Abstract
Similarities between lambda and rRNA transcription antitermination have led to suggestions that they involve the same Nus factors. However, direct in vivo confirmation that rRNA antitermination requires all of the lambda Nus factors is lacking. We have therefore analyzed the in vivo role of NusB and NusG in rRNA transcription antitermination and have established that both are essential for it. We used a plasmid test system in which reporter gene mRNA was measured to monitor rRNA antiterminator-dependent bypass of a Rho-dependent terminator. A comparison of terminator read-through in a wild-type Escherichia coli strain and that in a nusB::IS10 mutant strain determined the requirement for NusB. In the absence of NusB, antiterminator-dependent terminator read-through was not detected, showing that NusB is necessary for rRNA transcription antitermination. The requirement for NusG was determined by comparing rRNA antiterminator-dependent terminator read-through in a strain overexpressing NusG with that in a strain depleted of NusG. In NusG-depleted cells, termination levels were unchanged in the presence or absence of the antiterminator, demonstrating that NusG, like NusB, is necessary for rRNA transcription antitermination. These results imply that NusB and NusG are likely to be part of an RNA-protein complex formed with RNA polymerase during transcription of the rRNA antiterminator sequences that is required for rRNA antiterminator-dependent terminator read-through.
All rRNA operons in Escherichia coli have antiterminator sequences in their leader and spacer regions that allow RNA polymerase, modified with protein factors, to transcribe through Rho-dependent terminators of rRNA operons (1, 2, 5, 21). The identities of all of the protein factors have not yet been established, but the RNA sequences required have been determined (5, 37). The rRNA leader region antiterminator features include a region of dyad symmetry, referred to as boxB, and conserved sequences, boxA and boxC (21). The spacer regions do not contain the boxC feature (5). Studies with an entire rRNA operon on a plasmid show that leader region boxA mutations result in a 20 to 25% decrease in the amount of 16S and 23S rRNA (14). Spacer boxA mutations result in an additional 15% decrease of 50S subunits (28). These studies suggest that as a consequence of improper rRNA transcription antitermination (rRNA-AT), there is an increase in Rho-dependent termination that in turn results in a decrease of 16S and 23S rRNA. Mutational studies of the three rRNA antiterminator features, boxB, boxA, and boxC, identified boxA as the essential region for transcription antitermination and boxA alone as sufficient for in vivo read-through of Rho-dependent terminators (5). However, the other conserved features may have as-yet-unidentified functions in vivo. In addition, the boxA feature by itself stimulates an increase in the RNA polymerase transcription elongation rate from 35 to 65 nucleotides per s on the lacZ gene (38). Although there is no direct evidence, a relationship between an increase in RNA polymerase transcription elongation rate and terminator read-through may exist. Interestingly, Nus factors A, B, and G are necessary for the boxA-mediated increase in transcription elongation rate (37-39).
Present models suggest that the rRNA antiterminator is a site where RNA polymerase and protein factors interact, resulting in a transcription complex with altered terminator recognition and altered transcription elongation properties. Possible interacting factors at the rRNA antiterminator include those required for lambda N/nut antitermination, NusA, NusB, NusE (S10), and NusG, as well as ribosomal protein S4 (22, 38; for reviews, see references 9, 10, 27, and 29). Studies demonstrating a direct role for NusA in rRNA-AT used a nusA cold-sensitive mutant strain and found rRNA boxA-mediated changes in RNA polymerase activities to be defective (38). In vivo experiments demonstrating faulty rRNA expression in a nusB5 mutant strain also suggest a requirement for NusB in rRNA transcription (31). Additionally, in vitro experiments show that rRNA-AT activities fail in a NusB-depleted system (34). Although a direct role for NusE has not been described, a NusB-NusE heterodimer can bind rRNA boxA in vitro, implicating NusE in rRNA-AT (23, 25). NusG, as well as NusB, has been isolated from rRNA antiterminated complexes in vitro (19); however, there are no experimental data demonstrating the importance of NusG in rRNA-AT in vivo. NusG, an essential cellular protein, is known to be involved in multiple aspects of transcription, including influencing the activity of some Rho-dependent terminators and being required for lambda N/nut antitermination (11, 12, 19, 20, 35). The importance of NusG in Rho-dependent termination has been revealed by studies of the lac operon. There are two intragenic Rho-dependent terminators in lacZ. In the absence of NusG, Rho is virtually inactive at one terminator, while the other terminator is not significantly affected (7). Through interactions between NusG and Rho or RNA polymerase, NusG may modulate either Rho, RNA polymerase, or both to cause termination or antitermination (8, 19, 20, 35). These activities of NusG provide a logical need for its presence in the rRNA-AT system, a system that embraces both antitermination and Rho-dependent termination.
As described above, several lines of evidence suggest a role for all lambda Nus factors in rRNA-AT in E. coli, but up to now strong support for this suggestion has been lacking in vivo. In this work, we present direct evidence demonstrating an essential role for NusB and NusG in rRNA-AT in vivo by using a plasmid-borne reporter gene system to assess the role of these factors in terminator bypass. This assessment was done by examining the rRNA antiterminator-dependent read-through of a Rho-dependent terminator in the presence and absence of NusB and under conditions of NusG excess and depletion in vivo. We found that E. coli cells with nusB mutations or depleted of NusG had lost the ability to bypass terminators.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli MC4100 [araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB3501 deoC1 ptsF25 rbsR] (33) is the parent strain of the nusB-inactivated strain, IQ527 (MC4100 nusB::IS10 zba-525::Tn10). The nusB mutation is also called ssyB63 (32, 36). This strain generates larger, faster-growing colonies that have likely lost the insertion element and must be monitored carefully during use. The construction of plasmids pSL102, pSL103, pSL115, and pSL133 has been described previously (21). The pGB2(NusB) plasmid is a pGB2 derivative with a cloned 2.5-kb fragment containing the nusB gene (13). Plasmids pGB2 and pGB2(NusB) were a gift from D. Friedman. Strain RARNGT [RAR4100 nusG::kn F′(lacIq lacZ::Tn10 proAB+) pNG33] contains an insertion in the nusG gene and the plasmid pNG33, expressing the nusG gene from the arabinose operon promoter, PBAD (30, 39).
mRNA measurements of terminator read-through.
Overnight cultures of either MC4100 or IQ527 containing plasmid pSL102, pSL103, or pSL115 were grown at 37°C in Luria-Bertani (LB) medium with ampicillin (100 μg/ml). These strains containing an additional compatible plasmid, pGB2(NusB), were grown in LB medium with ampicillin (100 μg/ml) and spectinomycin (40 μg/ml). Cultures were inoculated at an optical density at 600 nm (OD600) of 0.04 and were grown to an OD600 of 0.6. Total cellular RNA was isolated and subjected to slot blot analysis (39). For the analysis, triplicate 0.25-μg amounts of each sample of denatured total RNA were used. These experiments were repeated at least three times.
For the NusB experiments, the following end-labeled oligonucleotide probes were used. Probe cat#1 (5′ TGCCATTGGGATATATCAACGGTGG 3′) is located at nucleotides 26 to 50 of the cat gene coding sequence and was used to detect transcripts beyond the Rho-dependent terminator. Probe bla#1 (5′ GGGAATAAGGGCGACACGGAAATG 3′) is located at nucleotides 13 to 36 of the bla gene coding sequence and was used to detect bla transcripts. The bla gene transcript was used to correct cat mRNA levels for incomplete cell disruption and possible variations in plasmid copy number (17).
For the NusG experiments, the following end-labeled oligonucleotide probes were used. Probe cat#2 (5′ CGAAGCTCGGCGGATTTGTCCTAC 3′) is located before the cat gene. Probe bla#2 (5′ GCCCGGCGTCAACACGGGATAATAC 3′) is located downstream of bla#1 at nucleotides 100 to 124 of the coding sequence. These new probes were used because pNG33 has the cat gene and part of the bla gene. The probes were end-labeled with [γ-32P]ATP (7,000 Ci/mmol; ICN, Costa Mesa, Calif.) and T4 polynucleotide kinase (New England, Biolabs, Beverly, Mass.). The membranes were prehybridized, hybridized, washed according to the procedure of Angelini et al. (4), scanned, and quantified with a Storm PhosphorImager (Amersham Biosciences Corp., Piscataway, N.J.) and the Image-Quant program.
Depletion of NusG from the cell.
Strain RARNGT containing pSL102, pSL103, or pSL115 was grown overnight at 37°C in LB medium containing 0.2% arabinose, kanamycin (50 μg/ml), and ampicillin (100 μg/ml). Cells were washed in LB medium to remove any arabinose and then inoculated at an OD600 of 0.02 in LB medium containing either 0.2% d-fucose, 0.2% d-glucose, kanamycin, and ampicillin (−Ara condition) or 0.2% arabinose, kanamycin, and ampicillin (+Ara condition). To maintain exponential growth, the cultures were diluted 1:10 after 3 and 5 h with the same fresh media. At 6 and 7 h, cultures with arabinose were diluted 1:10 and 1:5, respectively, with fresh media. Cell samples were removed at the indicated times for RNA preparation and Western analysis.
Western analysis.
The protocol described by Bollag and Edelstein (6) was followed. After sampling, cells were spun, resuspended in 3× sodium dodecyl sulfate (SDS)-loading dye, and boiled for 5 to 10 min. Samples prepared from 0.5 × 108 cells were loaded onto an SDS-12% polyacrylamide gel electrophoresis gel and subjected to electrophoresis for 45 min at 100 to 200 V. The proteins were then transferred onto a polyvinylidene difluoride membrane according to the manufacturer's instructions (Bio-Rad, Hercules, Calif.). The membrane was blocked for 1 h at room temperature by using 5% nonfat dry milk prepared in 1× TBS (30 mM Tris-Cl [pH 7.4], 150 mM NaCl). A 1:2,000 dilution of anti-NusG rabbit serum (a generous gift from Barbara Stitt) in blocking solution was added to the membrane. It was incubated for 1 h at room temperature and then washed with 1× TBSTT (1× TBS plus 0.05% Tween 20 and 0.2% Triton X-100) three times for 10 to 15 min. A 1:10,000 dilution of the secondary antibody, goat anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (Promega, Madison, Wis.) in blocking solution, was added to the membrane and incubated for 1 h at room temperature. The membrane was washed as before and then rinsed in 1× TBS for 5 min. The chemiluminescence substrate (Amersham Life Science, Arlington Heights, Ill.) was incubated with the membrane for 1 min, and then the membrane was exposed to X-ray film. To quantitate the amount of NusG detected in the cell lysates, 12.5, 25, and 50 ng of purified NusG were loaded on the same gel. After Western blotting and developing, the films were scanned with a densitometer (Amersham Biosciences), and the amount of NusG was quantitated with the Image-Quant program.
RESULTS
The basic protocol followed to measure rRNA-AT activity in the absence of Nus factors B and G was to comparatively examine mRNA transcript levels from three separate plasmid constructs (Fig. 1). The first construct contained a promoter gene and a reporter gene, the second also contained a terminator, and the third contained the terminator as well as the rRNA-AT sequences. Quantitation of reporter gene mRNA from all three constructs allowed a determination of terminator efficiency or bypass.
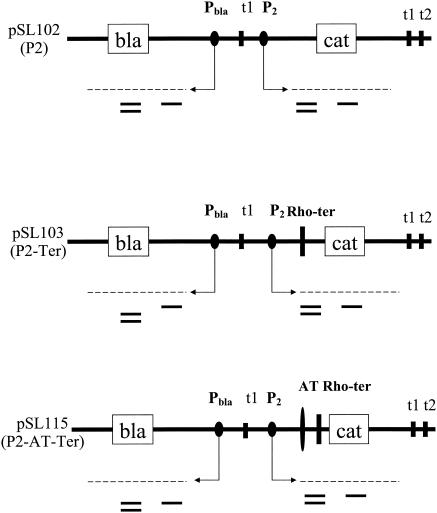
FIG. 1.
rRNA antiterminator test system. Details of the construction of these plasmids can be found in a report by Li et al. (21). Open boxes refer to the cat and bla genes. Arrows indicate the beginning and direction of transcription. Broken lines represent transcripts either starting at the P2 promoter and extending through the cat gene or starting at the bla promoter and extending through the bla gene. The short horizontal bars below the broken lines represent the cat#1 and bla#1 probes used for the NusB studies. The short horizontal double bars represent the cat#2 and bla#2 probes used for the NusG depletion studies. The P2 promoter is from the rrnG operon. Terminators t1 and t1 t2 are from the rrnB operon. pSL103 has an insertion of a Rho-dependent terminator (Rho-ter), a 567-bp HindIII fragment of the 16S rRNA gene from the rrnB operon cloned in the reverse orientation that fortuitously has Rho-dependent termination activity (21). pSL115 has an insertion of the antiterminator (AT) and the Rho-dependent terminator. The antiterminator is the 67-bp FnuDII-TaqI fragment from the leader region of the rrnG operon located between P2 and the 16S rRNA gene. It contains rRNA boxB, boxA, and boxC sequences (21).
In vivo role of NusB in rRNA-AT.
Previous measurements of rRNA levels in a nusB5 mutant strain showed a decrease in 16S and 23S rRNA levels relative to those in leader region transcripts, and it was concluded that the reduction was due to a defect in transcription antitermination (31). However, a direct link between antitermination and rRNA levels was not established. In the present work, we directly addressed the role of NusB in antitermination by using a defined rRNA-AT system and examining terminator bypass in the complete absence of NusB. We used a strain, IQ527, containing an insertional inactivation of the nusB gene (32, 36). By Western blot analysis, we confirmed the reported absence of the NusB protein in IQ527 (data not shown). We used this strain and a plasmid-borne reporter gene system described previously (2, 5, 21). Linear representations of the plasmids are shown in Fig. 1. The cat and bla mRNA levels were measured for each strain, and cat/bla ratios were determined. The cat/bla mRNA ratio for pSL102 reflects the amount of nonterminated transcription and was designated 100%. Rho-dependent terminator efficiency was determined by comparing the relative cat/bla mRNA transcription levels from pSL102 and pSL103. Likewise, terminator read-through was determined by comparing the cat/bla mRNA ratios for pSL103 and pSL115.
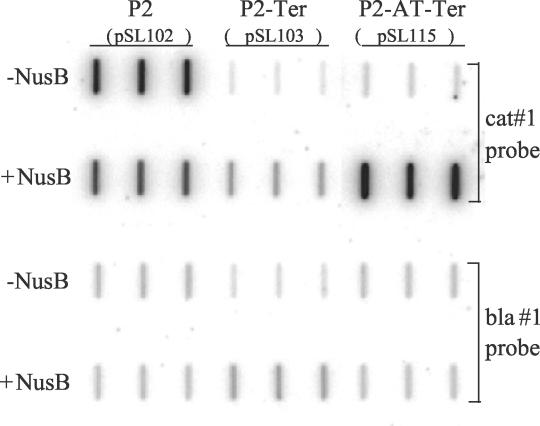
We measured the amount of terminator read-through in the nusB::IS10 strain and its parent (MC4100) in the presence or absence of a plasmid carrying the nusB gene, pGB2(NusB) (13). In the absence of NusB, the levels of terminator read-through were very low and similar with and without the rRNA-AT sequence (3 and 5%, respectively) (Table 1 and Fig. 2; compare P2 [rrnG operon P2 promoter]-Ter [a fragment of 16S RNA in the reverse orientation that contains an efficient Rho-dependent terminator] to P2-AT [a 67-bp fragment from the rrnG leader region containing rRNA antiterminator boxB, boxA, and boxC features]-Ter in the absence of NusB). Similarly, the presence or absence of NusB had no significant effect on terminator activity (3 or 4% in each case) (Table 1). However, in the presence of NusB and the antiterminator, terminator read-through was restored from 5 to 108% (Table 1). Because, in the absence of NusB, rRNA-AT-dependent terminator bypass was abolished, these results directly demonstrate the essential nature of NusB in rRNA-AT in vivo.
TABLE 1.
Role of NusB in rRNA-AT
| Test plasmid |
cat/bla mRNA (%)a
|
|
|---|---|---|
| WT | nusB::IS10 | |
| pSL102 (P2b) | 100 | 100 |
| pSL103 (P2-Terc) | 3 ± 0.8 | 3 ± 0.4 |
| pSL 115 (P2-ATd-Ter) | 191 ± 5.1 | 5 ± 1.5 |
| pSL102 + NusB | 100 | 100 |
| pSL103 + NusB | 3 ± 1.6 | 4 ± 0.6 |
| pSL115 + NusB | 233 ± 61 | 108 ± 6.5 |
cat/bla mRNA ratio normalized to that for pSL102. These experiments were repeated two or three times. WT, wild type.
P2, rrnG operon P2 promoter.
Ter, fragment of 16S rRNA in the reverse orientation that contains an efficient Rho-dependent terminator (21).
AT, 67-bp fragment from the rrnG leader region containing rRNA antiterminator boxB, boxA, and boxC features.
FIG. 2.
Effect of nusB inactivation on rRNA-AT. Shown are PhosphorImager scans of two duplicate mRNA slot blot membranes hybridized with either the cat#1 or the bla#1 probe. RNA samples are from strain IQ527 containing either pSL102, pSL103, or pSL115 and either pGB2 (−NusB) or pGB2(NusB) (+NusB). This figure shows one example of at least three experiments done with these strains. Triplicate amounts of each sample were loaded.
In vivo role of NusG in rRNA-AT.
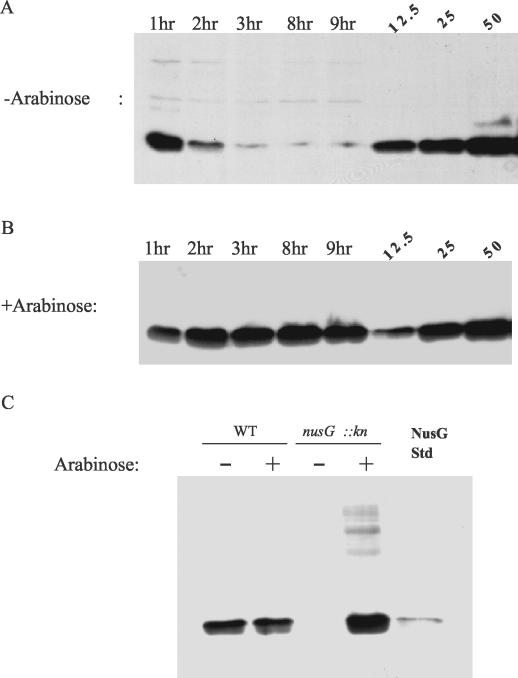
To address the question of whether or not NusG is required for rRNA-AT, we used a strain in which NusG could be depleted from the cell (35, 39). The strain used, RARNGT, contains a kanamycin resistance cassette inserted into the chromosomal copy of the nusG gene (35) and carries a plasmid expressing NusG from the arabinose promoter PBAD (39). Expression of the nusG gene in these cells requires the presence of arabinose in the media. Cells were isolated after 1 to 9 h of growth and analyzed for the presence of the NusG protein by Western blot analysis (Fig. 3A and B). As a control, we showed that the addition of arabinose had no effect on the expression of NusG in the parent strain (RAR4100) of RARNGT (Fig. 3C). The normal number of NusG molecules per cell in E. coli is 1 × 104 to 2 × 104 (20). In our strains, after 1 h of growth in the presence or absence of arabinose, there was approximately 1.5 × 104 molecules of NusG per cell, in good agreement with the level reported previously (Fig. 3A and B). After 3 h of growth in the presence of arabinose, there was an approximately two- to threefold increase in the amount of NusG (Fig. 3B). In the absence of arabinose, the cells were depleted of NusG after 3 h of growth (Fig. 3A). Quantitation of NusG after 8 or 9 h of growth without arabinose showed that there were only 300 to 600 molecules of NusG per cell (quantitation data not shown). The PBAD promoter may not be off entirely, even with the addition of glucose and fucose to the medium (see Materials and Methods), allowing this basal level of NusG to be produced.
FIG. 3.
Western blot analysis: depletion or expression of NusG in the cell. Aliquots of culture were removed at the indicated time, and an extract of 0.5 × 108 lysed cells was loaded on an SDS-polyacrylamide gel electrophoresis gel. Included on the gel were 12.5, 25, and 50 ng of a NusG standard. Quantitation of the NusG standard on a densitometer verified that these levels were in the linear range of the film used. At least three independent blots were used to quantitate the amount of NusG expressed at 1 h. (A) Example of a gel showing depletion of NusG up to 9 h of growth in the absence of arabinose. (B) Example of a gel showing depletion of NusG up to 9 h of growth in the presence of arabinose. (C) Control experiment with wild-type (RAR4100) and nusG::kn strain (RARNGT) cells in the presence or absence of arabinose.
Because NusG plays a role in both Rho-dependent termination and antitermination, we anticipated that NusG depletion might have a deleterious effect on the efficiency of the particular terminator used in our system, similar to the results seen with one of the intragenic Rho-dependent terminators in lacZ (7). The levels of cat/bla mRNA for P2-Ter were measured in the presence of arabinose and at timed intervals after arabinose removal. The results were normalized to mRNA levels from the plasmid containing only P2 and are shown in Table 2. In the presence of arabinose and with a two- to threefold increase in NusG, terminator read-through went from 14% at 1 h to 5% at 9 h (Table 2, P2-Ter, +Ara). When the cells grew in the absence of arabinose and NusG was depleted, terminator read-through followed the opposite trend, going from 12 to 31% (Table 2, P2-Ter, −Ara). These results indicate that NusG was needed for the maximal efficiency of this Rho-dependent terminator, but considerable termination still occurred when it was depleted.
TABLE 2.
Effect of NusG depletion on termination and antitermination
| Conditiona | Test plasmidb |
cat/bla mRNA (%) atc:
|
|||
|---|---|---|---|---|---|
| 1 h | 2 h | 3 h | 4 h | ||
| −Ara | pSL102 (P2) | 100 | 100 | 100 | 100 |
| pSL103 (P2-Ter) | 12 | 33 | 38 | 31 | |
| pSL115 (P2-AT-Ter) | 112 | 39 | 36 | 33 | |
| +Ara | pSL102 (P2) | 100 | 100 | 100 | 100 |
| pSL103 (P2-Ter) | 14 | 10 | 4 | 5 | |
| pSL115 (P2-AT-Ter) | 131 | 100 | 113 | 93 | |
−Ara, nusG gene not expressed in cells; +Ara, nusG gene expressed in cells.
See Table 1, footnotes b, c, and d, for a description of plasmid features.
cat/bla mRNA ratio normalized to that for pSL102. These experiments were repeated two or three times. cat/bla ratios varied from day to day, but the relative differences between the different plasmids remained the same.
When the rRNA-AT sequences were added to the plasmid system, there was no difference between the resulting system (P2-AT-Ter) and P2-Ter. After 9 h without arabinose, terminator read-through levels were similar for the two constructs, 31 and 33%, respectively (Table 2). However, in the presence of NusG, terminator read-through remained efficient. These results indicate that when NusG was depleted, the rRNA-AT system was completely unable to facilitate terminator bypass. From these data, we conclude that the rRNA-AT system is dependent on the presence of NusG for its activity.
DISCUSSION
Role of NusB in ribosomal antitermination.
Nuclear magnetic resonance experiments indicate that the N terminus of NusB likely functions as an rRNA boxA RNA-binding motif, suggesting that NusB may be an important link between the rRNA antiterminator and other factors in forming a stable transcription antitermination complex (3, 15). Previous studies of rRNA expression in a nusB5 mutant strain of E. coli demonstrated that wild-type NusB protein was necessary to obtain usual levels of rRNA products (31). However, direct in vivo demonstration of NusB activity in a defined rRNA-AT system has not previously been described. Using a strain with an insertional disruption of the nusB gene, we showed that rRNA-AT-dependent antitermination of a Rho-dependent terminator did not occur unless NusB was supplied to the cell. Although the nusB::IS10 strain has a prolonged doubling time, is cold sensitive, and has a diminished peptide elongation rate, the fact that it is viable demonstrates that a fully functional transcription antitermination system in rrn operons is not required for cell viability (32, 36). The mechanistic interactions of NusB resulting in antitermination are unknown. However, because NusB and NusE have been shown to bind to rRNA boxA in vitro and NusE has been shown to bind to RNA polymerase, the RNA-NusB/NusE subcomplex may interact with RNA polymerase and other factors to stabilize a transcription complex that is resistant to Rho-dependent terminators (19, 25, 26).
Role of NusG in ribosomal antitermination.
A role for NusG in antitermination was previously suggested by the identification of NusG in complexes formed in vitro on templates containing rRNA boxA (19); however, no previous in vivo experiments to address the requirement for NusG in rRNA-AT have been reported. We showed here that the presence of NusG is crucial for forming a functional rRNA-AT complex.
How NusG functions in rRNA-AT is not clear. Li et al. (19, 20) have suggested a model to explain the function of NusG in Rho-dependent antitermination in the lambda N/nut system. In this model, NusG interacts with RNA polymerase in the presence of the other antitermination factors and the nut sequence. Because NusG also binds to Rho (20), there is a high probability of transcript-bound Rho interacting with an antiterminating RNA polymerase containing NusG. This interaction could inhibit the translocation of Rho 5′ to 3′ along the transcript and therefore inhibit Rho-mediated termination, permitting RNA polymerase to continue transcribing (20, 24). Because rRNA-AT works efficiently to suppress Rho-dependent terminators (2), by analogy to the lambda model, NusG may also serve to inhibit Rho-dependent termination in the rRNA-AT system by interfering with Rho's termination activity.
Nus factors are required for increased transcription elongation rates and rRNA-AT.
It is now clear that the modified RNA polymerase capable of antitermination requires many of the same factors for terminator read-through and for increasing transcription speed. Vogel and Jensen (37) found that the transcription elongation rate of RNA polymerase increases from 45 to 65 nucleotides per s in the presence of rRNA boxA. They also found that this increase is dependent upon NusA and that NusA is required for terminator read-through (38). Zellars and Squires (39) found that NusB and NusG also increase the rate of transcription elongation in an rRNA boxA-dependent fashion. NusB increases the transcription elongation rate from 26 to 66 nucleotides per s, and NusG increases the rate from 37 to 66 nucleotides per s. The requirement of Nus factors for both rRNA-AT and the increased transcription elongation rate suggest that these two processes are intrinsically related. The speed at which the antiterminated RNA polymerase transcribes a terminator region may dictate how easily Rho can mediate termination. A model that relates both the read-through of a Rho-dependent terminator and an increase in the transcription elongation rate is supported by experiments studying RNA polymerase mutants with low and high transcription elongation rates and Rho mutants. Jin et al. (16) showed that a mutant RNA polymerase with a reduced transcription elongation rate can suppress a specific mutant Rho. They suggested that the ability of Rho to terminate is dependent upon the transcription speed of RNA polymerase. This hypothesis can be applied to rRNA-antiterminated RNA polymerases, whose increased rate of elongation may facilitate transcription through Rho-dependent terminators in the rRNA operons.
It is interesting that terminator read-through and an increased RNA polymerase transcription elongation rate may reveal separate facets of the same process. It is possible that the transcription elongation rate of an antiterminated RNA polymerase directly affects the level of terminator read-through, although this possibility has not been directly examined. An increase in the transcription elongation rate may be necessary for the synchronization of proper folding of the rRNA as it is being transcribed and the assembly process of the ribosomal subunits. Lewicki et al. (18) showed that an RNA polymerase with a very high transcription elongation rate, such as T7 RNA polymerase that transcribes five times faster than E. coli RNA polymerase, results in the formation of inactive ribosomes. These investigators suggested that the very rapid transcription of rRNA by T7 RNA polymerase uncouples the delicate relationship between transcription and ribosomal assembly (18). This result raises the possibility that the twofold antiterminator-dependent increase in the transcription elongation rate is necessary for the proper folding of the rRNA and the subsequent assembly of the ribosomal subunits (38, 40).
Acknowledgments
We thank David Friedman for the nusB-containing plasmids, Barbara Stitt for purified NusG protein and antibodies to NusG, and Max Gottesman for the nusG mutant strain SS287. We are grateful to Ciaran Condon and Boris Belitsky for helpful comments about the manuscript.
National Institutes of Health grant GM24751 to C.L.S. supported this work.
REFERENCES
- 1.Aksoy, S., C. L. Squires, and C. Squires. 1984. Evidence for antitermination in Escherichia coli rRNA transcription. J. Bacteriol. 159:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrechtsen, B., C. L. Squires, S. Li, and C. Squires. 1990. Antitermination of characterized transcriptional terminators by the Escherichia coli rrnG leader region. J. Mol. Biol. 213:123-134. [DOI] [PubMed] [Google Scholar]
- 3.Altieri, A. S., M. J. Mazzulla, H. Zhou, N. Costantino, D. L. Court, and R. A. Byrd. 1997. Sequential assignments and secondary structure of the RNA-binding transcriptional regulator NusB. FEBS Lett. 415:221-226. [DOI] [PubMed] [Google Scholar]
- 4.Angelini, G., N. Tanigaki, R. Tosi, and G. B. Ferrara. 1986. Southern blot and microfingerprinting analysis of two DR7 haplotypes. Immunogenetics 24:63-67. [DOI] [PubMed] [Google Scholar]
- 5.Berg, K., C. Squires, and C. L. Squires. 1989. Ribosomal RNA operon antitermination. Function of leader and spacer region boxB-boxA sequences and their conservation in diverse micro-organisms. J. Mol. Biol. 209:345-358. [DOI] [PubMed] [Google Scholar]
- 6.Bollag, D. M., and S. J. Edelstein. 1991. Protein methods, p. 181-211. Wiley-Liss, Inc., New York, N.Y.
- 7.Burns, C. M., and J. P. Richardson. 1995. NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc. Natl. Acad. Sci. USA 92:4738-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns, C. M., L. V. Richardson, and J. P. Richardson. 1998. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J. Mol. Biol. 278:307-316. [DOI] [PubMed] [Google Scholar]
- 9.Das, A. 1992. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J. Bacteriol. 174:6711-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das, A. 1993. Control of transcription termination by RNA-binding proteins. Annu. Rev. Biochem. 62:893-930. [DOI] [PubMed] [Google Scholar]
- 11.DeVito, J., and A. Das. 1994. Control of transcription processivity in phage λ: Nus factors strengthen the termination resistant state of RNA polymerase induced by N antiterminator. Proc. Natl. Acad. Sci. USA 91:8660-8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Downing, W. L., S. L. Sullivan, M. E. Gottesman, and P. O. Dennis. 1990. Sequence and transcriptional pattern of the essential E. coli secE-nusG operon. J. Bacteriol. 172:1621-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman, D., E. Olson, L. Johnson, D. Alessi, and M. Craven. 1990. Transcription-dependent competition for a host factor: the function and optimal sequence of the phage boxA transcription antitermination signal. Genes Dev. 4:2210-2222. [DOI] [PubMed] [Google Scholar]
- 14.Heinrich, T., C. Condon, T. Pfeiffer, and R. K. Hartmann. 1995. Point mutations in the leader boxA of a plasmid-encoded Escherichia coli rrnB operon cause defective antitermination in vivo. J. Bacteriol. 177:3793-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huenges, M., C. Rolz, R. Gschwind, R. Peteranderl, F. Berglechner, G. Richter, A. Bacher, H. Kessler, and G. Gemmecker. 1998. Solution structure of the antitermination protein NusB of Escherichia coli: a novel all-helical fold for an RNA-binding protein. EMBO J. 17:4092-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin, D. J., R. R. Burgess, J. P. Richardson, and C. A. Gross. 1992. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc. Natl. Acad. Sci. USA 89:1453-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klotsky, R. A., and I. Schwartz. 1987. Measurement of cat expression from growth-rate-regulated promoters employing beta-lactamase activity as an indicator of plasmid copy number. Gene 55:141-146. [DOI] [PubMed] [Google Scholar]
- 18.Lewicki, B. T. U., T. Margus, J. Remme, and K. H. Nierhaus. 1993. Coupling of rRNA transcription and ribosome assembly in vivo: formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J. Mol. Biol. 231:581-593. [DOI] [PubMed] [Google Scholar]
- 19.Li, J., R. Horwitz, S. McCracken, and J. Greenblatt. 1992. NusG, a new E. coli elongation factor required for processive antitermination of transcription by the N protein of phage λ. J. Biol. Chem. 267:6012-6019. [PubMed] [Google Scholar]
- 20.Li, J., S. W. Mason, and J. Greenblatt. 1993. Elongation factor NusG interacts with termination factor ρ to regulate termination and antitermination of transcription. Genes Dev. 7:161-172. [DOI] [PubMed] [Google Scholar]
- 21.Li, S., C. L. Squires, and C. Squires. 1984. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell 38:851-860. [DOI] [PubMed] [Google Scholar]
- 22.Mason, S. W., and J. Greenblatt. 1991. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 5:1504-1512. [DOI] [PubMed] [Google Scholar]
- 23.Mason, S. W., J. Li, and J. Greenblatt. 1992. Direct interaction between two Escherichia coli transcription antitermination factors, NusB and ribosomal protein S10. J. Mol. Biol. 223:555-566. [DOI] [PubMed] [Google Scholar]
- 24.Nehrke, K. W., and T. Platt. 1994. A quaternary transcription termination complex. Reciprocal stabilization by Rho factor and NusG protein. J. Mol. Biol. 243:830-839. [DOI] [PubMed] [Google Scholar]
- 25.Nodwell, J. R., and J. Greenblatt. 1993. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell 72:261-268. [DOI] [PubMed] [Google Scholar]
- 26.Nudler, E., E. Avetissova, V. Markovtsov, and A. Goldfarb. 1996. Transcription processivity: protein-DNA interactions holding together the elongation complex. Science 273:211-217. [DOI] [PubMed] [Google Scholar]
- 27.Nudler, E., and M. E. Gottesman. 2002. Transcription termination and anti-termination in E. coli. Genes Cells 7:755-768. [DOI] [PubMed] [Google Scholar]
- 28.Pfeiffer, T., and R. K. Hartmann. 1997. Role of the spacer boxA of Escherichia coli ribosomal RNA operons in efficient 23S rRNA synthesis in vivo. J. Mol. Biol. 265:385-393. [DOI] [PubMed] [Google Scholar]
- 29.Roberts, J. W. 1993. RNA and protein elements of E. coli and λ transcription antitermination complexes. Cell 72:653-655. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, 2nd ed., p. 7.76. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Sharrock, R. A., R. L. Gourse, and M. Nomura. 1985. Defective antitermination of rRNA transcription and de-repression of rRNA and tRNA synthesis in the nusB5 mutant of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:5275-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiba, K., K. Ito, and T. Yura. 1986. Suppressors of the secY24 mutation: identification and characterization of additional ssy genes in Escherichia coli. J. Bacteriol. 166:849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Squires, C. L., J. Greenblatt, J. Li, C. Condon, and C. Squires. 1993. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl. Acad. Sci. USA 90:970-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan, S. L., and M. Gottesman. 1992. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell 68:989-994. [DOI] [PubMed] [Google Scholar]
- 36.Taura, T., C. Ueguchi, K. Shiba, and K. Ito. 1992. Insertional disruption of the nusB (ssyB) gene leads to cold-sensitive growth of Escherichia coli and suppression of the secY24 mutation. Mol. Gen. Genet. 234:429-432. [DOI] [PubMed] [Google Scholar]
- 37.Vogel, U., and K. F. Jensen. 1995. Effects of the antiterminator BoxA on transcription elongation kinetics and ppGpp inhibition of transcription elongation in Escherichia coli. J. Biol. Chem. 270:18335-18340. [DOI] [PubMed] [Google Scholar]
- 38.Vogel, U., and K. F. Jensen. 1997. NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA. J. Biol. Chem. 272:12265-12271. [DOI] [PubMed] [Google Scholar]
- 39.Zellars, M., and C. Squires. 1999. Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol. Microbiol. 32:1296-1304. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, Y., J. J. Filter, D. L. Court, M. E. Gottesman, and D. I. Friedman. 2002. Requirement for NusG for transcription antitermination in vivo by the lambda N protein. J. Bacteriol. 184:3416-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]