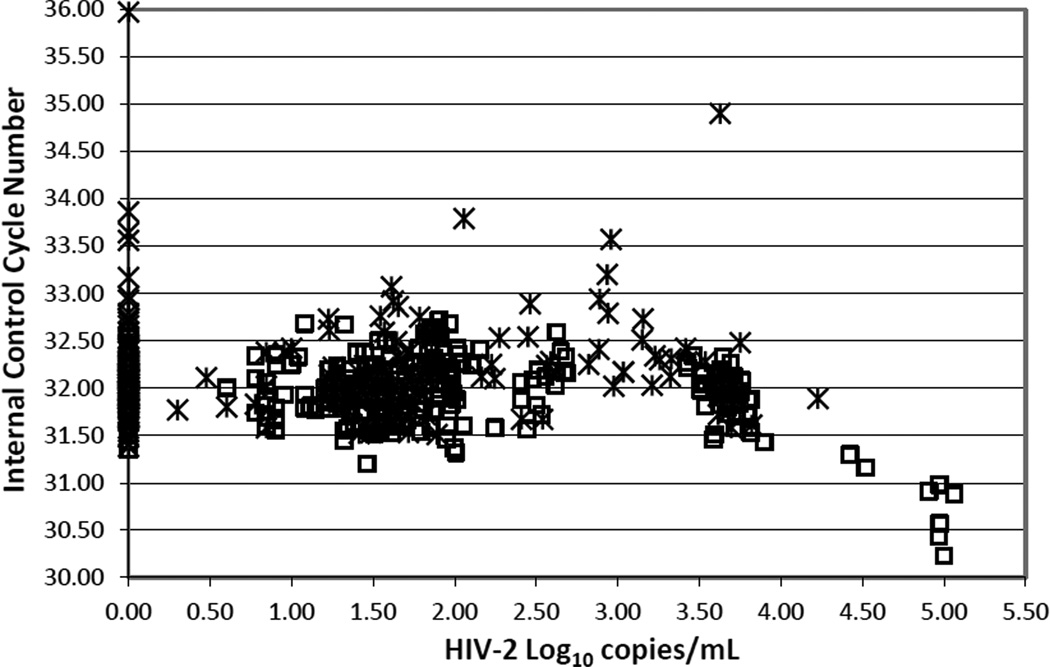
We developed a non-competitive IC to monitor for extraction efficiency and inhibitors of the HIV-2 assay. For 340 HIV-2 standards and 191 clinical EDTA plasma specimens, the mean (± SD) of IC CN was 32 (±0.55) (Figure 2). Samples with an IC CN greater than 3SD above the mean were considered for retesting at dilutions of 1:2 or 1:4 to reduce possible inhibitors. An assay run was rejected if IC CN of both positive and negative controls was greater than 3SD above the mean to account for possible problems with extraction efficiency. The apparent decrease in IC CN in samples above 4 log10 HIV-2 copies/mL resulted from the overlap of the emission fluorescence spectra for the 6-FAM and VIC dyes.
Figure 2.
HIV-2 viral loads in clinical plasma samples (crosses) and in HIV-2 NIH-Z and EHO standards (open squares) plotted against the corresponding cycle number (CN) for the internal RNA control. Clinical plasma samples included HIV-seronegative, HIV-seropositive, hepatitis B and hepatitis C– infected patients.
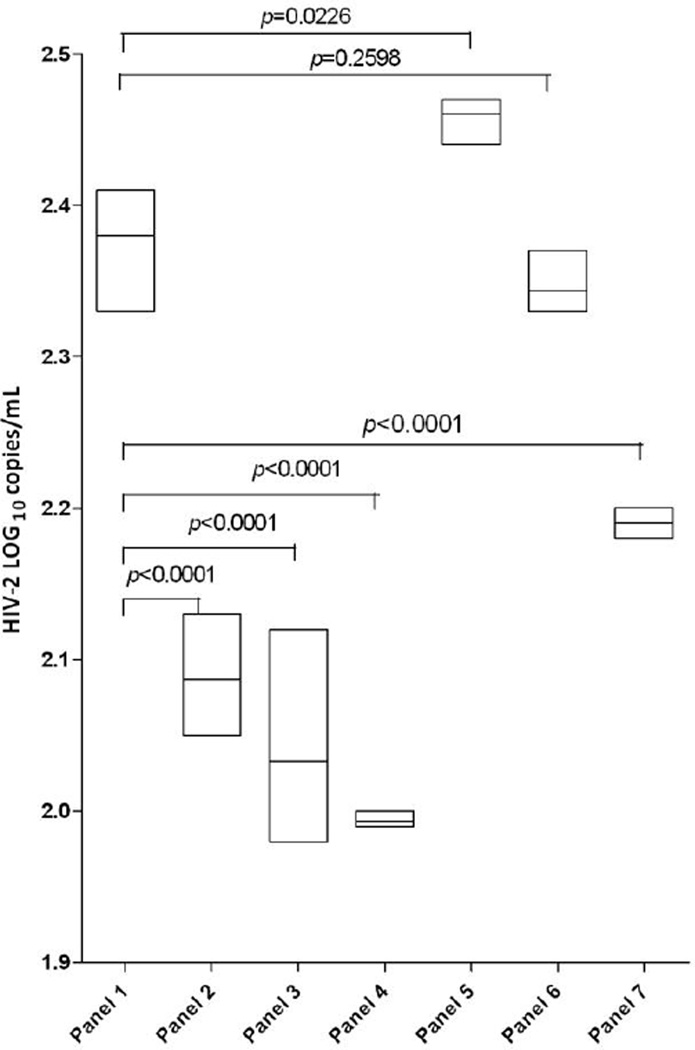
In order to determine the effect of common interfering substances found in blood from clinical samples we ran the Optichallenge Inhibition Panel through our assay. Panel samples were spiked with a range of HIV-2 virus levels in triplicate. As shown in Figure 3, a significant decrease in spiked HIV-2 RNA viral load detection was observed for hemolyzed plasma and icteric plasma but not for heparin or lipemic plasma compared to that for EDTA control plasma. Reduced inhibitory effects were observed in 4-fold diluted icteric plasma and 64-fold diluted hemolyzed (low) plasma (data not shown). Among the intrinsic substances in plasma that were tested, no interference was found for the following concentration: triglycerides (785mg/dL), bilirubin (7.5mg/dL) or hemoglobin (15mg/dL).
Figure 3.
Effect of interfering substances on measurements of HIV-2 RNA in plasma. Each box shows the highest, lowest and mean RNA values in triplicate samples. Panel 1, EDTA plasma; Panel 2, Hemolyzed plasma Low (0.5g/dL hemoglobin); Panel 3, Hemolyzed plasma Mid (1.0g/dL hemoglobin); Panel 4 Hemolyzed plasma Hi (1.95g/dL hemoglobin); Panel 5, Heparin plasma (19.85 USP/mL heparin); Panel 6, Lipemic plasma (785 mg/dL triglycerides); Panel 7, Icteric plasma (15mg/dL bilirubin). Mean RNA values were compared between the interfering substance and EDTA plasma using generalized linear models and Bonferroni corrections. A similar pattern of inhibition was observed for the IC CN for each panel (data not shown).
4.5 Distribution of HIV-2 plasma VL in Senegalese and US clinical samples
The HIV-2 plasma VL and CD4 counts for Senegalese IE and H2A study groups and US clinical plasma specimens had a similar range of HIV-2 VL (see Table 4). Among all HIV-2 seropositive specimens tested, 43 were Group A and 7 were Group B, as determined by the sequence genotyping of HIV-2 pol(protease, RT and/or integrase).22,26 Twenty-six (60%) of the Group-A samples had quantifiable HIV-2 VL ranging from 1.54–4.23 log10 RNA copies/mL. Four of seven (57%) Group-B samples had an HIV-2 VL between 1.81–3.73 log10copies/mL. In addition, one clinical US sample was determined to be an HIV-2 Group AB recombinant with an HIV-2 VL of 3.72 log10 RNA copies/mL.
Table 4.
Characteristics of HIV-2 clinical plasma samples from Senegal and the United States
| Sample source | Senegalese IE studya (n=55) |
Senegalese H2A studya (n=55) |
US clinicalb (n=52) |
|---|---|---|---|
| The status of ARV | ARV-naïve | ARV-experienced | ARV-naïve & experienced |
| Year samples collected CD4 cells | 2004–2006 | 2010–2011 | 2011–2012 |
| Mean cells/mm3 | 605 | 338 | N/A |
| No. (%) with counts of: | |||
| >500 | 30(55) | 7(13) | |
| 350–500 | 9(16) | 13(24) | |
| <350 | 16(29) | 35(64) | |
| HIV-2 plasma VL | |||
| Mean log10 copies/mL | 2.66 | 3.11 | 3.03 |
| No. (%) with VL of: | |||
| Not detected | 20(36) | 26(47) | 21(40) |
| Detected, <10 copies/mL | 5(9) | 2(4) | 2(4) |
| Quantifiable range, >10 copies/mL | 30(55) | 27(49) | 29(56) |
| Median log10 copies/mL (range) | 1.69(1.00–3.75) | 2.45(1.20–4.23) | 2.86(1.23–4.20) |
Single sample per individual patient.
Patients’ clinical information was limited.
5. Discussion
Although the prevalence HIV-2 infection is much lower compared to HIV-129, the diagnosis of HIV-2 infection is significant because of the different natural history and clinical management from HIV-1 infection. As such, there is a niche for a clinical HIV-2 VL assay to assist both timely diagnosis and monitoring of HIV-2 infected patients. The robotic specimen extraction on the Abbott m2000sp instrument and the LDA software of the m2000rt instrument provides a significant advantage for lower handling error and increased precision. Here, we showed the acceptable performance specifications for our HIV-2 RNA quantitative assay, which include linear range, precision, sensitivity, specificity and accuracy, following CLSI guidelines.20
The linear range of our assay (10–1,000,000 copies HIV-2 RNA/mL) encompasses the ranges typically reported for HIV-2 patient plasma VL.14,30–33 The sensitivity of the assay was 8 copies/mL (95% CI, 5–18), which is the most sensitive HIV-2 plasma RNA assay reported to date. To maintain the precision required to have a 90% power to detect a 5-fold difference in RNA copy number, the intra-assay SDs should be no greater than 0.15 log10 copies/mL.20 For the HIV-2 RNA assay, both the intra- and inter-assay precision (SDs) at nominal levels of 1.0, 1.7 and 3.7 log10 copies/mL met this criterion (Table 2).
We demonstrated the specificity of this assay by testing HIV-negative plasma from the US and Senegal, HIV-1, HBV, HCV and malaria, which all co-infect patients in African countries where HIV-2 infection is prevalent. Detection of HIV-2 RNA was not affected by the presence of HIV-1 RNA. The use of the OptiChallenge Interference Panel helped to evaluate HIV-2 RNA quantification in the presence of clinically important assay inhibitors. Of the panel members, heparin at 20 USP/mL, which is greater than the concentration (15 USP/mL) in the BD Vacutainers, was not inhibitory. Among the potential intrinsic substances in plasma that were tested, no interference was found for triglycerides (785mg/dL), bilirubin (7.5mg/dL) or hemoglobin (15mg/dL).
To evaluate the clinical performance of our assay, we tested HIV-2–seropositive plasma samples from ARV-naïve and ARV-experienced patients in Senegal and from ARV-treated patients residing in the US. All three groups yielded comparable ranges of HIV-2 VL (~1–4 log10 copies/mL). In addition, we tested seven clinical specimens obtained from patients infected with Group B HIV-2, and found that these exhibited a VL range similar to that observed for 43 Group A specimens. These data are concordant with an earlier study showing that the same HIV-2 primers and probes used in the present work provide accurate quantification of EM-counted Group A and B HIV-2 standards.19 Broad HIV-2 Group A and B amplification is likely due to the fact that only 9 of 1320 nucleotides (< 1%) were mismatched relative to the sequence primers/probe when compared to alignments of 24 HIV-2 complete genomes in the HIV Sequence Compendium 2010, including Group A and B strains.34
Taken together, our analysis shows that the HIV-2 assay developed in this study using the Abbott m2000sp/rt platform is sensitive and reliable for clinical monitoring of HIV-2 RNA VL in HIV-2– infected individuals.
Supplementary Material
Acknowledgements
We especially thank Abbott Molecular for supplying the m2000sp/rt platform. We also thank Silvio Cenzi, (Abbott Molecular) for advice and technical support, Lucia Torian (New Yory City Department of Health) for helpful discussion and patient samples, and Shu-Kuang Lee, (UW Department of Epidemiology) for the advice regarding statistical testing. The UW-Dakar HIV-2 Study Group includes: Macoumba Toure, Selly Ba, Ndeye Mery Dia Badiane, Louise Fortes, Cheikh T. Ndour, Jacques Ndour, Fatou Niasse, Fatima Sall, Balla Tall, Fatou Traore, Habibatou Diallo Agne, Ndeye Rokhaya Fall, Sophie Chablis, Marie Pierre Sy, Mame Dieumba, Bintou Diaw, Mbaye Ndoye, Khady Diop, Amadou Bale Diop, Cheikh Gueye, Boubacar Diamanka, Marianne Ndiaye, Mari, Josh Stern, Qinghua Feng, Dana Raugi, Charlotte Pan, Beruk Asfaw, Brad Church, Matt Coyne, Alexandra Hernandez, Kara Parker, Natalie Zheng, Julie MacElrath and Jim Mullins.
FUNDING: These studies were supported by grants to GSG & RWC from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (2R01-AI060466), the AIDS Clinical Trials Group Virology Specialty Laboratory (AI-38858), the University of Washington Royalty Research Fund, and the University of Washington Center for AIDS Research (CFAR)—an NIH-funded program (P30 AI027757) supported by the following NIH Institutes and Centers: NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, and NIA.
Abbreviations
- HIV-2
human immunodeficiency virus type 2
- VL
viral loads
- CN
cycle number
- IC
internal control
- LDA
laboratory-defined applications
- LLOD
lower limit of detection
- SD
standard deviation
- CV
coefficient of variation
- US
United States.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: Dr. Gottlieb had travel paid for by Abbott Molecular to present this data, in part, at an ACTG/Abbott Molecular meeting, Chicago IL. No other conflicts of interest.
REFERENCES
- 1.Clavel F, Guetard D, Brun-Vezinet F, Chamaret S, Rey MA, Santos-Ferreira MO, et al. Isolation of a new human retrovirus from West African patients with AIDS. Science. 1986;233(4761):343–346. doi: 10.1126/science.2425430. Epub 1986/07/18. [DOI] [PubMed] [Google Scholar]
- 2.Arien KK, Abraha A, Quinones-Mateu ME, Kestens L, Vanham G, Arts EJ. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M HIV 1 group O HIV-2 isolates. Journal of virology. 2005;79(14):8979–8990. doi: 10.1128/JVI.79.14.8979-8990.2005. Epub 2005/07/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Cock KM, Adjorlolo G, Ekpini E, Sibailly T, Kouadio J, Maran M, et al. Epidemiology and transmission of HIV-2. Why there is no HIV-2 pandemic. JAMA : the journal of the American Medical Association. 1993;270(17):2083–2086. doi: 10.1001/jama.270.17.2083. Epub 1993/11/03. [DOI] [PubMed] [Google Scholar]
- 4.Gottlieb GS, Hawes SE, Wong KG, Raugi DN, Agne HD, Critchlow CW, et al. HIV type 2 protease, reverse transcriptase, and envelope viral variation in the PBMC and genital tract of ARV-naive women in Senegal. AIDS research and human retroviruses. 2008;24(6):857–864. doi: 10.1089/aid.2008.0015. Epub 2008/06/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanki PJ, Travers KU, S MB, Hsieh CC, Marlink RG, Gueye NA, et al. Slower heterosexual spread of HIV-2 than HIV-1. Lancet. 1994;343(8903):943–946. doi: 10.1016/s0140-6736(94)90065-5. Epub 1994/04/16. [DOI] [PubMed] [Google Scholar]
- 6.Simon F, Matheron S, Tamalet C, Loussert-Ajaka I, Bartczak S, Pepin JM, et al. Cellular and plasma viral load in patients infected with HIV-2. AIDS. 1993;7(11):1411–1417. doi: 10.1097/00002030-199311000-00002. Epub 1993/11/01. [DOI] [PubMed] [Google Scholar]
- 7.Marlink R, Kanki P, Thior I, Travers K, Eisen G, Siby T, et al. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science. 1994;265(5178):1587–1590. doi: 10.1126/science.7915856. Epub 1994/09/09. [DOI] [PubMed] [Google Scholar]
- 8.Gottlieb GS, Sow PS, Hawes SE, Ndoye I, Coll-Seck AM, Curlin ME, et al. Molecular epidemiology of dual HIV-1/HIV-2 seropositive adults from Senegal, West Africa. AIDS research and human retroviruses. 2003;19(7):575–584. doi: 10.1089/088922203322230941. Epub 2003/08/12. [DOI] [PubMed] [Google Scholar]
- 9.Rayfield M, De Cock K, Heyward W, Goldstein L, Krebs J, Kwok S, et al. Mixed human immunodeficiency virus (HIV) infection in an individual: demonstration of both HIV type 1 and type 2 proviral sequences by using polymerase chain reaction. The Journal of infectious diseases. 1988;158(6):1170–1176. doi: 10.1093/infdis/158.6.1170. Epub 1988/12/01. [DOI] [PubMed] [Google Scholar]
- 10.Evans LA, Moreau J, Odehouri K, Seto D, Thomson-Honnebier G, Legg H, et al. Simultaneous isolation of HIV-1 and HIV-2 from an AIDS patient. Lancet. 1988;2(8625):1389–1391. doi: 10.1016/s0140-6736(88)90586-7. Epub 1988/12/17. [DOI] [PubMed] [Google Scholar]
- 11.Gunthard HF, Huber M, Kuster H, Shah C, Schupbach J, Trkola A, et al. HIV-1 superinfection in an HIV-2-infected woman with subsequent control of HIV-1 plasma viremia. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2009;48(11):e117–20. doi: 10.1086/598987. Epub 2009/04/23. [DOI] [PubMed] [Google Scholar]
- 12.Damond F, Worobey M, Campa P, Farfara I, Colin G, Matheron S, et al. Identification of a highly divergent HIV type 2 and proposal for a change in HIV type 2 classification. AIDS research and human retroviruses. 2004;20(6):666–672. doi: 10.1089/0889222041217392. Epub 2004/07/10. [DOI] [PubMed] [Google Scholar]
- 13.Santiago ML, Range F, Keele BF, Li Y, Bailes E, Bibollet-Ruche F, et al. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Tai Forest, Cote d'Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. Journal of virology. 2005;79(19):12515–12527. doi: 10.1128/JVI.79.19.12515-12527.2005. Epub 2005/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodes B, Toro C, Jimenez V, Soriano V. Viral response to antiretroviral therapy in a patient coinfected with HIV type 1 and type 2. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2005;41(2):e19–e21. doi: 10.1086/431204. Epub 2005/06/29. [DOI] [PubMed] [Google Scholar]
- 15.Gottlieb GS, Eholie SP, Nkengasong JN, Jallow S, Rowland-Jones S, Whittle HC, et al. A call for randomized controlled trials of antiretroviral therapy for HIV-2 infection in West Africa. AIDS. 2008;22(16):2069–2072. doi: 10.1097/QAD.0b013e32830edd44. discussion 73–4. Epub 2008/10/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan PA, Wakeman SE, Flanigan T, Cu-Uvin S, Kojic E, Kantor R. HIV-2 diagnosis and quantification in high-risk patients. AIDS research and therapy. 2008;5:18. doi: 10.1186/1742-6405-5-18. Epub 2008/08/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seyoum E, Wolday D, Mekonen T, Girma M, Meselle T, Kallander C, et al. Alternative approach to blood screening using the ExaVir reverse transcriptase activity assay. Current HIV research. 2005;3(4):371–376. doi: 10.2174/157016205774370438. Epub 2005/10/28. [DOI] [PubMed] [Google Scholar]
- 18.Damond F, Benard A, Ruelle J, Alabi A, Kupfer B, Gomes P, et al. Quality control assessment of human immunodeficiency virus type 2 (HIV-2) viral load quantification assays: results from an international collaboration on HIV-2 infection in 2006. Journal of clinical microbiology. 2008;46(6):2088–2091. doi: 10.1128/JCM.00126-08. Epub 2008/04/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damond F, Benard A, Balotta C, Boni J, Cotten M, Duque V, et al. An International Collaboration To Standardize HIV-2 Viral Load Assays: Results from the 2009 ACHIEV2E Quality Control Study. Journal of clinical microbiology. 2011;49(10):3491–3497. doi: 10.1128/JCM.02389-10. Epub 2011/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CLSI/NCCLS. Quantitative molecular methods for infectious diseases; Approved guideline, 2nd ed. CLSI document MM06-A2. 28. Vol. 23. Wayne, PA: Clinical and Laboratory Standards Institute; 2011. [Google Scholar]
- 21.Burd EM. Validation of laboratory-developed molecular assays for infectious diseases. Clinical microbiology reviews. 2010;23(3):550–576. doi: 10.1128/CMR.00074-09. Epub 2010/07/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottlieb GS, Badiane NM, Hawes SE, Fortes L, Toure M, Ndour CT, et al. Emergence of multiclass drug-resistance in HIV-2 in antiretroviral-treated individuals in Senegal: implications for HIV-2 treatment in resouce-limited West Africa. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2009;48(4):476–483. doi: 10.1086/596504. Epub 2009/01/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gandhi RT, Coombs RW, Chan ES, Bosch RJ, Zheng L, Margolis DM, et al. No Effect of Raltegravir Intensification on Viral Replication Markers in the Blood of HIV-1-infected Patients Receiving Antiretroviral Therapy. J Acquir Immune Defic Syndr. 2011 doi: 10.1097/QAI.0b013e31823fd1f2. Epub 2011/11/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClure J, Schmidt AM, Rey-Cuille MA, Bannink J, Misher L, Tsai CC, et al. Derivation and characterization of a highly pathogenic isolate of human immunodeficiency virus type 2 that causes rapid CD4+ cell depletion in Macaca nemestrina. Journal of medical primatology. 2000;29(3–4):114–126. doi: 10.1034/j.1600-0684.2000.290304.x. Epub 2000/11/21. [DOI] [PubMed] [Google Scholar]
- 25.Zheng NN, Kiviat NB, Sow PS, Hawes SE, Wilson A, Diallo-Agne H, et al. Comparison of human immunodeficiency virus (HIV)-specific T-cell responses in HIV-1- and HIV-2-infected individuals in Senegal. Journal of virology. 2004;78(24):13934–13942. doi: 10.1128/JVI.78.24.13934-13942.2004. Epub 2004/11/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottlieb GS, Smith RA, Dia Badiane NM, Ba S, Hawes SE, Toure M, et al. HIV-2 Integrase Variation in Integrase Inhibitor-Naive Adults in Senegal, West Africa. PloS one. 2011;6(7):e22204. doi: 10.1371/journal.pone.0022204. Epub 2011/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CLSI/NCCLS. Approved guideline, 2nd ed. CLSI document EP5-A2. Wayne, PA: Clinical and Laboratory Standards Institute; 2004. Evaluation of precision performance of quantitative measurement methods. [Google Scholar]
- 28.Holmes H, Berry N, Heath A, Morris C. Preparation and evaluation of the 1st international standard for the quantitation of HIV-2 RNA in plasma. Journal of virological methods. 2011;175(2):246–252. doi: 10.1016/j.jviromet.2011.05.025. Epub 2011/06/07. [DOI] [PubMed] [Google Scholar]
- 29.HIV-2 Infection Surveillance--United States, 1987–2009. MMWR Morbidity and mortality weekly report. 2011;60(29):985–988. Epub 2011/07/29. [PubMed] [Google Scholar]
- 30.Gottlieb GS, Sow PS, Hawes SE, Ndoye I, Redman M, Coll-Seck AM, et al. Equal plasma viral loads predict a similar rate of CD4+ T cell decline in human immunodeficiency virus (HIV) type 1- and HIV-2- infected individuals from Senegal, West Africa. The Journal of infectious diseases. 2002;185(7):905–914. doi: 10.1086/339295. Epub 2002/03/29. [DOI] [PubMed] [Google Scholar]
- 31.Damond F, Collin G, Descamps D, Matheron S, Pueyo S, Taieb A, et al. Improved sensitivity of human immunodeficiency virus type 2 subtype B plasma viral load assay. Journal of clinical microbiology. 2005;43(8):4234–4236. doi: 10.1128/JCM.43.8.4234-4236.2005. Epub 2005/08/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferns RB, Garson JA. Development and evaluation of a real-time RT-PCR assay for quantification of cell-free human immunodeficiency virus type 2 using a Brome Mosaic Virus internal control. Journal of virological methods. 2006;135(1):102–108. doi: 10.1016/j.jviromet.2006.02.005. Epub 2006/03/28. [DOI] [PubMed] [Google Scholar]
- 33.Ruelle J, Mukadi BK, Schutten M, Goubau P. Quantitative real-time PCR on Lightcycler for the detection of human immunodeficiency virus type 2 (HIV-2) Journal of virological methods. 2004;117(1):67–74. doi: 10.1016/j.jviromet.2003.12.006. Epub 2004/03/17. [DOI] [PubMed] [Google Scholar]
- 34.Kuiken C, Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, et al. HIV Sequence Compendium 2010 Published by Theoretical Biology and Biophysics Group. Los Alamos National Laboratory, NM; 2010. LA-UR 10-03684. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.