Abstract
Pederin belongs to a group of antitumor compounds found in terrestrial beetles and marine sponges. It is used by apparently all members of the rove beetle genera Paederus and Paederidus as a chemical defense against predators. However, a recent analysis of the putative pederin biosynthesis (ped) gene cluster strongly suggests that pederin is produced by bacterial symbionts. We have sequenced an extended region of the symbiont genome to gain further insight into the biology of this as-yet-unculturable bacterium and the evolution of pederin symbiosis. Our data indicate that the symbiont is a very close relative of Pseudomonas aeruginosa that has acquired several foreign genetic elements by horizontal gene transfer. Besides one functional tellurite resistance operon, the region contains a genomic island spanning 71.6 kb that harbors the putative pederin biosynthetic genes. Several decayed insertion sequence elements and the mosaic-like appearance of the island suggest that the acquisition of the ped symbiosis genes was followed by further insertions and rearrangements. A horizontal transfer of genes for the biosynthesis of protective substances could explain the widespread occurrence of pederin-type compounds in unrelated animals from diverse habitats.
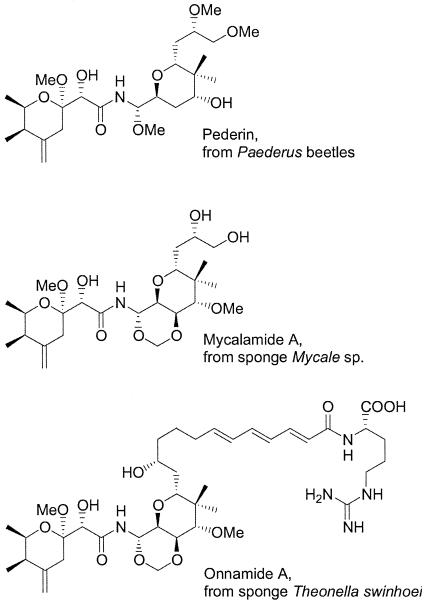
The pederin group of natural products consists of numerous members with potent antitumor and antiviral activities (Fig. 1) (15). Remarkably, these structurally highly similar substances have been isolated from two entirely unrelated animal groups. Pederin itself occurs in species of the beetle genera Paederus and Paederidus, while almost all other members are known only from marine sponges. In the beetles, pederin serves as a chemical defense against predators such as spiders (11). It is also the cause of Paederus dermatitis, a blistering inflammation that is contracted when the beetles are accidentally crushed on the human skin (20). In regions with a warm climate the beetles can appear in enormous numbers, resulting in notorious epidemics that have forced the evacuation of entire settlements (18, 25). Interestingly, they have also been proposed as a cause for 3 of the 10 biblical plagues (16).
FIG. 1.
Antitumor compounds of the pederin family, isolated from beetles and sponges.
Several lines of evidence clearly indicate that pederin is not produced by the beetles themselves but by as-yet-uncultured bacterial symbionts. Kellner and Dettner have demonstrated that within each examined beetle species pederin is synthesized in only about 90% of the females, who alone can transfer this trait to their offspring (10). According to 16S ribosomal DNA data, only these pederin-synthesizing females harbor a bacterium with the closest relationship to Pseudomonas aeruginosa (8). If eggs of such beetles are fed to nonproducing females, pederin is again synthesized in their offspring (7). Our search for the pederin biosynthesis genes, motivated by the prospect of generating an alternative biotechnological source of these rare antitumor compounds, has resulted in the isolation of a polyketide synthase cluster spanning 54 kb (19). This operon, designated the ped cluster, is present only in pederin-synthesizing females and their eggs and, architecturally, strikingly mirrors the chemical structure of pederin. Although the cluster had been isolated from beetle total DNA, its gene homology data and architecture imply that it is located on a bacterial genome. Notably, spot sequencing of regions adjacent to the ped cluster indicated the presence of open reading frames (ORFs) with remarkable homology to genes from P. aeruginosa, which is in perfect agreement with the 16S ribosomal DNA data provided by Kellner and Dettner (10).
Our preliminary sequence analysis had suggested that the ped cluster is part of a genomic island acquired by an P. aeruginosa-like bacterium. Genomic islands are large, horizontally transferred regions of DNA that can significantly enhance metabolic and colonizing capabilities and allow bacterial evolution in quantum leaps. Many of them play a crucial role in the acquisition of pathogenic traits in bacteria (5). However, in contrast to the large number of islands involved in pathogenicity, our knowledge about the biologically related symbiosis islands is restricted to a single example reported for rhizobia (23). In light of the absence of ped-type genes in P. aeruginosa and of the advantage of pederin biosynthesis for the survival of the beetle host, we suspected that the ped genes could be located on such an island. In the work presented here, we fully sequenced this locus to better understand its function in symbiosis. Additionally, we present a sequence analysis of regions outside the putative island to obtain further information on the biology of the as-yet-unculturable Paederus symbiont. These data corroborate that the ped cluster is a foreign element in the symbiont genome and provide insight into the evolution of pederin symbiosis.
MATERIALS AND METHODS
Isolation of bacteria from Paederus fuscipes.
100 P. fuscipes beetles collected at Aydin, Turkey, were ground in liquid nitrogen and resuspended in 5 ml of Luria-Bertani medium. The mixture was kept for 10 min on ice to let beetle residues settle. The supernatant was centrifuged at 100 × g for 10 min to sediment eukaryotic cells. The supernatant was then passed through a 40-μm-pore-size nylon filter (Millipore), and the bacteria were pelleted by centrifugation at 5,000 × g for 10 min. The purity of the bacterial fraction was checked by DAPI (4′,6′-diamidino-2-phenylindole) staining and microscopic inspection.
Preparation and screening of a P. fuscipes symbiont genomic library.
Genomic DNA from P. fuscipes bacteria was prepared by standard sodium dodecyl sulfate lysis and used for the construction of a cosmid library in the pWEB vector (Epicentre) according to the manufacturer's instructions. Two thousand clones were deposited in 96-well plate format and transferred to Hybond N+ membranes. Membranes were screened by using a fluorescein-labeled probe prepared from a 1.5-kb fragment of the ped cluster and the ECL labeling and detection system (Amersham). Adjacent genome regions were obtained by primer walking.
Cloning and sequencing of symbiont DNA adjacent to the ped locus.
Cosmids were sonicated, end repaired with BAL 31 and Klenow fragment, and size fractionated by gel electrophoresis to yield fragments of 1 to 2 kb. These were ligated into the EcoRV site of pBluescript II SK(−) (Stratagene) and end sequenced by using BigDye Terminator Ready Mix (Applied Biosystems) and an ABI3700 sequencer (Applied Biosystems). The order and direction of contigs were determined by restriction mapping and targeted subcloning of HindIII and EcoRI fragments. Sequence analysis was performed with the BLAST, PROSITE, Pfam, FRAMEPLOT, NNPP prokaryotic, and tRNASCAN-SE algorithms and the Lasergene DNASTAR package. Only regions covered at least eightfold by sequences are described in this work. All entries classified as pseudogenes were fully sequenced.
Expression of symbiont tellurite resistance genes in Escherichia coli.
The cosmid pPD5C10, containing the entire ter cluster, was transformed into E. coli XL1-Blue (Stratagene) by electroporation. Clones containing the cosmid were streaked out on Luria-Bertani plates containing various amounts of K2TeO3 (Aldrich) and carbenicillin at 100 μg/ml (Roth). E. coli containing the empty pWEB vector was used as a control.
Nucleotide sequence accession numbers.
The nucleotide sequences have been deposited at GenBank under accession numbers AY328003 to AY328024.
RESULTS
Construction and screening of a cosmid library prepared from bacteria of P. fuscipes beetles.
Previously identified genes had been isolated from a large metagenomic cosmid library of beetle total DNA (18). To further simplify screening and to obtain additional independent evidence that the ped region belongs to a bacterial genome, we constructed a new library from bacteria isolated from P. fuscipes, consisting of 2,000 clones. Screening of this library with a gene probe derived from the ped cluster yielded on average 1 positive in 130 clones, compared to 1 positive in 6,000 cosmids of the previous metagenomic library. This significant enrichment of ped genes clearly shows their bacterial origin.
Sequence analysis of a 106-kb region containing the ped cluster.
In order to determine the structure of the putative symbiosis island and neighboring regions, we sequenced the island in its entirety and an additional 34 kb to a coverage of 72% by at least eight overlapping sequences, thus providing sequence data from a region spanning approximately 106 kb. Database searches revealed homologies to 64 different genes (Table 1). Of these, however, 25 are represented only as pseudogenes that are truncated or contain frameshifts and stop codons. The large number of incomplete ORFs within and outside the putative island may indicate that the symbiont genome is undergoing degradation, as has been reported for other insect endosymbionts such as Buchnera aphidicola (2).
TABLE 1.
Genes and pseudogenes identified in the sequenced region, listed in the order of their position on the symbiont genome
| Putative gene producta | Sequence similarity (protein, origin)b | Identity/similarity (%) | % G + C | Protein accession number |
|---|---|---|---|---|
| CobQ, cobyric acid synthase | CobQ, PA1277 | 81/87 | 59 | AAG04666 |
| CopP, cobinamide kinase | CobP, PA1278 | 80/83 | 62 | AAG04667 |
| CobU, nicotinate-nucleotide dimethylbenzimidazole phosphoribosyltransferase | CobU, PA1279 | 75/81 | 62 | AAG04668 |
| (Trypsin domain protein) | PSPTO0934, Pseudomonas syringae (pv. tomato) | 55/79 | 52 | AAO54468 |
| Hypothetical protein | YPO0292, Yersinia pestis | 65/70 | 55 | CAC89155 |
| Hypothetical protein | YP064, Rhizobium etli | 40/58 | 54 | AAM54979 |
| Hypothetical protein | YPO0290, Y. pestis | 52/68 | 55 | CAC89153 |
| Hypothetical protein | Z1166, E. coli O157:H7 | 44/60 | 53 | AAG55720 |
| TerZ, tellurite resistance protein | TerZ, Proteus mirabilis | 54/71 | 57 | AAD47284 |
| TerA, tellurite resistance protein | TerA, E. coli O157:H7 | 58/74 | 51 | AAG55726 |
| TerB, tellurite resistance protein | TerB, E. coli O157:H7 | 48/65 | 51 | AAG55727 |
| TerC, tellurite resistance protein | TerC, Serratia marcescens | 69/84 | 51 | AAA86849 |
| TerD, tellurite resistance protein | TerD, Deinococcus radiodurans | 70/82 | 52 | AAF11773 |
| (TerD, tellurite resistance protein) | TerD, D. radiodurans | 64/78 | 56 | AAF11773 |
| (TerE, tellurite resistance protein) | TerE, E. coli O157:H7 | 54/69 | 50 | AAF36434 |
| TerF, tellurite resistance protein | SCO3939, Streptomyces coelicolor | 50/71 | 54 | CAA22211 |
| CysH, PAPS reductase | CysH, PA1756 | 85/90 | 55 | AAB53743 |
| (ThrH, homoserine kinase) | ThrH, PA1757 | ca. 60/75 | 49 | AAG05146 |
| AcoK, transcriptional regulator, LuxR subfamily | AcoK, PA1759 | 80/88 | 56 | AAG05148 |
| (AcoK, transcriptional regulator, LuxR subfamily) | AcoK, PA1760 | 81/86 | 57 | AAG05149 |
| (Hypothetical protein) | PA1749 | 81/89 | 52 | AAG05138 |
| DAHP synthase | PA1750 | 81/90 | 51 | AAG05139 |
| (Hypothetical protein) | PA1751 | 57/66 | 51 | AAG05140 |
| 2-Dehydropantoate 2-reductase | PA1752 | 73/82 | 51 | AAG05141 |
| CysB, LysR family transcriptional regulator | CysB, PA1754 | 92/95 | 56 | AAB53742 |
| Putative membrane protein | PA1360 | 81/88 | 56 | AAG04749 |
| Antiporter | NorM1, PA1361 | 82/86 | 58 | AAG04750 |
| PdxB, erythrose 4-phosphate dehydrogenase | PdxB, PA1375 | 76/84 | 59 | AAG04764 |
| AceK, isocitrate dehydrogenase kinase/phosphatase | AceK, PA1376 | 88/91 | 58 | AAG04765 |
| (Transposase) | Hypothetical 13.3-kDa protein, plasmid pEl1, Edwardsiella ictaluri | ca. 55/60 | 49 | AAF85957 |
| (Hypothetical integral membrane protein) | PA1689 | ca. 75 | 52 | AAD32694 |
| Dioxygenase | MLL1937, Mesorhizobium loti | 46/65 | 52 | BAB49189 |
| (Resolvase) | TnpR Tn5045 resolvase, plasmid pXAC33, Xanthomonas axonopodis cv. citri | ca. 60/70 | 53 | AAM39242 |
| (Hypothetical protein) | PA5264 | 41/55 | 52 | AAG08649 |
| (ABC transporter protein binding component) | PA4500 | 75/82 | 58 | AAG07884 |
| (Transposase) | XCC3722, retroviral integrase domain, Xanthomonas campestris pv. campestris | 49/55 | 66 | AAM42991 |
| PedA, methyltransferase | AdpE, Anabaena sp. strain 90 | 44/62 | 52 | CAC01607 |
| PedB, FMN-dependent oxidoreductase | MmpIII OR domain, Pseudomonas fluorescens | 54/73 | 53 | AAM12912 |
| PedC, acyltransferase | MmpIII AT domain, P. fluorescens | 38/55 | 49 | AAM12912 |
| PedD, acyltransferase | PksC, Bacillus subtilis | 52/68 | 52 | CAB13582 |
| PedE, methyltransferase | SnogM, Streptomyces nogalater | 38/55 | 48 | AAG42853 |
| PedF, polyketide synthase-peptide synthetase | PksK, B. subtilis | ca. 40/50 domain homology | 57 | AAA85144 |
| PedG, FAD-dependent oxygenase | SCO0122, S. coelicolor | 33/48 | 50 | CAB52349 |
| PedH, polyketide synthase-peptide synthetase | PksP, B. subtilis | ca. 40/50 domain homology | 56 | CAB13605 |
| (Transposase) | IS1421 transposase, Ralstonia solanacearum | ca. 70/80 | 58 | BAA97977 |
| (Cytochrome P450) | C-1027 ORF29, Streptomyces globisporus | 36/54 | 58 | AAL06684 |
| (Δ12 desaturase) | DesA, Gloeobacter ciolaceus | 37/51 | 58 | AAF61413 |
| Acyl carrier protein | HetM, Anabaena sp. strain PCC 7120 | 40/68 | 57 | AAA22001 |
| (Δ 9 desaturase) | RSP0792, R. solanacearum | 37/53 | 59 | CAD17943 |
| (Δ9 desaturase) | SCO3128, St. coelicolor | 41/56 | 60 | CAB95921 |
| (Acyl coenzyme A ligase) | SafB, Myxococcus xanthus | 40/59 | 58 | AAC44128 |
| (Hypothetical protein) | PA3290 | ca. 52/53 | 63 | AAG06678 |
| Helicase | ORF61, Pseudomonas resinovorans plasmid pCAR1 | 45/61 | 53 | BAC41688 |
| (Phage integrase) | Int, Shigella dysenteriae | 24/38 | 55 | AAF28112 |
| Hypothetical protein | PA5126 | 59/73 | 58 | AAG08511 |
| rRNA methylase | PA5127 | 85/91 | 55 | AAG08512 |
| (Transposase) | Actinobacillus actinomycetemcomitans | ca. 40/57 | 48 | BAA75251 |
| (Resolvase) | TnpT, Pseudomonas putida plasmid pWW0 | 66/86 | 52 | CAC86783 |
| (Transposase) | Avin4426, Azotobacter vinelandii | 60/80 | 46 | ZP_00092685 |
| (Transposase) | Avin4426, A. vinelandii | 60/80 | 46 | ZP_00092685 |
| Hypothetical protein | PA5314 | 84/91 | 55 | AAG08699 |
| Aldehyde dehydrogenase | PA5312 | 90/94 | 56 | AAG08697/PICK> |
| (MFS transporter) | PA5311 | 84/88 | 59 | AAG08696 |
| FAD-dependent oxidoreductase | PA5309 | 82/89 | 59 | AAG08694 |
Entries in parentheses are for degraded genes.
Proteins with PA numbers are from P. aeruginosa.
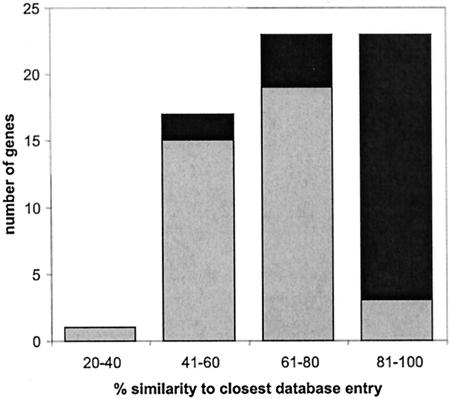
The identified homologies can be classified into two similarity groups (Fig. 2) arranged into clustered regions. The first group comprises 26 genes and exhibits a striking homology to genes from P. aeruginosa, which is usually 80 to 90% at the protein level and around 80% at the nucleotide level. This group encompasses almost all housekeeping genes. In previous 16S rRNA studies reported by Kellner, an uncultured bacterium with the closest relationship to P. aeruginosa, which is present only in beetles with high pederin content, was detected (8). The location of the pederin biosynthesis cluster beside genes with such extraordinary similarity to the P. aeruginosa genome strongly suggests that the pederin producer is identical to this beetle bacterium. Spot sequencing of two random cosmids of the library consistently revealed the presence of P. aeruginosa gene orthologs (data not shown). This persistence of P. aeruginosa-type genes throughout the genome, as opposed to an accidental local clustering close to the ped region, allows the conclusion that the symbiont is indeed a member of the genus Pseudomonas.
FIG. 2.
Similarity distribution of genes identified in the sequenced region. Black, deduced gene products with highest homology to proteins from P. aeruginosa. Grey, gene products resembling proteins from other bacteria. Similarity was assessed at the amino acid level.
The second homology group is only around 50 to 70% similar to known proteins and shows very weak or no homology to database entries at the nucleotide level. This group consists almost entirely of genes associated with secondary metabolism (ped genes), resistance, or gene transfer, i.e., genes that are commonly found to be horizontally acquired. These genes are mostly clustered into two regions: one centering around the ped genes (Fig. 3) and another with similarity to a tellurite resistance operon.
FIG. 3.
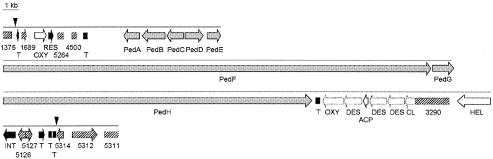
Map of the region containing the ped genes. Black vertical triangles mark the putative island borders. Stippled boxes and arrows indicate degraded ORFs. Genes in grey belong to the ped polyketide synthase system, of which pedF and pedH encode multidomain megaenzymes. Pseudogenes in black exhibit homology to IS elements, hatched elements represent orthologs of P. aeruginosa genes, and those in white are homologous to genes from other bacteria. T, transposase; OXY, oxygenase; RES, resolvase; DES, desaturase; ACP, acyl carrier protein; CL, acyl coenzyme A ligase; HEL, helicase; INT, phage integrase. Numbers correspond to the gene numbers in the P. aeruginosa genome.
P. aeruginosa orthologs.
The sequenced P. aeruginosa-like genes show an average G+C content of 55.7%, which is much lower than the 66.6% for P. aeruginosa (22). According to homology data, most of the identified P. aeruginosa-type genes outside the ped island are involved in the biosynthesis of amino acids and enzyme cofactors and appear to be complete and functional. These encode enzymes putatively involved in the biosynthesis of phenylalanine, tyrosine, and tryptophan (3-deoxy-d-arabionoheptulosonic acid 7-phosphate [DAHP] synthase); sulfur metabolism and cysteine biosynthesis (CysB and CysH); and the biosyntheses of vitamin B6 (erythrose-4-phosphate dehydrogenase), vitamin B12 (CobQ, CobP, and CobU), and coenzyme A (ketopantoate reductase). Other presumably intact genes are highly homologous to a regulator of the LuxR family (ortholog in P. aeruginosa, PA1759), a transport protein (PA1361), an isocitrate dehydrogenase kinase/phosphatase (PA1376) presumably controlling the glyoxylate shunt pathway, two oxidoreductases (PA5309 and PA5312), and two homologs of PA1360 and PA5314 with unknown function. In addition, six degraded genes which have intact orthologs in P. aeruginosa could be identified. Except for the putative regulator PA1760 and the homoserine kinase PA1757 nothing is known about the function of these genes.
The P. aeruginosa-like regions exhibit only very local synteny to the P. aeruginosa genome. Upstream of the tellurite operon, the three vitamin B12 biosynthesis genes are present in the same arrangement in P. aeruginosa (PA1277 to PA1279). Located between the tellurite operon and the putative ped island is a region with extensive homology to the P. aeruginosa genome between PA1750 and PA1760. Nine of these 11 genes were at least partially identified on the sequenced DNA. However, in addition to severe gene degradation and the deletion of two genes present in P. aeruginosa, the upstream and downstream halves of this region occupy switched positions compared to the orthologous sections in P. aeruginosa. The region further downstream is syntenous to the section encoding PA1360 to PA1376 but lacks 13 genes with mostly unknown functions. Two further P. aeruginosa orthologs are missing in a region otherwise highly similar to PA5309 to PA5314 downstream of the ped island.
The putative ped island.
Horizontally acquired genomic regions involved in pathogenicity or symbiosis are often found integrated at tRNA genes (5). However, no such gene could be detected at either side of the ped region. We have assigned the upstream end of the putative island to a transposase gene fragment located adjacent to the PA1376 homolog. The last gene at the downstream end with low similarity to P. aeruginosa genes is another fragmented insertion sequence (IS)-like gene adjacent to a PA5312 homolog. The low-homology region therefore spans 71.6 kb. Its average G+C content is 55.7% identical to that of the P. aeruginosa-like regions.
In addition to the ped genes described previously, a number of other homologies were detected on this section. However, of these, only five belong to intact ORFs. Notable is the large number of homologs to functions associated with horizontal gene transfer, a common feature of genomic islands. Three IS-like elements were identified at the upstream end and five were identified at the downstream end of the putative island, showing similarity to transposases, resolvases, and a phage integrase involved in horizontal acquisition of a Shiga toxin operon in Shigella dysenteriae (14). All of the IS-type genes are truncated at one or both ends. These homologies strongly suggest that the ped symbiosis genes are of foreign origin. Also interspersed within the region are heavily truncated orthologs of P. aeruginosa genes. Sequences resembling fragments of the putative ABC transporter component PA4500 and of PA1689, PA5264, and PA4500 with unknown function were found. In addition, apparently intact orthologs of the small hypothetical protein PA5126 and the rRNA methylase PA5127 are present. The mosaical architecture of the region, featuring numerous insertion sequences and orphan P. aeruginosa-like genes and pseudogenes, indicates that various integration and/or rearrangement processes have followed the acquisition of the ped genes. This characteristic is known for many other genomic islands.
Located immediately downstream of the ped genes is a pseudogene with two internal stop codons which exhibits highest similarity to cytochromes P450 catalyzing the oxygenation of polyketides. Examples are ORF29, which is involved in the biosynthesis of the enediyne antibiotic C-1027 (13), and the erythromycin hydroxylase EryF (1). Since pederin is a polyketide, this pseudogene could have once been a part of the ped cluster and participated in the biosynthesis of an ancestral pederin-type compound. Interestingly, the numerous members of the pederin group isolated from sponges almost always contain an additional P450-type oxygenation (15), which may be generated by an intact version of this gene.
A small degraded operon related to the biosynthesis of unsaturated fatty acids lies adjacent to the P450 pseudogene. It contains pseudogenes of an acyl coenzyme A ligase, a Δ12-desaturase and two Δ9-desaturases, and an intact acyl carrier protein gene. The only other complete non-P. aeruginosa-type ORFs in the ped region, apart from the ped genes themselves, encode a putative dioxygenase similar to asparagine β-hydroxylase and a protein with good homology to a DNA helicase from a Pseudomonas resinovorans plasmid.
The tellurite resistance operon.
The tellurite resistance operon region, with an unusually low G+C content of 51.4%, is located upstream of the ped island and contains seven genes with good homology at the amino acid level to the ter genes from Yersinia pestis, E. coli O157:H7, and other pathogens. Such genes are known to confer resistance against tellurite salts, which exhibit strong antibiotic properties (24). Upstream of the ter genes, four additional ORFs that have no counterparts in P. aeruginosa were identified. In most bacteria with ter operons, homologs to these ORFs can be found at the identical location, which suggests that they belong to the same operon. The exact function of ter clusters and their individual proteins is not well understood. Since tellurite salts are rare in nature, it is suspected that resistance against these compounds is only a secondary property (24).
Expression of the tellurite resistance genes in E. coli.
Our available sequence data indicated only one decayed gene in the ter region. To test whether the ter region is functional, we studied its properties in E. coli XL1-Blue. After transformation with pPD5C10, a cosmid carrying the ter operon, clones were streaked out on plates containing potassium tellurite. The ter cosmid resulted in resistance against this compound at a MIC of 500 μg/ml. The bacteria are black due to intracellular crystals of elementary tellurium (24). Control clones containing only the empty pWEB vector were unable to grow on tellurite-containing media. The ter genes are therefore functional in this strain.
DISCUSSION
The intention of our work was to gain insight into the biology of the uncultured Paederus symbiont and into the evolution of pederin symbiosis. A remarkable characteristic of the symbiont is the extremely high nucleotide homology of its genome to that of P. aeruginosa, a pathogenic bacterium that can be cultured easily. A dominant bacterium with the closest relationship to P. aeruginosa had previously also been detected by Kellner exclusively in pederin-positive beetles (8). Thus, according to these two lines of evidence, the pederin producer should be a member of the genus Pseudomonas.
Closer inspection of the analyzed 106-kb region revealed a number of characteristic differences from the P. aeruginosa genome. Notable is the degradation of several P. aeruginosa orthologs, which is in strong contrast to P. aeruginosa, with its low number of pseudogenes (12, 22). Further sequencing will be necessary to determine whether gene decay is indeed found genome wide or occurs only locally around the ped region. Degraded genomes have been observed in a number of other symbionts and pathogens (17). At the extreme end of such reductive processes are obligate intracellular symbionts such as B. aphidicola (26). These organisms have lost almost all dispensable coding and noncoding regions and are among the bacteria with the smallest known genomes. Although the Paederus symbiont seems to have lost a number of proteins, it apparently contains a large amount of noncoding DNA in the form of pseudogenes and large intergenic spacers. Such bacteria are speculated to occupy an intermediate position in the evolution towards a minimal genome (3, 21). Indeed, the occurrence of pederin only in the two closely related beetle genera Paederus and Paederidus suggests that pederin symbiosis is considerably less ancient than the family-wide association between B. aphidicola and aphids established about 200 million years ago (2). The more recent origin of pederin symbiosis is possibly also reflected by the fact that the association is unstable and not obligatory, since about 10% of the Paederus females found in nature are free of symbionts (9, 10).
Interestingly, most of the intact P. aeruginosa-type genes found in the region encode proteins putatively involved in amino acid and vitamin biosynthesis. For a number of symbiotic systems it has been shown that the supply of such compounds to the host is an important factor governing mutualism. An example is the provision of essential amino acids by B. aphidicola to aphids, which feed on a nutrient-poor diet (4). Although the vicinity of nutrient-providing genes to the ped cluster could argue for a similar role in Paederus beetles, the available data do not support a host dependence of symbiont nutrients. Symbiont-free beetles found in the field are virtually indistinguishable from producing specimens, which is probably a consequence of the fact that Paederus beetles, unlike sap-sucking insects, are carnivores with a balanced diet. However, symbiont metabolites could possibly augment the beetle diet and enhance fitness in times of limited nutrient supply.
Two regions with missing counterparts in the P. aeruginosa genome were identified. One is a functional tellurite resistance operon, whose lower G+C content indicates that it is of foreign origin. So far almost nothing is known about the actual role that such resistance genes play in bacterial fitness. It has been shown that ter clusters also confer resistance against bacteriophages and bacteriocins, i.e., antibacterial proteins (27). Determinants of tellurite resistance are known for a large number of pathogens and are commonly exploited to selectively isolate pathogenic bacteria (24). It is therefore speculated that these clusters protect the cell against host defense reactions via an unspecific mechanism. The identified ter region may enhance the survival of the symbiont in a similar way. Interestingly, the terE gene is not intact in the symbiont and is therefore apparently not needed for resistance. A similar finding has been documented by Whelan et al., studying a ter cluster from the plasmid R478 (27).
The second low-homology region containing the ped genes bears typical hallmarks of a symbiosis island. The ped cluster should represent one of the major factors governing pederin symbiosis, since female beetles use pederin to chemically defend themselves and their offspring against predators (11). The lack in P. aeruginosa of homologs in this region and the presence of a large number of degraded IS elements indicate that a P. aeruginosa-like bacterium has acquired the ped genes by horizontal transfer. Hence, this acquisition may have been a crucial event in the evolution of a symbiotically competent microorganism. According to the mosaical appearance of the ped island, the integration likely took place at a recombinatorial hotspot in the genome and was followed by further insertions and rearrangements.
The production of pederin-type compounds in sponges by a symbiont has not yet been demonstrated but is very likely, as complex polyketides are typically produced by bacteria. The existence of a highly mobile genomic element carrying genes important for symbiosis could explain why these metabolites are found widespread among unrelated animals living in entirely different environments. Such genes could be transferred between different bacteria by phage infection or other mechanisms. However, at least in the case of the Paederus symbiont, this island apparently has lost its mobility functions due to gene decay and seems to be a stable component of the genome. Similarly stabilized islands have been observed in several pathogens. It is believed that they are the result of an adaptive process, during which the island disposes of inessential genes to eventually become part of the core genome (5). In the symbiont, the degradation of almost all island genes except the ped cluster suggests a similar mechanism.
Secondary metabolites isolated from invertebrate animals but structurally resembling natural products from bacteria are remarkably widespread in nature. For many years, bacterial symbionts have been proposed to be the true source of these metabolites (6), but so far not a single producing bacterium has been isolated from any of these organisms. Since many of these compounds are promising drug candidates, there is a need for a better understanding of these symbiotic systems at the genetic level. These studies could ultimately result in solving the supply problem that exists with almost all invertebrate drug candidates, e.g., by developing a bacterial cultivation system or by heterologous expression of biosynthesis genes from symbionts in culturable bacteria. The pederin symbiont is the first mutualist producing a pharmacologically active natural product, whose putative biosynthesis gene cluster was cloned (19) and for which extensive genetic data now exist. Through comparative genomics we should gain a better understanding of symbiont genetics and improve our ability to generate environmentally sound supplies of symbiont drug candidates. In addition, comparative studies will also help to identify the factors governing pathogenesis and symbiosis in the Paederus symbiont and the closely related opportunistic pathogen P. aeruginosa.
Acknowledgments
We thank H. Başpinar and E. Şavk for helping with the collections of Turkish beetle specimens, D. Schnabelrauch for technical assistance, and R. L. L. Kellner, K. Dettner, and S.-J. Suh for valuable discussions. We are grateful to W. Boland for support and to C. Dale for suggestions on the manuscript.
This work was supported by research grant PI 430/1-1 from the Deutsche Forschungsgemeinschaft and by the Max Planck Society.
REFERENCES
- 1.Andersen, J. F., and C. R. Hutchinson. 1992. Characterization of Saccharopolyspora erythraea cytochrome P450 genes and enzymes, including 6-deoxyerythronolide-B hydroxylase. J. Bacteriol. 174:725-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann, P., L. Baumann, C. Y. Lai, D. Roubakhsh, N. A. Moran, and M. A. Clark. 1995. Genetics, physiology, and evolutionary relationships of the genus Buchnera—intracellular symbionts of aphids. Annu. Rev. Microbiol. 49:55-94. [DOI] [PubMed] [Google Scholar]
- 3.Dale, C., B. Wang, N. Moran, and H. Ochman. 2003. Loss of DNA recombinational repair enzymes in the initial stages of genome degradation. Mol. Biol. Evol. 20:1188-1194. [DOI] [PubMed] [Google Scholar]
- 4.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 5.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 6.Haygood, M. G., E. W. Schmidt, S. K. Davidson, and J. D. Faulkner. 1999. Microbial symbionts of marine invertebrates: opportunities for microbial biotechnology. J. Mol. Microbiol. Biotechnol. 1:33-43. [PubMed] [Google Scholar]
- 7.Kellner, R. L. L. 2001. Horizontal transmission of biosynthetic capabilities for pederin in Paederus melanurus (Coleoptera: Staphylinidae). Chemoecology 11:127-130. [Google Scholar]
- 8.Kellner, R. L. L. 2002. Molecular identification of an endosymbiotic bacterium associated with pederin biosynthesis in Paederus sabaeus (Coleoptera: Staphylinidae). Insect Biochem. Mol. Biol. 32:389-395. [DOI] [PubMed] [Google Scholar]
- 9.Kellner, R. L. L. 1999. What is the basis of pederin polymorphism in Paederus riparius rove beetles? The endosymbiotic hypothesis. Entomol. Exp. Appl. 93:41-49. [Google Scholar]
- 10.Kellner, R. L. L., and K. Dettner. 1995. Allocation of pederin during lifetime of Paederus rove beetles (Coleoptera: Staphylinidae): evidence for polymorphism of hemolymph toxin. J. Chem. Ecol. 21:1719-1733. [DOI] [PubMed] [Google Scholar]
- 11.Kellner, R. L. L., and K. Dettner. 1996. Differential efficacy of toxic pederin in deterring potential arthropod predators of Paederus (Coleoptera: Staphylinidae) offspring. Oecologia 107:293-300. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence, J. G., R. W. Hendrix, and S. Casjens. 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9:535-540. [DOI] [PubMed] [Google Scholar]
- 13.Liu, W., S. D. Christenson, S. Standage, and B. Shen. 2002. Biosynthesis of the enediyne antitumor antibiotic C-1027. Science 297:1170-1173. [DOI] [PubMed] [Google Scholar]
- 14.McDonough, M. A., and J. R. Butterton. 1999. Spontaneous tandem amplification and deletion of the Shiga toxin operon in Shigella dysenteriae 1. Mol. Microbiol. 34:1058-1069. [DOI] [PubMed] [Google Scholar]
- 15.Narquizian, R., and P. J. Kocienski. 2000. The pederin family of antitumor agents: structures, synthesis and biological activity, p. 25-56. In R. Mulzer and R. Bohlmann (ed.), The role of natural products in drug discovery, vol. 32. Springer-Verlag, Heidelberg, Germany. [DOI] [PubMed]
- 16.Norton, S. A., and C. Lyons. 2002. Blister beetles and the ten plagues. Lancet 359:1950. [DOI] [PubMed] [Google Scholar]
- 17.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1098. [DOI] [PubMed] [Google Scholar]
- 18.Penchenier, L., J. Mouchet, B. Cros, P. Legall, J. Y. Cosnefroy, P. Quezede, and J. Chandenier. 1994. Outbreaks of Paederus sabaeus in Central Africa. 1. Entomological and epidemiologic aspects. Bull. Soc. Pathol. Exot. 87:45-48. [PubMed] [Google Scholar]
- 19.Piel, J. 2002. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 99:14002-14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sendur, N., E. Savk, and G. Karaman. 1999. Paederus dermatitis: a report of 46 cases in Aydin, Turkey. Dermatology 199:353-355. [DOI] [PubMed] [Google Scholar]
- 21.Silva, F. J., A. Latorre, and A. Moya. 2001. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 17:615-618. [DOI] [PubMed] [Google Scholar]
- 22.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 23.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor, D. E. 1999. Bacterial tellurite resistance. Trends Microbiol. 7:111-115. [DOI] [PubMed] [Google Scholar]
- 25.Todd, R. E., S. L. Guthridge, and B. L. Montgomery. 1996. Evacuation of an Aboriginal community in response to an outbreak of blistering dermatitis induced by a beetle (Paederus australis). Med. J. Aust. 164:238-240. [DOI] [PubMed] [Google Scholar]
- 26.van Ham, R., J. Kamerbeek, C. Palacios, C. Rausell, F. Abascal, U. Bastolla, J. M. Fernandez, L. Jimenez, M. Postigo, F. J. Silva, J. Tamames, E. Viguera, A. Latorre, A. Valencia, F. Moran, and A. Moya. 2003. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA 100:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whelan, K. F., E. Colleran, and D. E. Taylor. 1995. Phage inhibition, colicin resistance, and tellurite resistance are encoded by a single cluster of genes on the IncHI2 Plasmid R478. J. Bacteriol. 177:5016-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]