Abstract
Six putative regulatory genes are located at the flank of the nystatin biosynthetic gene cluster in Streptomyces noursei ATCC 11455. Gene inactivation and complementation experiments revealed that nysRI, nysRII, nysRIII, and nysRIV are necessary for efficient nystatin production, whereas no significant roles could be demonstrated for the other two regulatory genes. To determine the in vivo targets for the NysR regulators, chromosomal integration vectors with the xylE reporter gene under the control of seven putative promoter regions upstream of the nystatin structural and regulatory genes were constructed. Expression analyses of the resulting vectors in the S. noursei wild-type strain and regulatory mutants revealed that the four regulators differentially affect certain promoters. According to these analyses, genes responsible for initiation of nystatin biosynthesis and antibiotic transport were the major targets for regulation. Data from cross-complementation experiments showed that nysR genes could in some cases substitute for each other, suggesting a functional hierarchy of the regulators and implying a cascade-like mechanism of regulation of nystatin biosynthesis.
Antibiotic production by Streptomyces bacteria has received much attention in recent years due to the problems associated with a constantly increasing incidence of multiresistant microbial pathogens. Considerable efforts are directed toward understanding the antibiotic biosynthetic pathways in Streptomyces spp. and manipulating the corresponding gene clusters in order to produce novel compounds with improved properties (17). In addition, remarkable progress is being made in dissecting the functions of the genes that regulate antibiotic production in streptomycetes (11). Coupling these two fields of research is of great importance both for a fundamental understanding of the antibiotic biosynthesis processes and for the rational engineering of novel antibiotic producers.
Biosynthesis of secondary metabolites, and in particular antibiotics, by Streptomyces bacteria is a complex process involving several levels of regulation. Many pleiotropic regulatory genes have been isolated from streptomycetes; in most cases, these genes affect antibiotic biosynthesis by influencing the expression of the pathway-specific regulatory genes (reviewed in reference 11). The latter genes are usually found physically linked to the structural antibiotic biosynthesis genes on the chromosomes of streptomycetes. Both positive and negative regulators directly affecting the expression of structural genes via binding to their promoter regions have been identified in antibiotic biosynthetic gene clusters. In some cases, it has been shown that expression of the pathway-specific regulatory genes is controlled by signaling molecules, such as A-factor, through the action of other regulators encoded by genes located outside of the biosynthetic gene clusters (22). Since antibiotic biosynthesis in Streptomyces spp. is linked to the process of cell differentiation (10), it is likely that expression of most of the pathway-specific regulators depends on some sort of signal transmitted via a complex network. Analysis of the regulatory genes in the antibiotic biosynthetic gene clusters is crucial for understanding the mechanisms of regulation, as well as for designing strategies for the construction of strains with enhanced antibiotic production.
Most of the detailed analyses of pathway-specific regulators described in the literature are concerned with the biosynthesis of nonmacrolide antibiotics such as actinorhodin, undecylprodigiosin, and daunorubicin (1, 32, 39). However, several studies where regulatory genes for macrolide antibiotic biosynthesis were analyzed have also been reported. At least some of these regulators must be rather special, since they control the expression of very large polyketide synthase (PKS)-encoding genes, which implies synthesis of unusually long mRNAs. The transcriptional activator SrmR encoded within the spiramycin biosynthetic gene cluster of Streptomyces ambofaciens has been shown to be required for the transcription of at least one of the PKS genes involved in assembly of the spiramycin macrolactone ring (14). The acyB2-encoded regulator of Streptomyces thermotolerans has been shown to activate the expression of the acyltransferase gene involved in biosynthesis of the macrolide antibiotic carbomycin (2). Five regulatory genes associated with the tylosin biosynthetic gene cluster of Streptomyces fradiae have been found (3). Gene inactivation experiments have confirmed differential roles for two regulators of the SARP (Streptomyces antibiotic regulatory protein) family, TylS and TylT, in controlling tylosin production (4). TylS was shown to control the expression of a global regulator (TylR) for the tylosin cluster, while TylT appeared not to be essential for antibiotic biosynthesis. In a separate report, the transcriptional repressor TylQ was found to play a central role in controlling tylosin biosynthesis in S. fradiae (31).
A detailed genetic analysis of PikD, the positive regulator for the pikromycin biosynthetic gene cluster in Streptomyces venezuelae, has recently been reported (40). PikD belongs to a LAL family of transcriptional regulators containing nucleotide triphosphate (NTP) binding motifs and a C-terminally located helix-turn-helix (HTH) motif of the LuxR type (12). Presumably, these functional features are responsible for the ability of LAL regulators to bind DNA and activate the transcription of target genes upon NTP hydrolysis. It was shown that PikD is required for pikromycin biosynthesis, and the ability of this protein to act as a transcriptional activator depends on the presence of functional NTP binding motifs.
The polyene macrolide antibiotic nystatin produced by Streptomyces noursei ATCC 11455 is widely used in treatments of fungal infections. Brautaset et al. have previously cloned and sequenced the entire nystatin biosynthetic gene cluster and located six putative regulatory genes within its flanking region (7). In the present work, we describe a comprehensive in vivo analysis of these genes by means of their inactivation in S. noursei, determination of targets by use of the xylE reporter system, and cross-complementation experiments.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Bacterial strains, plasmids, and recombinant phages used in this study are listed in Table 1. Some of the plasmids are described below. S. noursei strains were maintained on ISP2 agar medium (Difco, Detroit, Mich.) and grown in liquid Trypticase soy broth (TSB) medium (Oxoid) for DNA isolation. Escherichia coli strains were handled by standard techniques (26). Conjugation from E. coli ET12567(pUZ8002) to S. noursei and gene replacement were performed as described previously (13, 28). Nystatin production was assessed by high-performance liquid chromatography (25) of the dimethylformamide extracts of cultures from 500-ml shake flask fermentations in 100 ml of semidefined SAO-23 medium (28).
TABLE 1.
Bacterial strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Characteristics | Reference or sourcea |
|---|---|---|
| Strains | ||
| S. noursei | ||
| ATCC 11455 | WT strain, nystatin producer | ATCC |
| SR12 | WT with in-frame deletion in nysRI | This work |
| SR34 | WT with in-frame deletion in nysRII | This work |
| SR56 | WT with in-frame deletion in nysRIII | This work |
| NR4K | WT with Kmr insertion in nysRIV | This work |
| NR5D | WT with deletion in orf3 | This work |
| DNR609 | WT with Kmr insertion in orf2 | This work |
| E. coli | ||
| DH5α | General cloning host | BRL |
| JM110 | dam dcm strain | Promega |
| ET12567 (pUZ8002) | Strain for intergeneric conjugation | 13 |
| DASHII phage vector derivatives | ||
| N1 | Recombinant phage (nystatin gene cluster) | 7 |
| N40 | Recombinant phage (nystatin gene cluster) | 7 |
| N58 | Recombinant phage (nystatin gene cluster) | 7 |
| N69 | Recombinant phage (nystatin gene cluster) | 7 |
| N76 | Recombinant phage (nystatin gene cluster) | 7 |
| Recombinant plasmids | ||
| pGEM3Zf(−) | Cloning vector | Promega |
| pGEM7Zf(−) | Cloning vector | Promega |
| pGEM11Zf(−) | Cloning vector | Promega |
| pGEM7ZfErmE*li | pGEM7Zf(−) vector with ermE*p promoter | C. R. Hutchinson |
| pKTO2 | VWB-based Streptomyces integrative vector | 37 |
| pSET152 | E. coli-Streptomyces conjugative vector | 6 |
| pSOK101 | E. coli-Streptomyces conjugative vector | 41 |
| pSOK201 | E. coli-Streptomyces conjugative vector | 41 |
| pSOK804 | 2.3-kb SphI-HindIII fragment from pKTO2 ligated with a 3.0-kb SphI-HindIII fragment from pSET152 | This work |
| pSR12 | Vector for nysRI in-frame deletion | This work |
| pSR34 | Vector for nysRII in-frame deletion | This work |
| pSR56 | Vector for nysRIII in-frame deletion | This work |
| pL58KX5 | 5.0-kb KpnI-XbaI fragment from phage N58 cloned into pGEM3Zf(−) | This work |
| pNR4Km | 1.3-kb Kmr marker inserted into the ClaI site within the nysRIV gene in pL58KX5 after Klenow fill-in | This work |
| pNR4D | 6.3-kb KpnI-SphI fragment from pNR4Km ligated with a 3.0-kb KpnI-SphI fragment from pSOK101 | This work |
| pNR5D | Vector for orf3 inactivation | This work |
| pNR6 | 3.7-kb XmnI-FspI fragment from phage N69 with orf2 cloned into the SmaI site of pGEM3Zf(−) | This work |
| pNR6K | 1.3-kb Kmr marker inserted into the MluI site within orf2 in pNR6 after Klenow fill-in | This work |
| pLDR6K | 5.0-kb EcoRI-XbaI fragment from pNR6K ligated with a 3.0-kb EcoRI-XbaI fragment from pSOK804 | This work |
| pNRE2 | Vector for nysRI expression | This work |
| pC3A1 | Vector for nysRII expression | This work |
| pNR3T | Vector for nysRIII expression | This work |
| pNR4EL | Vector for expression of a 226-aa version of NysRIV | This work |
| pNR4ES | Vector for expression of a 210-aa version of NysRIV | This work |
| pIJ4081 | Vector with promoterless xylE gene | J. White |
| pGEM-XylE1 | pGEM3Zf(−)-based vector with promoterless xylE gene | This work |
| pAML8 | pSOK804-based vector with xylE under the control of nysHp | This work |
| pAML9 | pSOK804-based vector with xylE under the control of nysAp | This work |
| pAML10 | pSOK804-based vector with xylE under the control of nysDIIIp | This work |
| pAML11 | pSOK804-based vector with xylE under the control of nysRIp | This work |
| pAML12 | pSOK804-based vector with xylE under the control of nysIp | This work |
| pAML13 | pSOK804-based vector with xylE under the control of nysDIp | This work |
| pAML14 | pSOK804-based vector with xylE under the control of nysRIVp | This work |
ATCC, American Type Culture Collection; BRL, Bethesda Research Laboratories.
DNA manipulation and sequence analysis.
General techniques for DNA manipulation were used as described elsewhere (16, 26). DNA fragments were isolated from agarose gels with the QIAEX kit (QIAGEN, Hilden, Germany). Southern blot analysis was performed with a DIG High Prime labeling kit (Roche Biochemicals, Mannheim, Germany) according to the manufacturer's manual. Oligonucleotide primers were purchased from Amersham Pharmacia Biotech (Little Chalfont, Buckinghamshire, United Kingdom). Analyses of the amino acid sequences were performed by using the PSORT Prediction (http://psort.nibb.ac.jp/form.html), MEME (http://meme.sdsc.edu/meme/website/meme.html), Pfam (http://www.sanger.ac.uk/Software/Pfam/search.shtml), and MOTIF (http://motif.genome.ad.jp/) search engines.
Construction of plasmids for gene inactivation. (i) nysRI in-frame deletion vector.
A 1.37-kb DNA fragment designated SR1, encompassing the region upstream of nysRI and some of its coding region, was amplified from the phage N58 template by using primers SOS1 (5′-GCAATGAATTCCGTGGCTCG-3′) and SOS2 (5′-GGCTCTAGAGTCAG TAAGCCGGAAGAAC-3′) (restriction enzyme sites are underlined). A 1.50-kb DNA fragment designated SR2, encompassing the 3′ end of nysRI and the downstream region, was amplified from the N58 template by using primers SOS3 (5′-GCCTCTAGAGACCAGGACCGCCACCTCC-3′) and SOS4 (5′-GACAAGCTTCGGTGCTG CGGACGAGTTC-3′). The SR1 and SR2 PCR products were digested with the EcoRI/XbaI and XbaI/HindIII endonucleases, respectively, and ligated together with the 3.0-kb EcoRI-HindIII fragment from pSOK201, yielding the nysRI replacement vector pSR12. The in-frame deletion affecting the nysRI gene within the pSR12 plasmid eliminated the coding sequence for amino acids (aa) 13 to 943 in the NysRI protein, thus affecting all functional features predicted for this polypeptide.
(ii) nysRII in-frame deletion vector.
A 1.43-kb DNA fragment designated SR3, encompassing the region upstream of nysRII and some of its coding region, was amplified from the phage N58 template by using primers SOS5 (5′-GCAGAATTCGAGTCCGTGCTGCTCATCG-3′) and SOS6 (5′-GCACTGCAGGTGGTCGGTTGGTTCC-3′). A 1.52-kb DNA fragment designated SR4, encompassing the 3′ end of nysRII and the downstream region, was amplified from the N58 template by using primers SOS7 (5′-GGCCTGCAGAGCTGTACCTGCTCCTGG-3′) and SOS8 (5′-GACAAGCTTCCTGCCGCACCAACTCGAC-3′). The SR3 and SR4 PCR products were digested with the EcoRI/PstI and PstI/HindIII endonucleases, respectively, and ligated together with the 3.0-kb EcoRI-HindIII fragment from pSOK201, yielding the nysRII replacement vector pSR34. The in-frame deletion affecting the nysRII gene within the pSR34 construct eliminated the coding sequence for aa 14 to 936 in the NysRII protein, thus affecting most of this polypeptide, including the C-terminal HTH domain.
(iii) nysRIII in-frame deletion vector.
A 1.42-kb DNA fragment designated SR5, encompassing the region upstream of nysRIII and some of its coding region, was amplified from the phage N58 template by using primers SOS9 (5′-GACGAATTCAACTGGTCGCGCTGTTCTG-3′) and SOS10 (5′-GACCTGCAGTCAGGAGGAGCGAGGAGTC-3′). A 1.50-kb DNA fragment designated SR6, encompassing the 3′ end of nysRIII and the downstream region, was amplified from the N58 template by using primers SOS11 (5′-GCACTGCAGTGGAGAAGCACCTCACCAG-3′) and SOS12 (5′-GAGAAGCTTGAGTATTCGGAGGCCGCTC-3′). The SR5 and SR6 PCR products were digested with the EcoRI/PstI and PstI/HindIII endonucleases, respectively, and ligated together with the 3.0-kb EcoRI-HindIII fragment from pSOK201, yielding the nysRIII replacement vector pSR56. The in-frame deletion affecting the nysRIII gene within the pSR56 construct eliminated the coding sequence for aa 29 to 899 in the NysRIII protein, thus affecting all functional features predicted for this polypeptide.
(iv) nysRIV and orf2 insertional inactivation.
The plasmids constructed for insertional inactivation of nysRIV and orf2 were designated pNR4K and pLRD6K, respectively (see Table 1 for details).
(v) orf3 “frameshift” deletion.
A 1.3-kb DNA fragment from the S. noursei genome encompassing the 3′ ends of orf3 and orf2 was amplified by PCR with primers NR5D1 (5′-GCGAGCGGCCGCTTCACCCCGCAACTCA-3′) and NR5D2 (5′-CGCGAAGCTTGGCCGACTGCTCGACGTC-3′). The PCR product was digested with NotI and HindIII and then ligated with a 1.7-kb EcoRI-NotI DNA fragment from phage N58 (encompassing nysRIV and the N-terminal part of orf3) and a 3.0-kb EcoRI-HindIII fragment from pSOK201. The resulting plasmid, pNR5D, contained the S. noursei DNA fragment with a 43-bp deletion in the coding region of orf3. This deletion creates a frameshift mutation within the ORF3 coding region, subsequently leading to truncation of its product. As a result of this truncation, 165 C-terminal amino acid residues of orf3 were eliminated and replaced with 14 aa encoded by another reading frame (and thus unrelated to orf3).
Construction of plasmids for expression of regulatory genes from the ermE*p promoter. (i) nysRI expression vector.
A 0.6-kb DNA fragment representing a promoterless 5′ end of nysRI was PCR amplified from the phage N1 template by using primers NR1.1 (5′-CGCCGCATGCTGTTCTCACCCCACGT-3′) and NR1.2 (5′-GGCGCGACCGGTTCGGCCT-3′). The PCR product was digested with SphI/AgeI and then ligated together with a 2.8-kb AgeI-EcoRI DNA fragment from phage N1 into the pGEM7Zf(−) vector digested with SphI/EcoRI. From the resulting construct, a 3.4-kb SphI-HindIII fragment was isolated and ligated together with a 0.3-kb EcoRI-SphI fragment from pGEM7ZfErmE*li, containing the ermE*p promoter, into the EcoRI/HindIII-digested pSOK804 vector (for details, see Results, Table 1, and Fig. 2), resulting in the pNRE2 construct.
FIG. 2.
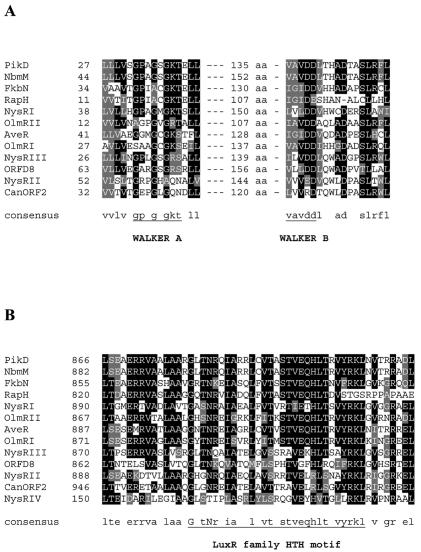
Amino acid sequence alignment. (A) Walker A and B NTP binding motifs in the N termini of LAL-family regulators. (B) LuxR-type HTH DNA binding motifs at the C termini of LAL-family regulators.
(ii) nysRII expression vector.
A 2.2-kb SalI-BclI fragment from phage N58 (representing the 3′ end of nysRII) was cloned into SalI/BamHI-digested pGEM11Zf(−). A 0.8-kb fragment representing the 5′ end of the nysRII gene was PCR amplified from the phage N58 template with primers NSR2.1 (5′-GCCGGCATGCGACGAACAGGACGAGAGGT-3′) and NSR2.3 (5′-GCCGTGGTCGACGAAGG-3′). The PCR fragment was digested with SphI/SalI and then ligated, together with a 2.2-kb SalI-HindIII fragment from the pGEM11Zf(−)-based construct, into the SphI-HindIII-digested pGEM3Zf(−) vector. From the latter, a 3.0-kb SphI-HindIII fragment was isolated and ligated, together with the 0.3-kb EcoRI-SphI ermE*p promoter fragment, into the EcoRI/HindIII-digested pSOK804 vector, resulting in the pC3A1 construct.
(iii) nysRIII expression vector.
A 2.8-kb SacI-NruI fragment from phage N58, encompassing 89 nucleotides (nt) upstream of the nysRIII start codon and a large portion of the coding region, was ligated together with a 0.5-kb NruI-EcoRI fragment from the same phage, representing the 3′ end of this gene, into pGEM3Zf(−). The nysRIII gene was excised from this construct as a 3.2-kb SphI-EcoRI fragment and ligated into pGEM7Zf(−). From the pGEM7Zf(−)-based construct the nysRIII gene was excised as a 3.2-kb SphI-HindIII fragment and ligated together with the 0.3-kb EcoRI-SphI ermE*p promoter fragment into the EcoRI/HindIII-digested pSOK804 vector, resulting in the pNTE3 construct.
(iv) nysRIV expression vectors.
The long (L) and short (S) versions of the nysRIV gene were PCR amplified from N58 recombinant phage DNA with primers NR4P3 (5′-CTCAGCATGCCGAAAGGATGGCG-3′) and NR4P5 (5′-AGGCAAGCTTCGGCGACACGGGCGT-3′) or primers NR4P4 (5′-CTCAGCATGCGTACGACCGGCGGG-3′) and NR4P5, respectively. The corresponding PCR products of 0.78 (NR4L) and 0.73 (NR4S) kb were digested with SphI and HindIII and then ligated, together with the 0.3-kb EcoRI-SphI fragment containing the ermE*p promoter, with the EcoRI-HindIII-digested pSOK804 vector, yielding vectors pNR4EL and pNR4ES, respectively.
PCR amplification of putative promoter regions.
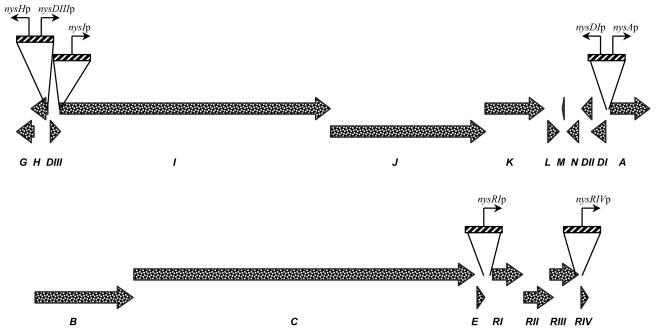
Seven intergenic regions from the nystatin biosynthetic cluster that might contain promoters have been amplified by PCR (see Fig. 4). A 315-bp DNA fragment designated nysHp and containing the region between the nysH and nysDIII genes was amplified from the N40 template by using primers NHP1 (5′-GCAGTCTAGAGAGGAACACCCCGGTTGAC-3′) and NHP2 (5′-GCAGAAGCTTGGCAAACCCTTCTCGAACAC-3′). In PCR a 315-bp intergenic fragment designated nysDIIIp was amplified from the N40 template by using primers ND31 (5′-GCAGTCTAGAGGCAA ACCCTTC TCGAACAC-3′) and ND32 (5′-GCAGAAGCTTGAGGAACACCCCGGTTGAC-3′). A 202-bp fragment encompassing the region between the nysDIII and nysI genes and designated nysIp was amplified from the same template with the help of primers NIP1 (5′-GCCAACTGGTAG CAGTTCTCAAGCTTTCG-3′) and NIP2 (5′-GCGGTCTAGACTCAACTCAACCCATCTCG-3′). The primers for nysAp, the intergenic region upstream of the nysA gene, were NSAP1 (5′-GCAGAAGCTTCGGTTACTTGGTCTCATGC-3′) and NSAP2 (5′-GCAGTCTAGAGCCTTGCTCACCCCTGCGG-3′); the 212-bp PCR product was amplified from the N76 template. A 212-bp fragment encompassing the region upstream of the nysDI gene and designated nysDIp was amplified from the N76 template by using primers ND11 (5′-GCAGTCTAGACGGTTACTTGGTCTCA TGC-3′) and ND12 (5′-GCAGAAGCTTGCCTTGCTCACCCCTGCGG-3′). The 351-bp nysRIp and nysRIVp DNA fragments upstream of the nysRI and nysRIV genes, respectively, were amplified from the N58 template by using primers NR11 (5′-GCAGAAGCTTGAGACGGCACCATGCCAC-3′) and NR12 (5′-GCAGTCTAGACACGCGTTCCTCCACGTG-3′) for the nysRIp fragment and primers NR41 (5′-GCAGAAGCTTGTCGTACGCCCGTCCGG-3′) and NR42 (5′-GCAGTCCAGAGAGACGCGCATCCTTTCGG-3′) for the nysRIVp fragment.
FIG. 4.
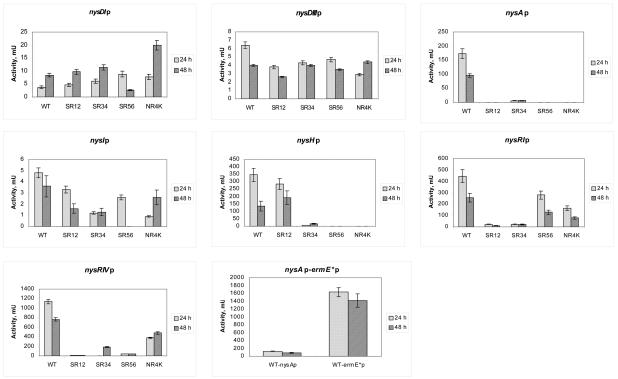
XylE activities in the protein extracts of WT S. noursei and regulatory mutants expressing xylE from different promoters. The last diagram shows a comparative analysis of xylE expression from the nysAp and ermE*p promoters in S. noursei ATCC 11455. Variations from the mean in each data series are represented by error bars.
Construction of xylE-based promoter probe vectors.
PCR-amplified fragments of intergenic regions were digested with the HindIII and XbaI endonucleases and ligated into the pGEM3Zf(−) vector. The 1.5-kb XbaI-BglII fragment with the promoterless xylE gene was excised from the pIJ4081 vector and subcloned into XbaI/BamHI-digested pGEM3Zf(−), resulting in pGEM-XylE1. The HindIII-XbaI promoter-containing fragments from constructs based on pGEM3Zf(−) were ligated, together with the 1.5-kb XbaI-EcoRI fragment containing the reporter gene xylE from pGEM-XylE1, into the HindIII/EcoRI-digested integrative vector pSOK804, resulting in seven constructs (Table 1). Each of the pSOK804-based constructs contains one of the seven intergenic regions upstream of the reporter gene xylE.
The resulting promoter-probe constructs were introduced by conjugation into the nysRI, nysRII, nysRIII, and nysRIV mutants for the XylE assay experiments.
Assay for XylE activity.
For quantitative enzymatic assays, protein extracts from S. noursei cultures were prepared. For the precultures, 20 ml of liquid TSB medium in 250-ml shake-flasks containing 3 g of 3-mm-diameter glass beads was inoculated with spore suspensions and incubated overnight at 30°C with shaking at 250 rpm. On the next day, 50 ml of MP5 medium (containing, per liter, 25 g of glycerol, 3 g of yeast extract, 2 g of NaCl, 0.2 g of K2HPO4, 0.2 g of MgSO4, and 0.02 g of FeSO4 [pH 7.2]) was inoculated with 1.5 ml of precultures and incubated at 30°C for 24 or 48 h with shaking at 250 rpm. Cells were harvested by centrifugation for 10 min at 5,000 rpm (Sorvall), washed with 10 ml of 20 mM phosphate buffer (pH 7.2), and resuspended in 10 ml of sample buffer (100 mM phosphate buffer [pH 7.5]-10% acetone [vol/vol]-20 mM EDTA [pH 8.0]). A 3-ml volume of cell suspension was sonicated for 2 min, and 10 μl of 10% Triton-100 was added per ml of extract. Extracts were placed on ice for 15 min and then centrifuged for 10 min at 15,000 rpm, and cell supernatants were used for XylE assays.
The reaction mixture for measurement of catechol dioxygenase activity consisted of 1.9 ml of assay buffer (100 mM phosphate buffer [pH 7.5], 0.2 mM catechol) preincubated at 37°C for 1 min and 100 ml of cell extract. The optical density at 375 nm was measured over 6 min.
Protein concentrations in extracts were measured according to the Bio-Rad Protein Assay method, by using bovine serum albumin as the standard. The catechol dioxygenase activity was calculated as the rate of change in optical density at 375 nm per minute per milligram of protein.
RESULTS
In silico analysis of putative regulatory genes and their deduced gene products.
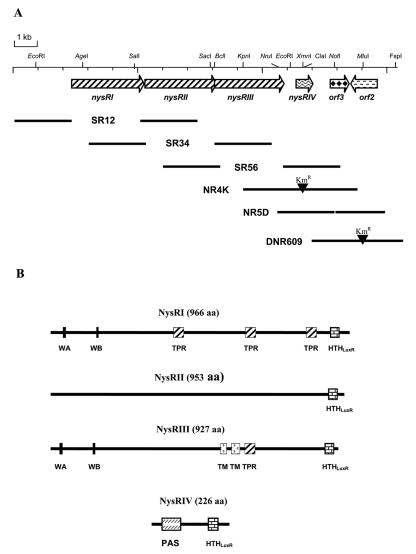
Three genes, designated nysRI, nysRII, and nysRIII, are located downstream of the nysE gene, encoding putative thioesterase, in the nystatin biosynthetic gene cluster of S. noursei ATCC 11455 (7) (Fig. 1A). The nysRIII gene's putative start codon overlaps by 11 nt with the 3′ end of the nysRII gene, suggesting that these two genes might be translationally coupled. Analysis of the deduced primary sequences of the proteins encoded by the nysRI, nysRII, and nysRIII genes revealed their significant similarity to each other and to a number of putative transcriptional activators of the LAL family (12) (data not shown). Several functional features were predicted for the NysRI, NysRII, and NysRIII polypeptides (Fig. 1B). Those include HTH DNA binding motifs of the LuxR type (15) located at the C termini of all three proteins (Fig. 2B) and Walker A and B NTP binding motifs (38) at the N termini of NysRI and NysRIII (Fig. 2A). In addition, two putative transmembrane regions were predicted in the central part of NysRIII, while tetratricopeptide repeats (TPR) (20) were detected in both the NysRI and NysRIII polypeptides (see Discussion).
FIG. 1.
(A) Organization of the regulatory gene locus associated with the nystatin biosynthetic gene cluster in S. noursei ATCC 11455 (GenBank accession number AF263912). The only restriction enzyme sites indicated are those used for vector construction as described in Materials and Methods and Table 1. Heavy solid lines represent DNA fragments used in gene replacement experiments. (B) Putative functional features predicted for the NysRI, NysRII, NysRIII, and NysRIV proteins. WA and WB, Walker A and B NTP binding motifs; PAS, PAS-like domain.
The nysRIV gene, previously designated orf4 (7), is located 404 nt downstream of nysRIII. The start codon for nysRIV has been reassigned, according to a better match of an upstream sequence (AGGA) to the consensus Shine-Dalgarno sequence (30), and is likely to be located 48 nt upstream of the start codon originally proposed (7). Thus, nysRIV presumably encodes a 226-aa rather than a 210-aa protein (see below). A database search showed a high degree of NysRIV sequence identity (63%) to the regulatory protein PteR encoded within the pentaene macrolide antibiotic gene cluster of Streptomyces avermitilis (23). Detailed sequence analysis revealed the presence of a PAS-like domain at the N terminus of NysRIV (33) and a putative C-terminal HTH motif of the LuxR type (Fig. 1B and 2B).
orf3, located downstream of nysRIV, encodes a protein of 253 aa similar to transcriptional repressors of the DeoR family (36). orf2, which is transcribed in the direction opposite that of all the other putative regulatory genes, encodes a 354-aa polypeptide similar to transcriptional regulators of the AsnC type (19).
Inactivations of the regulatory genes and their effects on nystatin biosynthesis.
Disruption of the nysRI gene, described previously, has led to complete elimination of nystatin biosynthesis in the S. noursei mutant NRD2 (7). However, judging from the operon-like organization of the nysRI, nysRII, and nysRIII genes (Fig. 1A), this mutation very likely had a polar effect. In order to determine the individual roles of these three genes in the regulation of nystatin biosynthesis, we constructed in-frame deletion mutants. The deletions were generated via selection of a second crossover event after integration of the pSR12, pSR34, and pSR56 gene replacement vectors (see Materials and Methods) into the genome of the S. noursei wild-type (WT) strain. The resulting mutant strains, SR12, SR34, and SR56 (see Fig. 1A for genotypes), were analyzed for nystatin production.
Mutant SR12 (ΔnysRI) produced nystatin at a severely reduced level, i.e., 0.5% of that in the WT (Table 2). This result confirmed the assumed polar effect of the nysRI disruption in the NDR2 mutant, since not even traces of nystatin could be detected upon fermentation of the latter (7). Nystatin production in mutants SR34 (ΔnysRII) and SR56 (ΔnysRIII) was reduced by 93 and 91%, respectively, compared to that in the WT strain (Table 2).
TABLE 2.
Nystatin production by recombinant S. noursei strains with inactivated or overexpressed regulatory genes
| Strain or background (genotype) | Complementation | Nystatin production (% of WT)a |
|---|---|---|
| SR12 (ΔnysRI) | 0.5 | |
| pNRE2 (ermE*p::nysRI) | 59 | |
| SR34 (ΔnysRII) | 7 | |
| pC3A1 (ermE*p::nysRII) | 100 | |
| SR56 (ΔnysRIII) | 9 | |
| pNRT3 (ermE*p::nysRIII) | 100 | |
| NR4K (nysRIV::Kmr) | 2 | |
| pNR4ES (ermE*p::nysRIV-S) | 2.5 | |
| pNR4EL (ermE*p::nysRIV-L) | 57 | |
| ATCC 11455 (WT) | pSOK804 | 100 |
| pNRE2 (ermE*p::nysRI) | 104 | |
| pC3A1 (ermE*p::nysRII) | 121 | |
| pNRT3 (ermE*p::nysRIII) | 100 | |
| pNR4EL (ermE*p::nysRIV) | 136 |
Values are means from three independent experiments. In general, variations were within 7% of the mean.
The nysRIV gene was inactivated by insertion of the Kmr gene into its coding sequence via a gene replacement procedure using plasmid pNR4D (Table 1; Fig. 1A). The resulting NR4K mutant produced nystatin at a level of ca. 2% of WT production (Table 2).
orf3 and orf2 were inactivated by deletion and disruption with the Kmr cassette by using plasmids pNR5D and pLDR6K, respectively, creating frameshift mutations (see Materials and Methods) (Table 1). Neither the orf3 nor the orf2 mutation had a significant effect on nystatin biosynthesis, suggesting that these genes are not directly involved in the regulation of nystatin biosynthesis, at least under the conditions tested (data not shown).
Complementation of the nysRI, nysRII, nysRIII, and nysRIV mutants by expression of the regulatory genes from the ermE*p promoter.
The pSOK804 plasmid vector, containing an integration function (an integrase gene and AttP) from the streptomycete temperate phage VWB (37) and part of the pSET152 vector (6), was constructed (Table 1). Plasmid pSOK804 was able to integrate site-specifically into one site in the genome of S. noursei (data not shown), at a frequency about 2 orders of magnitude higher than that for the pSET152 vector previously used for gene expression in S. noursei (41). pSOK804-based integration vectors were assembled for the expression of the nysRI, nysRII, nysRIII, and nysRIV genes in S. noursei (see Materials and Methods and Table 1). To circumvent potential problems related to self-regulation of these genes' endogenous promoters, we chose to use the constitutive ermE*p promoter (5) for their expression. Five integrative expression vectors were constructed (see Materials and Methods and Table 1) and used for complementation of the corresponding S. noursei mutants. The results of these experiments are summarized in Table 2. Nystatin synthesis was either partly or fully restored in the SR12, SR34, SR56, and NR4K mutants upon introduction of the vectors expressing the respective regulatory genes, suggesting that the mutations did not have polar effects. Only vector pNR4EL, expressing the longer, 226-aa version of NysRIV, was able to complement NR4K, thus corroborating the new assignment of the nysRIV start codon (see above).
The vectors used in complementation experiments were also introduced into WT S. noursei in order to test whether potential overexpression of the regulators might increase nystatin production. Interestingly, while no effect was observed with nysRI and nysRIII, additional expression of nysRII from ermE*p provided for a 21% increase in nystatin production (Table 2). Expression of nysRIV from the pNR4EL vector in the WT S. noursei strain had the strongest positive effect on nystatin synthesis: the resulting recombinant strain produced nystatin at a level 36% above that of the WT (pSOK804) (Table 2).
Promoter activity studies with the regulatory mutants.
Although definitive roles in controlling nystatin biosynthesis were established for LAL-family regulators and NysRIV by the experiments described above, their individual contributions to the process, as well as the target genes, remained unknown. To address these questions, seven putative promoter regions for the structural and regulatory genes from the nystatin cluster (Fig. 3) were cloned upstream of a promoterless xylE reporter gene (see Materials and Methods). Since we deduced that the nysH-nysDIII and nysDI-nysA intergenic regions contain divergent promoters, these regions were cloned in two alternative orientations to allow the assessment of both promoters. The reporter cassettes were cloned into the pSOK804 integrative vector, and the resulting plasmids were introduced into the S. noursei WT strain and the regulatory mutants. XylE activity assays of crude extracts prepared from the recombinant strains were used to monitor relative expression levels from the various promoters (Fig. 4).
FIG. 3.
Putative promoter regions in the nystatin biosynthesis gene cluster used in these studies.
In the WT background, XylE activity could be detected for all promoter-probe vectors. Data from the XylE assay (Fig. 4) showed that expression from the promoter for the putative mycosamine transferase gene nysDI (and probably for the cotranscribed nysDII and nysN genes) was 2 times higher in the SR56 and NR4K mutants than in the WT background at the 24-h time point. Interestingly, this pattern was changed after 48 h: XylE activity in the SR56 mutant was reduced to ca. 30% of that in the WT, while that in the NR4K mutant continued to increase. nysDIIIp, the promoter for a putative GDP-mannose dehydratase gene, depended only weakly on the regulators: the XylE activity measured for the nysDIIIp::xylE construct in the regulatory mutants was ca. 20 to 50% lower than that in the WT. This difference was most visible at the 24-h time point and was less profound after 48 h.
Expression from the nysAp promoter for the PKS loading module gene nysA (and presumably for the cotranscribed nysB and nysC genes) was very strongly dependent on all four regulators. Essentially no XylE activity was observed in the corresponding protein extracts except for the SR34 mutant, where only very low XylE activity (ca. 3% of the WT level) was detected. Compared to the promoters for mycosamine biosynthesis and attachment genes, nysAp provided for ca. 30-times-higher XylE expression in the WT.
nysIp, the promoter presumably driving the expression of the NysI, NysJ, and NysK PKS proteins, responsible for further elongation and termination of synthesis of the nystatin polyketide chain, showed limited dependence on the presence of the regulators. The strongest effects observed were those in the SR34 and NR4K mutants at 24 h, where XylE activity due to expression from nysIp was ca. 60 to 70% lower than that in the WT background. Interestingly, nysIp seemed to be dependent on the NysRIII regulator at 48 h, while no such trend could be observed when XylE activity in the protein extract from the 24-h culture was measured (Fig. 4). Also, the XylE activity in the NR4K mutant almost reached the level of that in the WT at 48 h.
According to the XylE assay, the promoter for the transporter gene nysH (and presumably for the cotranscribed nysG gene) was essentially independent of NysRI, while its activity was greatly diminished in the nysRII, nysRIII, and nysRIV mutants (see Discussion and Fig. 4).
nysRIp (the promoter region upstream of the regulatory gene nysRI) showed only moderate dependence on the NysRIII and NysRIV regulators, since the XylE activities in the corresponding mutants were diminished by ca. 50 to 60% from that in the WT. At the same time, nysRIp was strongly dependent on NysRI and NysRII (Fig. 4). This result suggested that NysRI regulates its own expression and that NysRII is involved in this process as well. The nysRIp promoter seemed to be very strong and was superseded only by nysRIVp, which provided the highest level of XylE expression demonstrated in these experiments. The activity of the nysRIVp promoter was greatly affected in all three LAL regulatory mutants, while NysRIV seemed to be moderately autoregulating its own expression, as XylE activity in the NR4K mutant was diminished by ca. 60%.
In order to gain more insight into the results of the complementation experiments described in the preceding section, a control experiment designed to assess the efficiency of the ermE*p promoter in S. noursei was performed. In this experiment, XylE activity was measured in protein extracts from the WT strain expressing xylE from the nysAp and ermE*p promoters. Apparently, ermE*p provided for a much more efficient (ca. 12-times-higher) expression of xylE than nysAp (Fig. 4). ermE*p appears to be the strongest of the promoters investigated in this study.
Cross-complementation experiments.
The xylE promoter fusion experiments provided important clues on the target genes controlled by the four NysR regulators in the nystatin gene cluster. However, the possible hierarchy of the regulators remained obscure. In order to gain deeper insight into the mechanism of regulation of nystatin biosynthesis, cross-complementation experiments were carried out. The idea behind these studies was to test which of the regulatory genes, when expressed from the heterologous ermE*p promoter, could substitute for each other in the regulatory mutants.
Accordingly, pSOK804-based expression vectors containing four pathway-specific regulatory genes were introduced into the SR12, SR34, SR56, and NR4K mutants, and nystatin production by recombinant strains was assessed (Table 3). Nystatin production in the SR12 mutant could be restored to the same extent (ca. 60% of the WT level) by introduction of any of the four regulatory genes. Interestingly, both nysRII and nysRIII were able to complement the SR12 mutant, while no cross-complementation was observed between the nysRII and nysRIII genes, suggesting that these regulatory genes can be placed on the same hierarchy level (see Discussion). nysRIV was able to restore nystatin biosynthesis to a significant level (60 to 87% of the WT) in all regulatory mutants.
TABLE 3.
Restoration of nystatin production in cross-complementation experiments with the regulatory mutantsa
| Mutant | Nystatin production (% of WT)b with complementation by:
|
|||
|---|---|---|---|---|
| pNRE2 (ermE*p::nysRI) | pC3A1 (ermE*p::nysRII) | pNRT3 (ermE*p::nysRIII) | pNR4EL (ermE*p::nysRIV) | |
| SR12 (ΔnysRI) | 60 | 58 | 68 | 62 |
| SR34 (ΔnysRII) | 5.4 | 100 | 5.6 | 77 |
| SR56 (ΔnysRIII) | 8.5 | 11 | 98 | 87 |
| NR4K (nysRIV::Kmr) | 2.5 | 2.0 | 2.5 | 60 |
See the text for details.
Values are means from two independent experiments. Variations were within 11% of the mean.
DISCUSSION
The results obtained in the gene inactivation experiments clearly show that at least four regulatory genes control nystatin production in S. noursei. The nysRI, nysRII, nysRIII, and nysRIV genes are required for efficient nystatin biosynthesis and probably represent transcriptional activators for the gene cluster. Clearly, nysRII and nysRIII are not as important in this respect as nysRI, since their inactivation has less profound effects on antibiotic production. This notion is exemplified by the fact that, according to the xylE fusion experiments, NysRI regulates an endogenous promoter for its own gene, which is also likely to drive the expression of nysRII and nysRIII. The operon-like structure of nysRI-nysRII-nysRIII and the apparent polar effect of nysRI disruption imply that these three genes might be transcribed from the same promoter located upstream of nysRI. nysRIp is the second strongest (after nysRIVp) of the nys promoters and remains at least partially active in all regulatory mutants. Both nysRIp and nysRIVp seem to be weaker than the ermeE*p promoter when used for the expression of xylE in WT S. noursei (Fig. 4). This fact, along with the results from the complementation experiments (Table 2), suggests that a high level of expression of the regulatory genes alone does not ensure a high level of nystatin production, since only partial complementation was observed when nysRI and nysRIV were expressed from ermE*p. It seems plausible, therefore, that the mechanism governing gene expression in the nystatin biosynthetic cluster requires coordinated expression of the regulatory genes, which is provided through the intrinsic regulatory genes' promoters.
Remarkably, XylE expression from the nysRIp promoter is strongly reduced only in ΔnysRI and ΔnysRII mutants, suggesting that transcription of the nysRI-nysRII-nysRIII genes is autoregulated. Taking the above into consideration, it is conceivable that in both of these mutants, neither of the regulators is efficiently expressed. It seems, however, that the basal level of expression of NysRII and NysRIII in the ΔnysRI mutant is sufficient to activate the nysHp promoter (Fig. 4). The latter appears to be strongly dependent on the availability of the NysRII, NysRIII, and NysRIV regulators.
The NysRI, NysRII, and NysRIII proteins seem to derive from a common ancestor, since they contain homologous regions and their domain organization, especially for NysRI and NysRIII, is similar (Fig. 1B). Proteins with significant similarity to NysRI, NysRII, and NysRIII are found mostly in actinomycetes and are proposed to be transcriptional activators for antibiotic biosynthesis, lipase, and cholesterol oxidase genes (18, 21, 27, 29). Genes encoding proteins similar to NysRI to -III are also located in the biosynthetic gene cluster for the polyene antibiotic candicidin (9). Most of these proteins might be considered members of the LAL subfamily of transcriptional regulators proposed by De Schrijver and De Mot (12), on the basis of their size and the presence of the N-terminal NTP binding and C-terminal LuxR HTH motifs. The NTP binding motifs in the transcriptional regulator PikD have been shown to be required for its activity (40). Therefore, it seems likely that NysRI and NysRIII also require NTP binding and hydrolysis for their function. The presence of the TPRs, which are implicated in protein-protein interactions (35), in NysRI and NysRIII suggests that these proteins might interact with other proteins. It is well documented that protein-protein interactions play an important role in transcriptional control in bacteria (34).
All experimental data obtained for the nysRIV gene (see below) point to its central role in controlling nystatin biosynthesis. Indeed, very little nystatin is produced by the nysRIV disruption mutant, while expression of nysRIV from the ermE*p promoter results in significant stimulation of nystatin production in the WT strain. Also, nysRIV can complement all nysR regulatory mutants. Detection of a PAS-like domain within NysRIV suggests that this protein might respond to the energy levels in the cell. PAS domains are found in many signaling proteins, where they serve as signal sensor domains (24). Promoter probe studies clearly demonstrate that nysRIVp is strongly downregulated in all three LAL regulator mutants. However, the nysRIVp promoter appears to be the strongest of the seven nys promoters studied, and even in the absence of LAL regulators, its activity is at the level of the nysDIIIp and nysIp promoters. The latter fact suggests that a relatively high level of nysRIV transcription is required for this gene's product to exert a positive effect on nystatin biosynthesis.
The xylE fusion experiments with the promoters for the nystatin structural genes provided the first clues on the regulatory mechanism controlling nystatin biosynthesis in S. noursei. Apparently, promoters driving the expression of nysDIII and nysDI-nysDII-nysN (presumably cotranscribed) are only weakly dependent on the NysR regulators.
Regulation of the promoters driving the expression of PKS genes, nysIp and nysAp, is strikingly different. First, the level of XylE expression from the nysAp promoter in the WT strain seems to be at least 35 times higher than that from nysIp. This is not surprising, since nysA encodes the loading module of the nystatin PKS, expression of which is pivotal for initiation of biosynthesis (8). The mutations in the nysR regulatory genes have much stronger effects on nysAp than on nysIp (see Fig. 4), suggesting that initiation of nystatin biosynthesis promoted by the nysA gene product is the primary target for the regulators.
Cross-complementation experiments helped to establish a hierarchy among the four NysR regulators of the nystatin gene cluster. Differences in the degree of complementation observed in these experiments could most probably be attributed to the expression of the genes in trans from the heterologous promoter. The ability of nysRII and nysRIII to complement the ΔnysRI mutant implies that NysRII and NysRIII can each substitute for NysRI and that expression of these proteins in the SR12 mutant is severely affected. It is not clear, however, why both nysRII and nysRIII can complement the ΔnysRI mutant, since deletion of either of these genes has a detrimental effect on nystatin biosynthesis, and these genes cannot substitute for each other in cross-complementation experiments (Table 3). The answer to this question probably lies in the plausibility of concerted action of the regulators or might be that such complementation is due to the use of a nonnatural constitutive promoter, ermE*p, in the complementation experiments.
Based on the data from promoter analysis and cross-complementation experiments, the following tentative model can be suggested. Expression of the LAL regulatory operon would start with NysRI, which positively regulates its own promoter. However, it seems that NysRII is required for efficient transcription from nysRIp, while NysRIII is not essential (Fig. 4). It is thus logical to assume that NysRI and NysRII function in concert as autoregulators of the LAL operon, ensuring its efficient transcription. NysRII seems to play a pivotal role here, as expression of this protein can alleviate the effect of ΔnysRI mutation. Since no cross-complementation is observed in the case of nysRII and nysRIII, it seems likely that their products are both required for efficient nysRIV expression. Since nysRIV can complement all regulatory mutants, but none of the other regulators can complement NR4K, nysRIV most probably directly controls the expression of nystatin biosynthetic genes.
Acknowledgments
We thank A. M. Lian for conducting initial promoter studies and R. Aune for performing the fermentations. We are also grateful to L. Van Mellaert, C. R. Hutchinson, and J. White for providing vectors pKTO2, pGEM7ZfErmE*Li, and pIJ4081, respectively.
This work was supported by the Research Council of Norway, SINTEF, and Alpharma AS.
REFERENCES
- 1.Arias, P., M. A. Fernandez-Moreno, and F. Malpartida. 1999. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J. Bacteriol. 181:6958-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arisawa, A., N. Kawamura, H. Tsunekawa, K. Okamura, H. Tone, and R. Okamoto. 1993. Cloning and nucleotide sequences of two genes involved in the 4"-O-acylation of macrolide antibiotics from Streptomyces thermotolerans. Biosci. Biotechnol. Biochem. 57:2020-2025. [DOI] [PubMed] [Google Scholar]
- 3.Bate, N., A. R. Butler, A. R. Gandecha, and E. Cundliffe. 1999. Multiple regulatory genes in the tylosin biosynthetic cluster of Streptomyces fradiae. Chem. Biol. 6:617-624. [DOI] [PubMed] [Google Scholar]
- 4.Bate, N., G. Stratigopoulos, and E. Cundliffe. 2002. Differential roles of two SARP-encoding regulatory genes during tylosin biosynthesis. Mol. Microbiol. 43:449-458. [DOI] [PubMed] [Google Scholar]
- 5.Bibb, M. J., J. White, J. M. Ward, and G. R. Janssen. 1994. The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome binding site. Mol. Microbiol. 14:533-545. [DOI] [PubMed] [Google Scholar]
- 6.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 7.Brautaset, T., O. N. Sekurova, H. Sletta, T. E. Ellingsen, A. R. Strøm, S. Valla, and S. B. Zotchev. 2000. Biosynthesis of the polyene antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem. Biol. 7:395-403. [DOI] [PubMed] [Google Scholar]
- 8.Brautaset, T., S. E. F. Borgos, H. Sletta, T. Ellingsen, and S. B. Zotchev. 2003. Site-specific mutagenesis and domain substitutions in the loading module of the nystatin polyketide synthase, and their effects on nystatin biosynthesis in Streptomyces noursei. J. Biol. Chem. 278:14913-14919. [DOI] [PubMed] [Google Scholar]
- 9.Campelo, A. B., and J. A. Gil. 2002. The candicidin gene cluster from Streptomyces griseus IMRU 3570. Microbiology 148:51-59. [DOI] [PubMed] [Google Scholar]
- 10.Champness, W. C., and K. F. Chater. 1994. Regulation and integration of antibiotic production and morphological differentiation in Streptomyces spp., p. 61-93. In P. J. Piggot, C. P. Moran, and P. Youngman (ed.), Regulation of bacterial development. American Society for Microbiology, Washington, D.C.
- 11.Chater, K. F., and M. J. Bibb. 1997. Regulation of bacterial antibiotic production. Bio/Technology 15:57-106. [Google Scholar]
- 12.De Schrijver, A., and R. De Mot. 1999. A subfamily of MalT-related ATP-dependent regulators in the LuxR family. Microbiology 145:1287-1288. [DOI] [PubMed] [Google Scholar]
- 13.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 14.Geistlich, M., R. Losick, J. R. Turner, and R. N. Rao. 1992. Characterization of a novel regulatory gene governing the expression of a polyketide synthase gene in Streptomyces ambofaciens. Mol. Microbiol. 6:2019-2029. [DOI] [PubMed] [Google Scholar]
- 15.Henikoff, S., J. C. Wallace, and J. P. Brown. 1990. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 183:111-132. [DOI] [PubMed] [Google Scholar]
- 16.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces, a laboratory manual. John Innes Foundation, Norwich, United Kingdom.
- 17.Hutchinson, C. R. 1997. Antibiotics from genetically engineered microorganisms, p. 683-702. In W. R. Strohl (ed.), Biotechnology of antibiotics. Marcel Dekker, Inc., New York, N.Y.
- 18.Ikeda, H., T. Nonomiya, M. Usami, T. Ohta, and S. Omura. 1999. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA 96:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koelling, R., and H. Lother. 1985. AsnC: an autogenously regulated activator of asparagine synthetase A transcription in Escherichia coli. J. Bacteriol. 164:310-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marck, C., O. Lefebvre, C. Carles, M. Riva, N. Chaussivert, A. Ruet, and A. Sentenac. 1993. The TFIIIB-assembling subunit of yeast transcription factor TFIIIC has both tetratricopeptide repeats and basic helix-loop-helix motifs. Proc. Natl. Acad. Sci. USA 90:4027-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molnar, I., and Y. Morooka. 1993. Nucleotide sequence analysis of a region upstream of the cholesterol oxidase-cytochrome P450 operon of Streptomyces sp. SA-COO revealing repeating units coding for putative transmembrane and DNA-binding proteins. J. Ferment. Bioeng. 76:257-264. [Google Scholar]
- 22.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 23.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponting, C. P., and L. Aravind. 1997. PAS: a multifunctional domain family comes to light. Curr. Biol. 7:674-677. [DOI] [PubMed] [Google Scholar]
- 25.Raatikainen, O. 1991. High performance liquid chromatography of heptaene polyenes: assay of heptaene produced by Streptomyces griseoviridis. J. Chromatogr. 588:356-360. [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Schwecke, T., J. F. Aparicio, I. Molnar, A. Konig, L. E. Khaw, S. F. Haydock, M. Oliynyk, P. Caffrey, J. Cortes, J. B. Lester, G. A. Bohm, J. Staunton, and P. F. Leadlay. 1995. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc. Natl. Acad. Sci. USA 92:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekurova, O., H. Sletta, T. E. Ellingsen, S. Valla, and S. B. Zotchev. 1999. Molecular cloning and analysis of a pleiotropic regulatory gene locus from the nystatin producer Streptomyces noursei ATCC 11455. FEMS Microbiol. Lett. 177:297-304. [DOI] [PubMed] [Google Scholar]
- 29.Servin-Gonzalez, L., C. Castro, C. Perez, M. Rubio, and F. Valdez. 1997. bldA-dependent expression of the Streptomyces exfoliatus M11 lipase gene (lipA) is mediated by the product of a contiguous gene, lipR, encoding a putative transcriptional activator. J. Bacteriol. 179:7816-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stratigopoulos, G., and E. Cundliffe. 2002. Expression analysis of the tylosin-biosynthetic gene cluster: pivotal regulatory role of the tylQ product. Chem. Biol. 9:71-78. [DOI] [PubMed] [Google Scholar]
- 32.Tang, L., A. Grimm, Y. X. Zhang, and C. R. Hutchinson. 1996. Purification and characterization of the DNA-binding protein DnrI, a transcriptional factor of daunorubicin biosynthesis in Streptomyces peucetius. Mol. Microbiol. 22:801-813. [DOI] [PubMed] [Google Scholar]
- 33.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Titgemeyer, F., and W. Hillen. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie Leeuwenhoek 82:59-71. [PubMed] [Google Scholar]
- 35.Tzamarias, D., and K. Struhl. 1995. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 9:821-831. [DOI] [PubMed] [Google Scholar]
- 36.Valentin-Hansen, P., P. Hoejrup, and S. Short. 1985. The primary structure of the DeoR repressor from Escherichia coli K-12. Nucleic Acids Res. 13:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Mellaert, L., L. Mei, E. Lammertyn, S. Schacht, and J. Anne. 1998. Site-specific integration of bacteriophage VWB genome into Streptomyces venezuelae and construction of a VWB-based integrative vector. Microbiology 144:3351-3358. [DOI] [PubMed] [Google Scholar]
- 38.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White, J., and M. J. Bibb. 1997. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J. Bacteriol. 179:627-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson, D. J., Y. Xue, K. A. Reynolds, and D. H. Sherman. 2001. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 183:3468-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zotchev, S. B., K. Haugan, O. Sekurova, H. Sletta, T. E. Ellingsen, and S. Valla. 2000. Identification and partial characterization of the gene cluster governing biosynthesis of a novel antibacterial polyketide-derived antibiotic in Streptomyces noursei ATCC 11455. Microbiology 146:611-619. [DOI] [PubMed] [Google Scholar]