Abstract
Streptomyces coelicolor is the prototype for the investigation of antibiotic-producing and differentiating actinomycetes. As soil bacteria, streptomycetes can metabolize a wide variety of carbon sources and are hence vested with various specific permeases. Their activity and regulation substantially determine the nutritional state of the cell and, therefore, influence morphogenesis and antibiotic production. We have surveyed the genome of S. coelicolor A3(2) to provide a thorough description of the carbohydrate uptake systems. Among 81 ATP-binding cassette (ABC) permeases that are present in the genome, we found 45 to encode a putative solute binding protein, an essential feature for carbohydrate permease function. Similarity analysis allowed the prediction of putative ABC systems for transport of cellobiose and cellotriose, α-glucosides, lactose, maltose, maltodextrins, ribose, sugar alcohols, xylose, and β-xylosides. A novel putative bifunctional protein composed of a substrate binding and a membrane-spanning moiety is likely to account for ribose or ribonucleoside uptake. Glucose may be incorporated by a proton-driven symporter of the major facilitator superfamily while a putative sodium-dependent permease of the solute-sodium symporter family may mediate uptake of galactose and a facilitator protein of the major intrinsic protein family may internalize glycerol. Of the predicted gene clusters, reverse transcriptase PCRs showed active gene expression in 8 of 11 systems. Together with the previously surveyed permeases of the phosphotransferase system that accounts for the uptake of fructose and N-acetylglucosamine, the genome of S. coelicolor encodes at least 53 potential carbohydrate uptake systems.
Streptomycetes represent a major fraction of the bacterial soil population (20). They contribute substantially to carbon recycling by degrading a whole variety of biopolymers that stem from dead plant and animal material (16). These organic compounds, which include xylan, chitin, and cellulose, are broken down by exoenzymes. The products are funneled into the cell by specific carbohydrate importers that usually recognize mono- and disaccharides. The recent publication of the genome of the model organism Streptomyces coelicolor A3(2) revealed two interesting features concerning carbon utilization (7), that is, (i) a huge number of 172 genes encoding secreted proteins, such as hydrolases, chitinases, cellulases, lipases, nucleases, and proteases, and (ii) the occurrence of 81 ATP-binding cassette (ABC) permeases that are possibly used for the uptake of sugars, oligopeptides, and nucleosides as well as for drug export (46, 56). The numerousness of exoenzymes and ABC systems is 5- to 10-fold higher than that of other bacteria, underlining the broad metabolic capacity of streptomycetes (7).
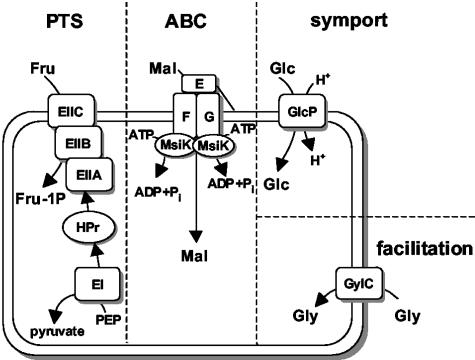
Canonical carbohydrate-specific ABC systems in gram-positive bacteria are oligoprotein assemblies: a membrane-anchored substrate-binding protein is exposed to the outside of the cell, scavenging for a specific substrate molecule (10) (Fig. 1). Two membrane-integral proteins usually form the transport channel, which typically consists of 12 transmembrane segments. The energy for uptake is delivered by an intracellular ATPase, which in streptomycetes is usually not encoded within the same operon (18, 39, 42). This has been demonstrated by the characterization of the msiK gene, which encodes the ATPase subunit of several ABC sugar porters.
FIG. 1.
Schematic overview of carbohydrate uptake systems of S. coelicolor. Representative sugars for the depicted transport systems are fructose (Fru) for PTS, maltose (Mal) for ABC, glucose (Glc) for symport, and glycerol (Gly) for facilitated diffusion. Abbreviations: E, MalE; F, MalF; G, MalG; EIIA, enzyme IIA; EIIB, enzyme IIB; EIIC, enzyme IIC; PEP, phosphoenolpyruvate.
Among the carbohydrate transport systems that have been characterized at the molecular level, ABC transporters have been shown to translocate maltose and maltodextrins in S. coelicolor, cellobiose, cellotriose, and trehalose in Streptomyces reticuli, cellobiose and xylobiose in Streptomyces lividans, and chitobiose and N-acetylglucosamine in Streptomyces olivaceoviridis (18, 39-43, 57, 64). In S. coelicolor, fructose, N-acetylglucosamine, and possibly two further carbohydrates are transported by the phosphotransferase system (PTS) (29, 30, 33, 54). A homologous PTS specific for N-acetylglucosamine was also found in S. olivaceoviridis (61).
In this report, we provide a compilation of carbohydrate uptake systems present in S. coelicolor. In addition to the few already known transport systems, we identified new potential ones by similarity searches of the genome DNA-protein data bank with well-characterized prokaryotic and eukaryotic carbohydrate permeases. To define the possible substrate specificity, each system was evaluated by the analysis of the adjacent metabolic and regulatory genes. Newly identified genes were subjected to reverse transcriptase PCR (RT-PCR) to determine whether they are expressed or not.
MATERIALS AND METHODS
Computer analyses and screening strategies.
All data are based upon sequence comparisons made by using the freely accessible genome data of S. coelicolor A3(2) (www.sanger.ac.uk/Projects/S_coelicolor). Sequences were sampled and subjected to similarity searches (BLAST) at the websites of the National Center for Biotechnology Information at the National Institutes of Health, Bethesda, Md., at www.ncbi.nlm.nih.gov, and of The Wellcome Trust Sanger Institute, Hinxton, Cambridge, United Kingdom, at www.sanger.ac.uk/Projects/S_coelicolor. Default settings were used, however, without filtering low-complexity regions. Sequence alignments were conducted with CLUSTALW, applying predefined algorithms at www.ebi.ac.uk/clustalw of The European Bioinformatics Institute at The European Molecular Biology Laboratory. Protein family signature sequences were obtained from the Institute of Bioinformatics, Geneva, Switzerland, at www.expasy.org/prosite. Clone Manager 5 of Scientific & Educational Software, Durham, N.C., was applied to process and analyze DNA sequence data.
To detect ABC systems involved in carbohydrate import, regions adjacent to genes encoding paralogues of the well-characterized sugar binding protein MalE of Escherichia coli and MalE of S. coelicolor were analyzed (10, 57). In most cases, the juxtaposed genes bore high similarity to genes encoding membrane proteins of the ABC type and were thus considered a putative carbohydrate permease operon. The flanking genes were analyzed whether they encode possible metabolic enzymes, regulators, or ATP binding proteins. In the case that the gene products of such a locus correlated with the respective binding protein in terms of substrate specificity as determined by BLAST analysis, we considered the permease to be the one for the particular carbohydrate. Other types of carbohydrate transport systems, which include the major facilitator superfamily (MFS), the solute-sodium symporter (SSS) family, and the major intrinsic protein (MIP) family, were selected by similarity analysis of well-characterized proteins found in other organisms. The sequences were obtained from the recently updated transporter classification database at The University of California at San Diego (http://tcdb.ucsd.edu/tcdb) (9).
RT-PCR.
Cells of S. coelicolor M145 A3(2) (genotype SCP1− SCP2−, prototroph) were grown for 12 to 18 h under static conditions and were subsequently subjected to 24 h of vigorous shaking at 28°C. The growth medium was tryptic soy broth without dextrose (Difco) with or without 50 mM test carbon source. As an exception, cells tested for glcP1, glcP2, and fruA expression were grown on mineral medium with 50 mM glycerolglucose, or fructose, respectively (34).
Total RNA was prepared as described previously (30). The One-Step RT-PCR kit (Qiagen) was used in combinations with gene-specific oligonucleotides, which were as follows (forward and reverse): for cebE1, TTCGAGGACATGTCCAAGG and ATGAACTTGTTCCAGTCACC; for cebE2, AAGGAGATCACCGAGACC and TTGTCGTCGTAGTACTGC; for bxlE1, AAGATCGAACAGCAGATCGTCG and ACGGACGGCAGGAAGTTGTCC; for bxlE2, AGAACGACGCCTACAAGACG and ACGGCATTGCGTAGATCTTGC; for xylF, TACGAGAAGTTCGACAAGC and TTGTCGAACGAGGTGTAGG; for lacG, TTCGCCCTGGTGTTCCTCG and ATGGAGGCGAGCACCAGGTCG; for smoE, ATCAAGGTGAACTTCACC and AAGGTGTTCACGACCGTCG; for rbsH, TCCTCATCTCGTACGGGAAGCTGC and ATGGCGAGCTTCTGCCGCTTCACC; for rbsE2, ACACCTTCTGGGACATCGTCC and TCGAAGGTCTTCTCCACTCC; for rbsE3, TACTTCACCGTCGCCGACAAGG and TTGACGTACTTGCGCATGTCG. For 16S rRNA, oligonucleotides 16SrRNA1 and 16SrRNA2 were used (30). The assay mixture contained, in a 20-μl volume, 100 ng of RNA and 5 pmol of each primer. Four microliters of the reaction mixture was taken at appropriate PCR cycles (cycles 21 to 36, depending on the appearance of signal in the linear range), and amplification products were separated and visualized on a 1% agarose gel. RT-PCR experiments without prior reverse transcription were performed to assure exclusion of DNA contamination. The quality of the RNA preparations was controlled by the presence of equal amounts of 16S rRNA, which is constitutively expressed. The functionality of oligonucleotides was checked by PCR of chromosomal DNA, giving amplification products of equal intensity for each pair. Data were verified in two independent experiments.
RESULTS AND DISCUSSION
Fifty-three carbohydrate transport systems were detected in the genome of S. coelicolor (Tables 1 and 2). Table 1 gives an overview of gene clusters whose functions are known from experimental investigation, for which a possible substrate could be proposed based on unambiguous similarity data, or for which homologous systems of high similarity have been studied in other organisms. These 22 systems include 14 permeases of the ABC transporter family, four PTS-specific permeases, two copies of a putative sugar porter of the MFS, one member of the SSS family, and one facilitator of the MIP family (14, 32, 33, 36, 46, 49). In the following sections, we describe these systems in detail. Table 2 lists 32 putative gene loci, which, however, were less well definable concerning the possible substrate. This table contains only genes for ABC-type porters. Thus, 87% of all found carbohydrate transporters in S. coelicolor appear to be ATP-driven systems.
TABLE 1.
List of carbohydrate transport systems of S. coelicolor
| Family and substrate(s) | Permease gene(s) | Cosmid location(s) | Representative homologue | Source(s) or reference(s) |
|---|---|---|---|---|
| ABC family | ||||
| Maltodextrins | malEFG | SC10B7.24c to -26c | malEFG, E. coli | 10, 44, 58 |
| Cellobiose, cellotriose | ceb1EFG | 2SCC13.03 to -05 | cebEFG, S. reticuli | 41, this work |
| ceb2EFG | SC5F1.09 to -11 | |||
| β-Xylosides | bxl1EFG | SC1H10.17 to -19 | bxlEFG, S. lividans | Accession no. AF043654, this work |
| bxl2EFG | SCF85.18 to -20 | |||
| α-Glucosides | agl1EFG | SC1H10.02.c | aglEFG, S. lividans | 59 |
| SC8F11.37c and -38c | ||||
| α-Glucosides | agl2EFG | SCE50.06 to -08 | aglEFG, S. melilotii | 63 |
| Xylose | xylFGH | SC7B7.06 to -08 | xylH, E. coli | 51 |
| Chitobiose | ngcEFG | SC7B7.02 to -04 | ngcEFG, S. olivaceoviridis | 64 |
| Lactose | lacGFE | SC6D11.04 to -06c | lacEFG, A. radiobacter | 13, 62, this work |
| Sugar alcohols | smoEFG | SC17.16 to -18 | smoEFG, R. sphaeroides | 50, this work |
| Ribose | rbsH | SCC57A.16 to -19 | rbsCB, E. coli | 35, this work, this work |
| rbsE2F2 | SCAH10.22 to -24 | |||
| rbsE3F3G | SCF43.19c to -21c | |||
| PTS family | ||||
| Fructose | fruA | SCE22.13c | IIFru, E. coli | 30 |
| N-Acetylglucosamine | nagE2 | SCE19A.07 | IICNag, E. coli | 29 |
| malX2 | SCE19A.05c | IIBNag, E. coli | ||
| crr | SC1A8A.10 | IIAGlc, E. coli | ||
| Unknown | nagE1 | SCE19A.06 | IINag, E. coli | 33 |
| Unknown | malX1 | SC51A.12 | IIMalX, E. coli | 33 |
| SSS family, galactose | galP | SCE66.18 | SglT, V. parahaemolyticus | 1, 38, this work |
| MFS family, glucose | glcP1 | SC7A1.22 | GlcP, Synechocystis sp. strain PCC6803 | 65, this work |
| glcP2 | SC9A4.15 | GlcP, Synechocystis sp. strain PCC6803 | 65, this work | |
| MIP family, glycerol | gylC | SC152.01 | glpF, E. coli | 14 |
TABLE 2.
Putative binding protein-dependent carbohydrate transport systems of S. coelicolora
| Cosmid location | E | F | G | C | ATPase |
|---|---|---|---|---|---|
| SC2H4.13 | Mal | Mal | Mal, Mdx | β-Mannosidase | |
| SC3F9.04 | Rib | Rib | Oxidoreductase | Ara, Rib | |
| SC4A7.32 | Unspec | Ara, Xyl | Aldose epimerase | Ara, Xyl | |
| SC6A11.06c | Tre, Mal | Mal, Mdx | Lac, Gly3P | Phosphofructokinase | |
| SC7B7.06 | Xyl | Xyl | Chitinase | Unspec | |
| SC7E4.29 | Mal | Unspec | Mdx | β-hexosaminidase, Glm-6P-deaminase | |
| SC8A6 | Mal | Mal | Mal | β-Glucosidase | |
| SC6D10.01 | Tre, Mal | Lac | Lac | Sugar hydrolase | |
| SC8F11.05c | Rib | Gal | Rib | ||
| SC9A4.29c | Unspec | Unspec | Unspec | ||
| SCBAC17A6.22c | Unspec | Mdx | Unspec | ||
| SCBAC17A6.36 | Unspec | Unspec | Unspec | Hydrolase | |
| SCC24.05c | Unspec | Unspec | Unspec | α-l-Arabinofuranosidase | |
| SCD95A.19 | Unspec | Tagatose-6P kinase, glucose kinase | |||
| SCE59.05c | Unspec | Unspec | Lac | N-Acetylglucosaminidase, UDP-Nag transferase | |
| SCE65.20 | Mal | Mal | Mal, Cdx | β-Galactosidase, racemase | |
| SCE134.06c | Tre, Mal | Unspec | Unspec | Oxidoreductase | |
| SCF1.15 | Unspec | Unspec | Lac | α-Galactosidase | |
| SCF11.11 | Unspec | Tre, Mal | Pal, Lac | β-N-acetylglucosaminidase | |
| SCF11.18 | Unspec | Lac | Mal, Lac | α-Galactosidase | |
| SCF41.11 | Unspec | Lac | Mal | Oxidoreductase | |
| SCF51A.31 | Unspec | Unspec | Sbt, Mtl | ||
| SCF91.20 | Lac | Gly3P | Lac | Gly3P-dehydrogenase | |
| SCG22.02 | Unspec | Tre, Mal, Lac | Pal, Mal | α-,β-Glucosidases | |
| SCG22.11c | Lac, Ara | Lac, Ceb | Ceb, Ara | β-Glucoronidase | |
| β-Galactosidase | |||||
| SC139.29 | Mal, Tre | Mal, Lac | Tre, Mal | Xylanase, xylosidase | |
| SCJ4.46 | β-Xyl | Lac | α-Glc | ||
| SCF1.15 | Unspec | Lac, Gly3P | Unspec | α-Galactosidase | |
| SCL2.29c | Unspec | Lac | Unspec | α-Galactosidase | |
| SCM10.02c | Mal | ||||
| SCM11.07 | Unspec | Lac, Mdx, Gly3P | Lac, Ceb | α-Mannosidase |
Abbreviations: Ara, arabinose; ATPase, ABC-type ATPase; C, catabolic enzyme; Cdx, cyclodextrin; Ceb, cellobiose; E, periplasmic sugar-binding protein; F and G, ABC-type membrane protein; Glm, glucosamine; Gly3P, glycerol-3-phosphate; α-Glc, α-glucoside; Lac, lactose; Mdx, maltodextrin; Mal, maltose; Mtl, mannitol; Nag, N-acetylglucosamine; Pal, palatinose; Rib, ribose; Sbt, sorbitol; Tre, Trehalose; Unspec, unspecific; Xyl, xylose; β-Xyl, β-xyloside.
ABC-type carbohydrate transporters.
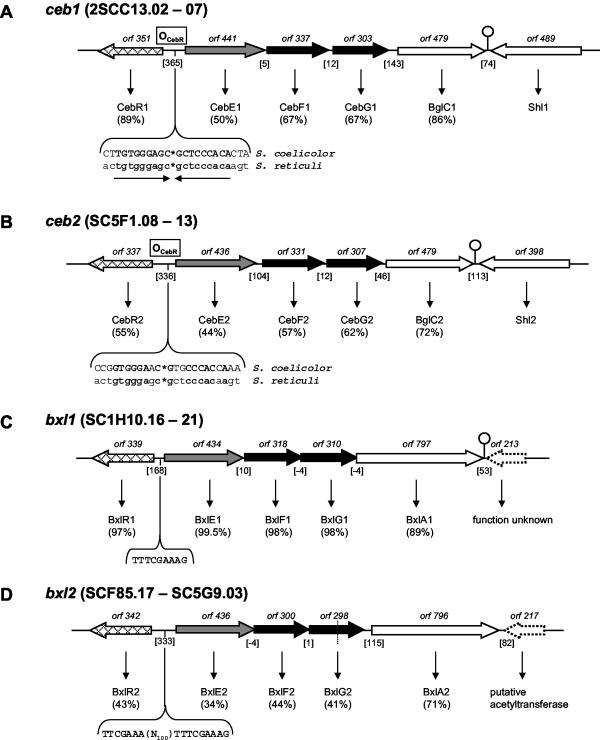
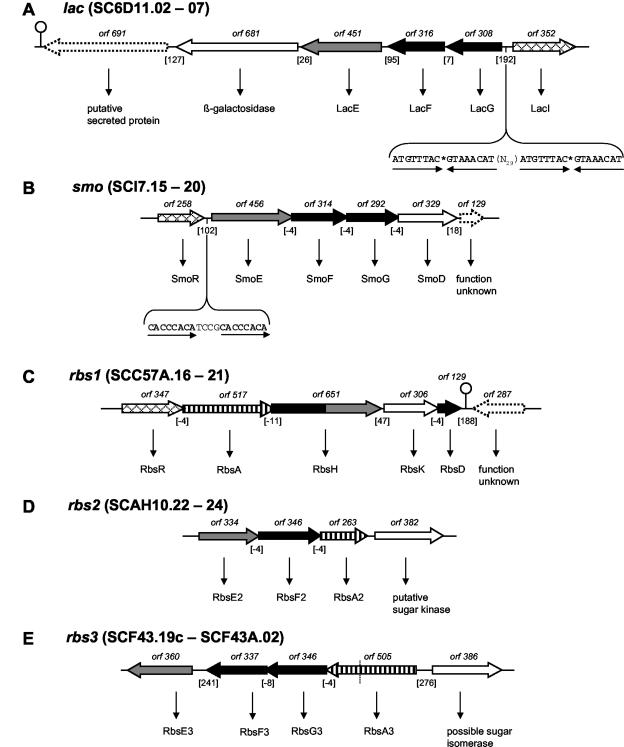
When we started our analysis, there was only one ABC-type carbohydrate permease, MalEFG, described for S. coelicolor that transports maltodextrins and maltose (44, 57). As a first step in the identification of further permeases, we screened the genome for the presence of such systems that were described for other Streptomyces species (18, 39, 41, 42, 59, 64). This led to the identification of genes for two potential cellobiose- and cellotriose-specific permeases, ceb1 and ceb2, two potential β-xyloside-specific permeases, bxl1 and bxl2, two putative gene clusters that could account for uptake of α-glucosides, agl1 and agl2, and a gene locus containing genes for two adjacent ABC systems, one of which has been shown to translocate N-acetylglucosamine and chitobiose (Fig. 2 and 3).
FIG. 2.
(A) ceb1 system. The depicted region includes six orfs, cebE1, cebF1, cebG1, bglC1, and shl1, comprising the ABC transport system and the first step of cellobiose utilization flanked by cebR1 and the probable secreted sugar hydrolase-encoding shl1. A palindromic region of 18 bp upstream of cebE1, shown to be part of the ceb operator (OCebR) in S. reticuli, was detected (the half-sites are separated by asterisks). ORF numbers denote the lengths of the gene products. Genes that encode membrane proteins (generally with the suffix F or G) are shown in black (membrane-spanning proteins); genes that encode substrate-binding proteins are shown in grey (with the suffix E). White arrows represent metabolic genes, probably functionally correlated to the respective system. Vertically striped arrows denote genes for ATP binding and/or hydrolyzing protein. Cross-hatched arrows indicate regulatory genes. Genes of unknown function and/or presumably not involved in the respective system are represented by dotted arrows. Putative regulatory elements are shown in uppercase type and boldface type where in consensus with comparable sequences or to emphasize direct or inverted repeats therein. Dyad repeats are drawn as a stem-loop sign. Dotted lines represent cosmid borders. The numbers of intergenic base pairs are given in brackets, the location of an ORF on the cosmid is designated by SCXX.nn, with “c” denoting complementarity. (B) ceb2 locus. (C and D) bxl1 and bxl2 loci. (E and F) agl1 and agl2 loci. RIP, right imperfect palindrome; PBS, potential binding sequence (60). Abbreviations and designations for panels B to F are the same as for panel A. For further explanations, see the text.
FIG. 3.
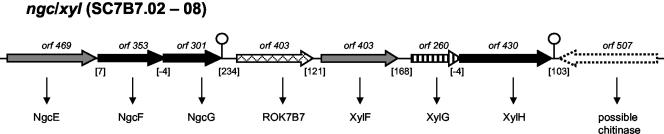
Two EFG-like gene arrangements are shown that represent the ngc/rok7B7/xyl gene locus. The conventions of presentation and designations are described in the legend to Fig. 2A.
Cellobiose and cellotriose.
BLAST searches with the ceb genes of S. reticuli revealed two similar gene loci, ceb1 and ceb2, within the S. coelicolor genome with an identical gene order, cebREFGbglCΩshl, and 47 to 66% identical proteins (exception, Shl1 and Shl2 with 11% identity) among each other (Fig. 2A and B) (41). Both gene clusters encode a divergently transcribed regulator, CebR of the LacI family, a sugar-binding protein, CebE, two integral membrane proteins, CebF and CebG, the β-glucosidase BglC, and a putative sugar hydrolase, Shl. The latter gene is not part of a putative operon, since it is convergently positioned. The deduced proteins share 50 to 86% and 44 to 72% amino acid identities, respectively, to an ABC transporter for cellobiose and cellotriose encoded by the well-characterized ceb operon of S. reticuli. The S. reticuli system has been extensively characterized. Genes cebEFG are induced by cellobiose, the encoded permease is a high-affinity uptake system for cellobiose and cellotriose, and since a cebE mutation abolishes this function, this ceb operon is the only system for these substrates (41). A palindromic sequence of 18 bp was found 128 bp upstream of both cebE1 and cebE2. This cis element has been reported as the CebR binding site OCebR in S. reticuli (40). It is perfectly conserved in system one, whereas system two shares 72% DNA sequence identity.
β-Xylosides.
Two operons were found that share high similarity with the bxl genes of S. lividans (accession no. AF043654), encoding possible β-xyloside BxlEFG permeases of the ABC type (Fig. 2C and D). The metabolic enzyme β-d-xylosidase, BxlA, is encoded by the adjacent downstream gene. The gene cluster bxl1 is almost identical to the S. lividans system, with 89 to 99% protein identity, whereas the bxl2 gene cluster exhibits the same gene order but is more distantly related, with protein identity values from 34 to 71%. The putative regulators BxlR1 and BxlR2, encoded within these loci, belong, as in the case of the ceb operons, to the LacI family. A common palindromic element of low GC content (TTTCGAAA) was detected in the bxlR-bxlE intergenic regions of both systems. This sequence is situated 7 bp in front of bxlE1. The element occurs twice at positions bp 154 and 63 upstream of bxlE2. This sequence is also found in the intergenic region between bxlR and bxlE of S. lividans, and at the 5′ end of xylanase genes (xln) in various streptomycetes. A regulatory role has therefore been postulated (accession no. AF043654).
α-Glucosides.
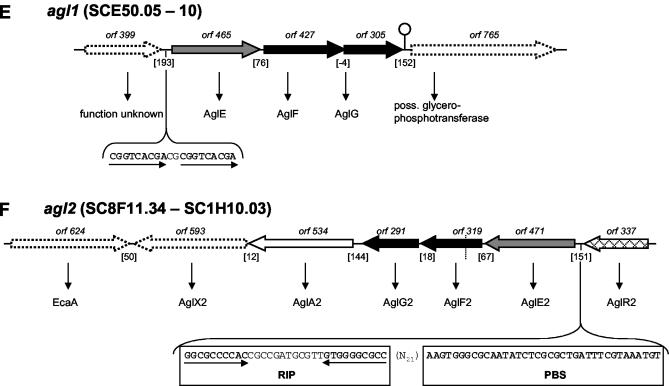
We identified two gene clusters, agl1 and agl2, that encode potential ABC transporters for α-glucosides. agl1EFG encodes a permease that bears considerable similarity (35, 38, and 46% protein identity, respectively) to AglEFG encoded by the aglEFGAK operon of Sinorhizobium meliloti (Fig. 2E) (63). Heterologous expression of the latter genes in Ralstonia eutrophus promoted growth on all tested α-glucosides, which were sucrose, maltose, and trehalose. Thus, the agl1 locus may encode a permease with a broad specificity for α-glycosidic di- and trisaccharides. No genes for a catabolic α-glucosidase or a regulator were found in the vicinity of agl1EFG. However, S. coelicolor has at least three α-glucosidases that share about 56% identical amino acids among each other. aglA1 is associated with the malEFG operon (57), aglA2 is part of the agl2 gene cluster (see below), and the third one is encoded by SCH63.27.
A second gene cluster, agl2REFGAX, was found on cosmid SC8F11 (Fig. 2F). The gene products exhibit 98 to 100% identity to aglEFGAX of S. lividans, which has been proposed to encode a transporter for an α-glucoside (59). However, the authors were unable to determine the substrate, although this has been addressed thoroughly by assaying the purified AglA α-glucosidase for 24 α-glycosidic compounds (59). The S. lividans agl cluster is regulated by the LacI-like regulator RDR (regulator gene within the right-directed repeat of AUD1), which is located upstream of aglE together with the cis elements RIP (right imperfect palindrome) and PBS (potential binding sequence). RDR is part of the highly amplifiable 11.4-kb element AUD1 that encodes two further related regulators, LDR and MDR (regulator genes within the left- and middle-directed repeat of AUD1). These regulators are involved in the massive amplification of AUD1 to more than 100 copies (59). Hence, RDR has a dual regulatory function in S. lividans. Since the AUD1 element also precedes the agl2 gene cluster in S. coelicolor, the systems are most likely homologous and should, therefore, be subject to identical regulation.
The ngc-xyl locus.
In S. olivaceoviridis, N-acetylglucosamine and N,N′-diacetylchitobiose are transported by the ABC permease NgcEFG (64). The system has been characterized by extensive biochemical analysis of NgcE, which binds N-acetylglucosamine and other chitin-degradation products. Inactivation of the gene locus demonstrated further the specificity of NgcEFG. The S. coelicolor gene cluster with the highest similarity was found on cosmid SC7B7 (Fig. 3). It comprises two consecutive EFG gene arrangements interrupted by a regulator gene of the ROK family (53). The first EFG gene set shares protein identities of 31 to 48% with ngcEFG of S. olivaceoviridis and was therefore designated ngcEFG. However, the genes do not encode a functional permease for N-acetylglucosamine because S. coelicolor exclusively uses the PTS for the uptake of this carbon source (29). Thus, ngcEFG may be a good candidate for a chitobiose transport system. The ROK regulator shows, with 94%, a >50% higher identity to NgcR of S. olivaceoviridis than the ngcEFG-encoded products. Thus, it can be considered a true homologue concerning the physiological role. We have tentatively designated the gene rok7B7, since it is not clear yet which of the adjacent operons is controlled by the encoded protein.
The second gene cluster includes genes whose products have similarity (34 to 47% protein identity) to the predicted xylose ABC permease XylFGH of E. coli (51). With the same gene order as found in E. coli, the genes encode a solute-binding protein, an ATPase, and a membrane protein. Thus, in contrast to the other ABC systems described above, this permease comprises the required ATPase gene but only one membrane-integral protein. A putative chitinase gene in the opposite orientation follows downstream. xylFGH may encode a xylose permease. S. coelicolor can readily use this carbon source, and gene homologues for the metabolic enzymes xylose isomerase, XylA, and xylulokinase, XylB, are located on the overlapping cosmids 2SCG11 and SCG11A. The putative xylA (2SCG11.03c) and xylB (SCG11A.01) genes are divergently transcribed. A possible ROK family regulator gene (SCG11A.02) is situated downstream of xylB, which could thus be involved in the regulation of xylA, xylB, and xylFGH.
ABC systems for lactose, ribose, sugar alcohols, and other carbohydrates.
Further potential ABC-type porters for carbohydrates were classified under the assumption that the gene locus must encode a periplasmic binding protein (E), which is usually not found in noncarbohydrate ABC-type systems. A BLASTP query yielded around 30 paralogues of S. coelicolor MalE, which served as a sequence template of an ABC solute binding-protein (57). An iterative search of a far distant paralogue resulted in the nearly identical set of proteins, presumably containing all ABC carbohydrate binding proteins that are present in S. coelicolor. Amino acid identities ranged from 23 to 35%. A phylogenetic tree of MalE paralogues revealed a high degree of radial symmetry with no remarkable clustering (data not shown). A signature sequence of 18 amino acids sharing the consensus pattern (GAP)-(LIVMFA)-(STAVDN)-X4-(GSAV)-(LIVMFY)2-Y-(ND)-X3-(LIVMF)-X-(KNDE) could be observed with fairly good agreement in the N-terminal third of the proteins (52). In particular, MalE and the products of open reading frame (ORF) SCD95A.19 and ORF SC6D11.04c matched exactly to this signature sequence.
Lactose.
An assembly of genes around ORF SC6D11.04c may encode a lactose-specific ABC system because the products of SC6D11.04c to SC6D11.06c exhibit 24, 29, and 31% protein identity to LacEFG of Agrobacterium radiobacter, respectively (13, 62) (Fig. 4A). Interestingly, the system has an unusual reversed gene order, GFE. The permease gene cluster is flanked by a putative β-galactosidase (SC6D11.03c) that shows 43% protein identity to the β-galactosidase BgaB of Bacillus stearothermophilus (15). A probable regulator of the LacI family with an identity of 35% to LacI of E. coli was found to precede lacG and was thus designated LacI (12). Between these two ORFs, two identical copies of a perfect 16-bp palindrome are located as possible cis regulatory elements. They are spaced by 29 bp, with the closer one positioned 38 bp upstream of lacG.
FIG. 4.
The lac, smo, rbs1, rbs2, and rbs3 loci are shown. The conventions of presentation and designations are described in the legend to Fig. 2A.
Sugar alcohols.
Sugar alcohols are reduced forms of aldoses or ketoses. While mannitol is a very good carbon source for S. coelicolor, growth on sorbitol is barely detectable. A gene cluster on cosmid SCI7 was found that could encode a permease for mannitol or another sugar alcohol (Fig. 4B). Designated smoREFGD, this cluster contains smoR, a putative regulator gene of the operon, smoE, the solute binding protein-encoding gene, smoF and smoG, genes coding for membrane translocators, and smoD, a putative alcohol dehydrogenase-encoding gene. The deduced protein product of smoR is 39% identical to the galactitol regulator GatR from Klebsiella oxytoca and is categorized as a member of the DeoR family (7a). Identities of the other deduced proteins to the corresponding system in Rhodobacter sphaeroides are 43% for the periplasmic sorbitol binding protein SmoE and 41% for sorbitol-mannitol transport inner membrane proteins SmoF and SmoG (accession no. AFO18073) (50). SmoD exhibits 30% identity to GutB, a sorbitol dehydrogenase present in Bacillus subtilis (27). Two direct repeats of 8 bp, spaced by four nucleotides and 54 bp upstream of smoE, might be the cis sites for SmoR.
Putative ribose permease with novel domain structure.
A putative ABC system for ribose uptake was discovered on cosmid SCC57A (Fig. 4C). Most remarkably, the permease gene rbsH encodes a unique fusion protein composed of a membrane-spanning moiety and an extracellular solute-binding moiety, which both exhibit 47% protein identity to the ribose permease RbsC and to the ribose-binding protein RbsB of E. coli (35). Another noteworthy feature is the presence of an ATPase gene (rbsA) (8). The gene cluster further comprises a ribokinase gene (rbsK), whose product bears 41% protein identity to the ribokinase of E. coli, and rbsD, which encodes a small protein (129 amino acids) showing 33 and 44% identity to the E. coli and B. subtilis high-affinity RbsD ribose transport proteins (6, 17, 31). A probable LacI family-type regulator, RbsR, is encoded in the beginning of the gene cluster. RbsR shares 37% identity to its E. coli counterpart (24).
Two further gene clusters were found that potentially could encode a ribose ABC permease (Table 1). The assumed gene products of SCAH10.22 to -24 (rbs2EFA) are a substrate-binding lipoprotein, an integral membrane protein, and an ATPase, each with remarkable similarities to ribose ABC systems of other bacteria. SCF43.19 to -22 (rbs3EFGA) may encode a third ribose-like permease. Despite the primary similarity to ribose systems, these loci might also be involved in the transport of ribose-containing substrates, such as nucleoribosides, that streptomycetes are estimated to feed on.
The presence of ATPase genes in all three putative ribose uptake systems is in obvious contrast to the majority of the other ABC systems described here.
Less-well-definable ABC porters for carbohydrate.
Thirty-one gene clusters were detected that could encode further ABC-type porters for carbohydrates (Table 2). All but two contain genes for a solute-binding protein with at least one adjacent permease gene. Four of them comprise a gene for an ABC-type ATPase, which implies that most ABC porters must share a common ATPase. It was not possible to suggest a probable substrate for the encoded permeases because similarity analyses revealed a substrate heterogeneity among each gene cluster. Yet, their presence underlines the fact that S. coelicolor has an astonishing number of carbohydrate transporters at its disposal, predominantly of the ABC type. The system to which SC7E4.29 belongs has been described as the dasRABC regulon of Streptomyces griseus, which encodes an ABC transporter that is involved in a glucose-dependent differentiation process (48). The transporter is expressed towards the commencement of aerial hypha formation and during sporulation. However, our analysis did not provide an unambiguous hint as to a possible substrate, and dasABC was therefore classified as a less-well-definable system.
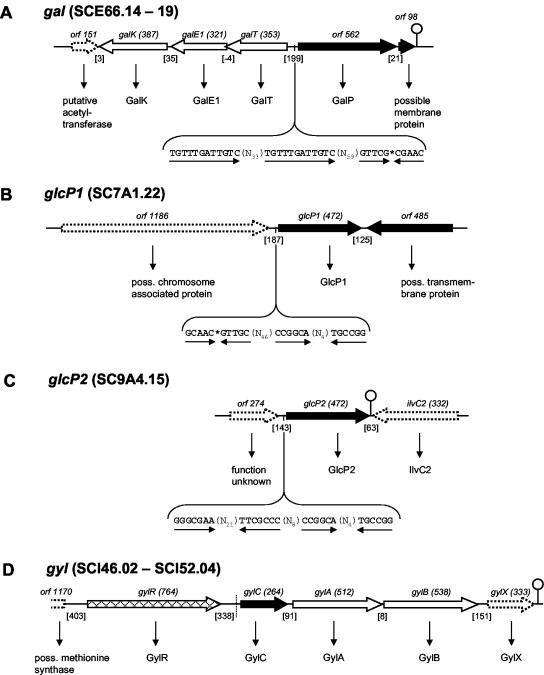
Putative galactose-Na+ symporter of the SSS family.
Galactose can be readily utilized by S. coelicolor. Genes for the utilization of galactose have been identified in S. lividans, and the homologues of S. coelicolor have been noted (1, 7). The products of the galKE1T operon in S. lividans mediate the funneling of galactose into glycolysis via phosphorylation (GalK), epimerization (GalE1) and transfer of an uridylyl group, mediated by GalT. The promoter region of the galKEIT operon has been analyzed with respect to its role in glucose repression (23). The galactose transporter-encoding gene, however, has so far not been elucidated. This could be galP, an orf that is divergently orientated upstream of galT. GalP shares 28% similarity with the Na+-galactose symporter SglS of Vibrio parahaemolyticus (38, 55) (Fig. 5A). The postulated galactose permease belongs to the SSS family and appears to be the only member of this family within the S. coelicolor genome that serves as a carbohydrate permease (36). Possible regulatory elements are a 10-bp palindrome 14 bp upstream of galP as well as two 12-bp direct repeats separated by 33 bp and located 85 bp in front of galP.
FIG. 5.
The gal, glcP1, glcP, and gyl loci are shown. The conventions of presentation and designations are described in the legend to Fig. 2A.
Glucose permease of the MFS.
Glucose is a preferred carbon source of S. coelicolor, and many reports have been published that deal with the mechanism of glucose repression (2, 19, 22). However, a glucose transporter has not yet been identified. When the genome was screened with the non-PTS glucose permeases GlcP of Synechocystis sp. strain PCC6803 and Glf of Zymomonas mobilis, a homologous protein with 51 and 33% identity, respectively, was detected and designated GlcP (61, 65). Interestingly, the corresponding gene occurs in two copies on the chromosome, glcP1 (SC7A1.22) and glcP2 (SC9A4.15), which exhibit 99% DNA sequence identity to each other and encode identical gene products (Fig. 5B and C). DNA sequence similarity is extended to 39 bp (except two nucleotides) in the upstream area and only 4 bp in the downstream region. A common dyad repeat is located 18 bp upstream of both glcP genes. Furthermore, a 10-bp palindromic region is present 70 bp upstream of glcP1, and another dyad sequence is exclusively found 41 bp in front of glcP2. The adjacent genes do not encode potential metabolic or regulatory genes that may be functionally associated.
BLAST searches with other sugar permeases of the MFS, such as the xylose permease XylE and arabinose permease AraE of E. coli, revealed that S. coelicolor has more than 50 members of this family. However, no further genes besides glcP1 and glcP2 were detected that encode an obvious permease for carbohydrates.
Glycerol permease of the MIP family.
To complete the analysis on carbohydrate uptake systems, it should be noted that glycerol is efficiently metabolized by the genes of the gylRCABX region (14) (Fig. 5D). Deletion of gylR demonstrated that substrate induction and catabolite repression of the gyl operon are mediated through GylR. GylC shares 49% amino acid identity to GlpF of B. subtilis (5). GylA and GylB exhibit 52 and 32% identity to glycerol kinase GlpK and glycerol-3-phosphate dehydrogenase GlpD of E. coli, respectively (4, 26). GylX shares similarity with proteins of unknown function present in other actinomycetes and archaea. We observed that glycerol transport is inducible by this substrate and repressible by glucose (unpublished data). Furthermore, we demonstrated that the specific repressor GylR, a member of the IclR family, binds within the upstream region (400 bp) of gylC (unpublished data) (37). There are three potential palindromic binding sites of 8 to 10 bp for GylR.
Which carbohydrate genes are expressed and what is the true substrate?
The substrate definition of the well-definable carbohydrate transport systems that are compiled in Table 1 has been derived from available molecular genetic, biochemical, and in silico data. From transcriptional and growth analyses of malEFG and from the mutation of malE and the adjacent regulatory gene malR, it is obvious that this ABC permease is inducible by amylose and maltotriose (44). It transports not only maltotriose with the highest specificity but also maltose and other maltodextrins. The regulator MalR reacts best on maltopentaose and confers substrate induction and glucose repression (44, 58). From the published data on the glycerol-, galactose-, fructose-, and N-acetylglucosamine-utilizing systems, it is rather evident that GylC, GalP, FruA, and NagE2 are the respective permeases (14, 23, 29, 30, 47).
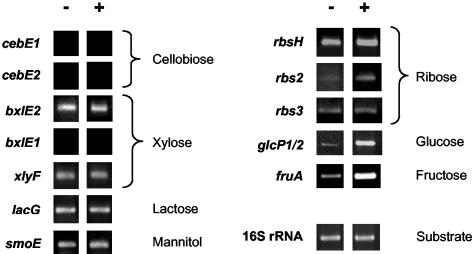
For the remaining systems of Table 1, we suggested their possible function solely on the basis of in silico data. To support our findings, we performed RT-PCR experiments to examine whether the particular system is expressed and whether it is induced by the suggested substrate. Therefore, we measured the mRNA levels of a representative gene of the respective operon (see Materials and Methods for details). As can be seen from Fig. 6, cebE1, cebE2, and bxlE1 were not expressed, which indicates that these are silent genes or that their expression occurs only under very specific growth conditions. mRNA of bxlE2 was readily detectable. Since it may transport β-xylosides, we checked whether the presence of xylose could induce bxlE2. This was not the case. Due to cost restriction, we did not try induction by the more likely inducer xylobiose (18). Gene expression of xylF, lacE, and smoE was as well detectable by RT-PCR. However, the addition of xylose, lactose, or mannitol to the growth medium, respectively, did not reveal induction of gene expression. Thus, these systems are constitutively expressed in the complex rich medium we used. In correlation with the mRNA lacE data, we measured that lactose uptake is also constitutive (our unpublished data). Of the three potential ribose permease systems, we found that they are all expressed. Here, the amounts of rbsE2 mRNA were elevated when ribose was present in the growth medium, which strengthens the suggestion that the rbs2 gene locus encodes a ribose-specific permease. Expression of glcP was clearly glucose dependent and therefore confirmed our suggestion. It should be noted that we could not distinguish between glcP1 and glcP2 gene activity, due to the almost identical sequence conservation. RT-PCR of fruA (Fig. 6) and crr (IIA domain of the PTS permease for N-acetylglucosamine) further revealed the specificity of the suggested systems (29, 30, 33). Due to the uncertainty of a possible substrate and cost restriction (chitobiose), we refrained from analyzing the agl and ngc loci.
FIG. 6.
Expression profiling of proposed carbohydrate uptake systems. For each system, RNA from S. coelicolor A3(2) grown without (left column, −) or in the presence of (right column, +) the indicated carbon source was used for RT-PCR. As an example of undoubted substrate induction, transcriptional analysis of fruA (encoding the specific fructose-PTS enzyme IIABC) is depicted (30).
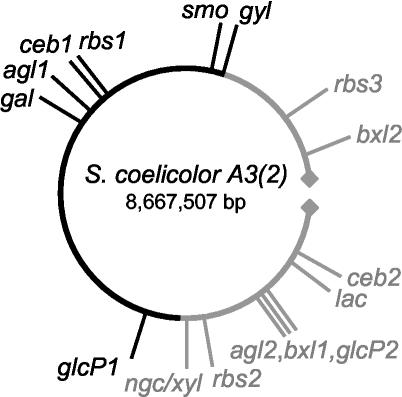
Chromosomal distribution.
Different from the genomes of most other bacteria, the chromosome of S. coelicolor is linear. A central core appears to be flanked by two arms, which are subject to more-intensive genetic variation. These arms range from 0 to about 1.5 Mbp (left arm) and from about 6.4 Mbp to the end of the chromosome (right arm) (7). The genetic localization of the newly identified carbohydrate uptake systems shows a distribution both in the core and in the variable region (Fig. 7). It is remarkable that ceb1, glcP1, and rbs1 are located in the core region, whereas the ceb2, glcP2, rbs2, and rbs3 systems lie within the variable region. This supports the thesis that the variable region harbors nonessential and duplicate genes (7).
FIG. 7.
Genetic map of S. coelicolor. A depiction of the positions of the gene clusters of newly identified transport systems (bxl2, rbs3, smo, rbs1, ceb1, agl1, gal, glcP1, gyl, ngc/xyl, rbs2, agl2, bxl1, glcP2, lac, and ceb2) is shown. The constant region of the chromosome is shown in black; the variable ends are in grey. The direction of the chromosome is counterclockwise.
This report provides a comprehensive survey of the carbohydrate permeases present in S. coelicolor. We were able to collect a total of 53 potential systems and conclude the following. (i) The huge number of detected carbohydrate uptake systems reflects perfectly the lifestyle of streptomycetes. They live in the soil and feed primarily on complex long-chain carbohydrates from plants, insects, and fungi. These polysaccharides are broken down by exoenzymes and internalized as mono- and disaccharides. (ii) It seems that S. coelicolor may predominantly use permeases of the ABC type for the internalization of carbohydrates, a feature that has recently been found in the archaean Sulfolobus solfataricus as well (11). It can be inferred that α-glucosides, lactose, maltodextrins, maltose, ribose, β-xylosides, xylose, and sugar alcohols are likely to be transported by ABC systems, but only the maltose-maltodextrin system MalEFG has been experimentally proven (44, 57). Another 31 ABC systems are present that may contribute to the uptake of further carbohydrates. Genes encoding the required ATPase for ABC permease function were only within the permease gene cluster in four cases. This finding, which is unusual among bacteria, supports the results from previous studies with S. reticuli and S. lividans, where the ATPase MsiK assists several ABC permeases (18, 42). (iii) Fructose, N-acetylglucosamine, and probably two yet-unidentified sugars are transported by PTSs (29, 30, 33). The fructose and N-acetylglucosamine PTSs are the only uptake systems for the respective substrate in S. coelicolor. They are unequivocally characterized by mutation and extensive biochemical analysis (29, 30). (iv) S. coelicolor probably has just one porter of the SSS family (galactose), the MFS (glucose), and the MIP family (glycerol) at its disposal. It is noteworthy that a potential permease of the preferred carbon source glucose is encoded by two identical gene copies. Research on the glycerol and galactose operons strongly suggests that, although not biochemically characterized, the permease gene is carried within the respective operon (14). (v) Three systems are present in duplicated form. It appears that the two potential permeases for cellobiose and cellotriose, Ceb1 and Ceb2, are not expressed. The genes encoding the potential β-xyloside permease Bxl2 are expressed, whereas Bxl1 may be silent. glcP expression was clearly glucose inducible. Due to the high sequence identity, it was not possible to determine to which extent the glcP1 or glcP2 allele is transcribed. However, it should be noted that for ceb, bxl, and glcP, both loci putatively encode isofunctional transporters that may be expressed at different growth phases or under different growth conditions, as is true also for anabolic genes, such as that for glycogen synthesis (45). (vi) A gene encoding a regulator was found in most gene clusters. Just the regulators, ROK7B71, SmoR, and GylR, belong to the ROK, DeoR, and IclR families, respectively. All others are members of the LacI family of bacterial regulators. This group appears to be rather heterogeneous, which holds true also for the primary sequence of the α-helix-turn-α-helix motifs (28). Therefore, the respective cognate operators can display pronounced variation, thus diminishing undesired cross-recognition. Nevertheless, we have included in our analysis the presence of putative cis active elements that may be recognized by these regulatory proteins. A divergent orientation of repressor and metabolic genes indicates that the metabolic operon and the repressor itself might be simultaneously controlled by one or more copies of a regulator binding site in between. Such autogenous regulation has been found in divergeons for sucrose and arabinose utilization (21, 25). (vii) The presence of metabolic genes within a permease gene cluster occurred at a rate of 74%. It indicates that a number of catabolic enzymes are not coordinately regulated with the respective permease. This is reasonable when low- and high-affinity transport systems for the same substrate are expressed independently of an abundant or scarce supply of carbon source or when the catabolic enzyme is required for metabolism of different substrates. Glucose kinase, for instance, should be present whenever free internal glucose from glucose-containing saccharides is delivered and is thus independently regulated (3, 22).
This study documents that our molecular understanding of streptomycete nutrition concerning the intake of carbohydrates is still at the very beginning. We hope that the compilation presented here may serve as a guide to stimulate further investigation of S. coelicolor and other relevant Streptomyces species.
Acknowledgments
We thank Miriam König and Andreas W. Thomae for assistance and fruitful discussions.
This work was supported through grant SFB473 and Graduiertenkolleg 40 of the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Adams, C. W., J. A. Fornwald, F. J. Schmidt, M. Rosenberg, and M. E. Brawner. 1988. Gene organization and structure of the Streptomyces lividans gal operon. J. Bacteriol. 170:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angell, S., C. G. Lewis, M. J. Buttner, and M. J. Bibb. 1994. Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Mol. Gen. Genet. 244:135-143. [DOI] [PubMed] [Google Scholar]
- 3.Angell, S., E. Schwarz, and M. J. Bibb. 1992. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol. Microbiol. 6:2833-2844. [DOI] [PubMed] [Google Scholar]
- 4.Austin, D., and T. J. Larson. 1991. Nucleotide sequence of the glpD gene encoding aerobic sn-glycerol 3-phosphate dehydrogenase of Escherichia coli K-12. J. Bacteriol. 173:101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beijer, L., R. P. Nilsson, C. Holmberg, and L. Rutberg. 1993. The glpP and glpF genes of the glycerol regulon in Bacillus subtilis. J. Gen. Microbiol. 139:349-359. [DOI] [PubMed] [Google Scholar]
- 6.Bell, A. W., S. D. Buckel, J. M. Groarke, J. N. Hope, D. H. Kingsley, and M. A. Hermodson. 1986. The nucleotide sequences of the rbsD, rbsA, and rbsC genes of Escherichia coli K12. J. Biol. Chem. 261:7652-7658. [PubMed] [Google Scholar]
- 7.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 7a.Brinkkötter, A., A. Shakeri-Garakani, and J. W. Lengeler. 2002. Two class II D-tagatose-bisphosphate aldolases from enteric bacteria. Arch. Microbiol. 177:410-419. [DOI] [PubMed] [Google Scholar]
- 8.Buckel, S. D., A. W. Bell, J. K. Rao, and M. A. Hermodson. 1986. An analysis of the structure of the product of the rbsA gene of Escherichia coli K12. J. Biol. Chem. 261:7659-7662. [PubMed] [Google Scholar]
- 9.Busch, W., and M. H. Saier, Jr. 2002. The transporter classification (TC) system, 2002. Crit. Rev. Biochem. Mol. Biol. 37:287-337. [DOI] [PubMed] [Google Scholar]
- 10.Ehrmann, M., R. Ehrle, E. Hofmann, W. Boos, and A. Schlösser. 1998. The ABC maltose transporter. Mol. Microbiol. 29:685-694. [DOI] [PubMed] [Google Scholar]
- 11.Elferink, M. G., S. V. Albers, W. N. Konings, and A. J. Driessen. 2001. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol. 39:1494-1503. [DOI] [PubMed] [Google Scholar]
- 12.Farabaugh, P. J. 1978. Sequence of the lacI gene. Nature 274:765-769. [DOI] [PubMed] [Google Scholar]
- 13.Greenwood, J. A., A. Cornish, and C. W. Jones. 1990. Binding-protein-dependent lactose transport in Agrobacterium radiobacter. J. Bacteriol. 172:1703-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hindle, Z., and C. P. Smith. 1994. Substrate induction and catabolite repression of the Streptomyces coelicolor glycerol operon are mediated through the GylR protein. Mol. Microbiol. 12:737-745. [DOI] [PubMed] [Google Scholar]
- 15.Hirata, H., T. Fukazawa, S. Negoro, and H. Okada. 1986. Structure of a beta-galactosidase gene of Bacillus stearothermophilus. J. Bacteriol. 166:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodgson, D. A. 2000. Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv. Microb. Physiol. 42:47-238. [DOI] [PubMed] [Google Scholar]
- 17.Hope, J. N., A. W. Bell, M. A. Hermodson, and J. M. Groarke. 1986. Ribokinase from Escherichia coli K12. Nucleotide sequence and overexpression of the rbsK gene and purification of ribokinase. J. Biol. Chem. 261:7663-7668. [PubMed] [Google Scholar]
- 18.Hurtubise, Y., F. Shareck, D. Kluepfel, and R. Morosoli. 1995. A cellulase/xylanase-negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol. Microbiol. 17:367-377. [DOI] [PubMed] [Google Scholar]
- 19.Kwakman, J. H., and P. W. Postma. 1994. Glucose kinase has a regulatory role in carbon catabolite repression in Streptomyces coelicolor. J. Bacteriol. 176:2694-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locci, R. 1986. Streptomycetes and related genera, p. 2492-2541. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md.
- 21.Luesink, E. J., J. D. Marugg, O. P. Kuipers, and W. M. de Vos. 1999. Characterization of the divergent sacBK and sacAR operons, involved in sucrose utilization by Lactococcus lactis. J. Bacteriol. 181:1924-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahr, K., G. P. van Wezel, C. Svensson, U. Krengel, M. J. Bibb, and F. Titgemeyer. 2000. Glucose kinase of Streptomyces coelicolor A3(2): large-scale purification and biochemical analysis. Antonie Leeuwenhoek 78:253-261. [DOI] [PubMed] [Google Scholar]
- 23.Mattern, S. G., M. E. Brawner, and J. Westpheling. 1993. Identification of a complex operator for galP1, the glucose-sensitive, galactose-dependent promoter of the Streptomyces galactose operon. J. Bacteriol. 175:1213-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauzy, C. A., and M. A. Hermodson. 1992. Structural and functional analyses of the repressor, RbsR, of the ribose operon of Escherichia coli. Protein Sci. 1:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mota, L. J., L. M. Sarmento, and I. de Sa-Nogueira. 2001. Control of the arabinose regulon in Bacillus subtilis by AraR in vivo: crucial roles of operators, cooperativity, and DNA looping. J. Bacteriol. 183:4190-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muramatsu, S., and T. Mizuno. 1989. Nucleotide sequence of the region encompassing the glpKF operon and its upstream region containing a bent DNA sequence of Escherichia coli. Nucleic Acids Res. 17:4378. [PMC free article] [PubMed] [Google Scholar]
- 27.Ng, K., R. Ye, X. C. Wu, and S. L. Wong. 1992. Sorbitol dehydrogenase from Bacillus subtilis. Purification, characterization, and gene cloning. J. Biol. Chem. 267:24989-24994. [PubMed] [Google Scholar]
- 28.Nguyen, C. C., and M. H. Saier, Jr. 1995. Phylogenetic, structural and functional analyses of the LacI-GalR family of bacterial transcription factors. FEBS Lett. 377:98-102. [DOI] [PubMed] [Google Scholar]
- 29.Nothaft, H., D. Dresel, A. Willimek, K. Mahr, M. Niederweis, and F. Titgemeyer. 2003. The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J. Bacteriol. 185:7019-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nothaft, H., S. Parche, A. Kamionka, and F. Titgemeyer. 2003. In vivo analysis of HPr reveals a fructose-specific phosphotransferase system that confers high-affinity uptake in Streptomyces coelicolor. J. Bacteriol. 185:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Reilly, M., K. Woodson, B. C. Dowds, and K. M. Devine. 1994. The citrulline biosynthetic operon, argC-F, and a ribose transport operon, rbs, from Bacillus subtilis are negatively regulated by Spo0A. Mol. Microbiol. 11:87-98. [DOI] [PubMed] [Google Scholar]
- 32.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parche, S., H. Nothaft, A. Kamionka, and F. Titgemeyer. 2000. Sugar uptake and utilisation in Streptomyces coelicolor: a PTS view to the genome. Antonie Leeuwenhoek 78:243-251. [DOI] [PubMed] [Google Scholar]
- 34.Parche, S., R. Schmid, and F. Titgemeyer. 1999. The phosphotransferase system (PTS) of Streptomyces coelicolor identification and biochemical analysis of a histidine phosphocarrier protein HPr encoded by the gene ptsH. Eur. J. Biochem. 265:308-317. [DOI] [PubMed] [Google Scholar]
- 35.Park, Y., Y. J. Cho, T. Ahn, and C. Park. 1999. Molecular interactions in ribose transport: the binding protein module symmetrically associates with the homodimeric membrane transporter. EMBO J. 18:4149-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reizer, J., A. Reizer, and M. H. Saier, Jr. 1994. A functional superfamily of sodium/solute symporters. Biochim. Biophys. Acta 1197:133-166. [DOI] [PubMed] [Google Scholar]
- 37.Reverchon, S., W. Nasser, and J. Robert-Baudouy. 1991. Characterization of kdgR, a gene of Erwinia chrysanthemi that regulates pectin degradation. Mol. Microbiol. 5:2203-2216. [DOI] [PubMed] [Google Scholar]
- 38.Sarker, R. I., Y. Okabe, M. Tsuda, and T. Tsuchiya. 1996. Sequence of a Na+/glucose symporter gene and its flanking regions of Vibrio parahaemolyticus. Biochim. Biophys. Acta 1281:1-4. [DOI] [PubMed] [Google Scholar]
- 39.Schlösser, A. 2000. MsiK-dependent trehalose uptake in Streptomyces reticuli. FEMS Microbiol. Lett. 184:187-192. [DOI] [PubMed] [Google Scholar]
- 40.Schlösser, A., T. Aldekamp, and H. Schrempf. 2000. Binding characteristics of CebR, the regulator of the ceb operon required for cellobiose/cellotriose uptake in Streptomyces reticuli. FEMS Microbiol. Lett. 190:127-132. [DOI] [PubMed] [Google Scholar]
- 41.Schlösser, A., J. Jantos, K. Hackmann, and H. Schrempf. 1999. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl. Environ. Microbiol. 65:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlösser, A., T. Kampers, and H. Schrempf. 1997. The Streptomyces ATP-binding component MsiK assists in cellobiose and maltose transport. J. Bacteriol. 179:2092-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlösser, A., and H. Schrempf. 1996. A lipid-anchored binding protein is a component of an ATP-dependent cellobiose/cellotriose-transport system from the cellulose degrader Streptomyces reticuli. Eur. J. Biochem. 242:332-338. [DOI] [PubMed] [Google Scholar]
- 44.Schlösser, A., A. Weber, and H. Schrempf. 2001. Synthesis of the Streptomyces lividans maltodextrin ABC transporter depends on the presence of the regulator MalR. FEMS Microbiol. Lett. 196:77-83. [DOI] [PubMed] [Google Scholar]
- 45.Schneider, D., C. J. Bruton, and K. F. Chater. 2000. Duplicated gene clusters suggest an interplay of glycogen and trehalose metabolism during sequential stages of aerial mycelium development in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 263:543-553. [DOI] [PubMed] [Google Scholar]
- 46.Schneider, E. 2001. ABC transporters catalyzing carbohydrate uptake. Res. Microbiol. 152:303-310. [DOI] [PubMed] [Google Scholar]
- 47.Seno, E. T., and K. F. Chater. 1983. Glycerol catabolic enzymes and their regulation in wild-type and mutant strains of Streptomyces coelicolor A3(2). J. Gen. Microbiol. 129:1403-1413. [DOI] [PubMed] [Google Scholar]
- 48.Seo, J. W., Y. Ohnishi, A. Hirata, and S. Horinouchi. 2002. ATP-binding cassette transport system involved in regulation of morphological differentiation in response to glucose in Streptomyces griseus. J. Bacteriol. 184:91-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, C. P., and K. F. Chater. 1988. Cloning and transcription analysis of the entire glycerol utilization (gylABX) operon of Streptomyces coelicolor A3(2) and identification of a closely associated transcription unit. Mol. Gen. Genet. 211:129-137. [DOI] [PubMed] [Google Scholar]
- 50.Stein, M. A., A. Schäfer, and F. Giffhorn. 1997. Cloning, nucleotide sequence, and overexpression of smoS, a component of a novel operon encoding an ABC transporter and polyol dehydrogenases of Rhodobacter sphaeroides Si4. J. Bacteriol. 179:6335-6340. [DOI] [PMC free article] [PubMed]
- 51.Sumiya, M., E. O. Davis, L. C. Packman, T. P. McDonald, and P. J. Henderson. 1995. Molecular genetics of a receptor protein for D-xylose, encoded by the gene xylF, in Escherichia coli. Receptors Channels 3:117-128. [PubMed] [Google Scholar]
- 52.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Titgemeyer, F., J. Reizer, A. Reizer, and M. H. Saier, Jr. 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140:2349-2354. [DOI] [PubMed] [Google Scholar]
- 54.Titgemeyer, F., J. Walkenhorst, J. Reizer, M. H. Stuiver, X. Cui, and M. H. Saier, Jr. 1995. Identification and characterization of phosphoenolpyruvate:fructose phosphotransferase systems in three Streptomyces species. Microbiology 141:51-58. [DOI] [PubMed] [Google Scholar]
- 55.Turk, E., O. Kim, J. le Coutre, J. P. Whitelegge, S. Eskandari, J. T. Lam, M. Kreman, G. Zampighi, K. F. Faull, and E. M. Wright. 2000. Molecular characterization of Vibrio parahaemolyticus vSGLT: a model for sodium-coupled sugar cotransporters. J. Biol. Chem. 275:25711-25716. [DOI] [PubMed] [Google Scholar]
- 56.van Veen, H. W., and W. N. Konings. 1998. The ABC family of multidrug transporters in microorganisms. Biochim. Biophys. Acta 1365:31-36. [DOI] [PubMed] [Google Scholar]
- 57.van Wezel, G. P., J. White, M. J. Bibb, and P. W. Postma. 1997. The malEFG gene cluster of Streptomyces coelicolor A3(2): characterization, disruption and transcriptional analysis. Mol. Gen. Genet. 254:604-608. [DOI] [PubMed] [Google Scholar]
- 58.van Wezel, G. P., J. White, P. Young, P. W. Postma, and M. J. Bibb. 1997. Substrate induction and glucose repression of maltose utilization by Streptomyces coelicolor A3(2) is controlled by malR, a member of the lacl-galR family of regulatory genes. Mol. Microbiol. 23:537-549. [DOI] [PubMed] [Google Scholar]
- 59.Volff, J. N., and J. Altenbuchner. 2000. The 1-kb-repeat-encoded DNA-binding protein as repressor of an alpha-glucosidase operon flanking the amplifiable sequence AUD1 of Streptomyces lividans. Microbiology 146:923-933. [DOI] [PubMed] [Google Scholar]
- 60.Wang, F., X. Xiao, A. Saito, and H. Schrempf. 2002. Streptomyces olivaceoviridis possesses a phosphotransferase system that mediates specific, phosphoenolpyruvate-dependent uptake of N-acetylglucosamine. Mol. Genet. Genomics 268:344-351. [DOI] [PubMed] [Google Scholar]
- 61.Weisser, P., R. Kramer, H. Sahm, and G. A. Sprenger. 1995. Functional expression of the glucose transporter of Zymomonas mobilis leads to restoration of glucose and fructose uptake in Escherichia coli mutants and provides evidence for its facilitator action. J. Bacteriol. 177:3351-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams, S. G., J. A. Greenwood, and C. W. Jones. 1992. Molecular analysis of the lac operon encoding the binding-protein-dependent lactose transport system and beta-galactosidase in Agrobacterium radiobacter. Mol. Microbiol. 6:1755-1768. [DOI] [PubMed] [Google Scholar]
- 63.Willis, L. B., and G. C. Walker. 1999. A novel Sinorhizobium meliloti operon encodes an alpha-glucosidase and a periplasmic-binding-protein-dependent transport system for alpha-glucosides. J. Bacteriol. 181:4176-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao, X., F. Wang, A. Saito, J. Majka, A. Schlösser, and H. Schrempf. 2002. The novel Streptomyces olivaceoviridis ABC transporter Ngc mediates uptake of N-acetylglucosamine and N, N′-diacetylchitobiose. Mol. Genet. Genomics 267:429-439. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, C. C., M. C. Durand, R. Jeanjean, and F. Joset. 1989. Molecular and genetical analysis of the fructose-glucose transport system in the cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 3:1221-1229. [DOI] [PubMed] [Google Scholar]