Abstract
Streptococcus agalactiae is a major cause of invasive infections in human newborns. To satisfy its growth requirements, S. agalactiae takes up 9 of the 20 proteinogenic amino acids from the environment. Defined S. agalactiae mutants in one or several of four putative peptide permease systems were constructed and tested for peptide uptake, growth in various media, and expression of virulence traits. Oligopeptide uptake by S. agalactiae was shown to be mediated by the ABC transporter OppA1-F, which possesses two substrate-binding proteins (OppA1 and OppA2) with overlapping substrate specificities. Dipeptides were found to be taken up in parallel by the oligopeptide permease OppA1-F, by the dipeptide ABC transporter DppA-E, and by the dipeptide symporter DpsA. Reverse transcription-PCR analysis revealed a polycistronic organization of the genes oppA1-F and dppA-E and a monocistronic organization of dpsA in S. agalactiae. The results of quantitative real-time PCR revealed a medium-dependent expression of the operons dppA-E and oppA1-F in S. agalactiae. Growth of S. agalactiae in human amniotic fluid was shown to require an intact dpsA gene, indicating an important role of DpsA during the infection of the amniotic cavity by S. agalactiae. Deletion of the oppB gene reduced the adherence of S. agalactiae to epithelial cells by 26%, impaired its adherence to fibrinogen and fibronectin by 42 and 33%, respectively, and caused a 35% reduction in expression of the fbsA gene, which encodes a fibrinogen-binding protein in S. agalactiae. These data indicate that the oligopeptide permease is involved in modulating virulence traits and virulence gene expression in S. agalactiae.
Streptococcus agalactiae has a limited capacity to synthesize amino acids and must acquire from the environment 9 of the 20 proteinogenic amino acids for growth (38). The amino acid requirements of S. agalactiae can be satisfied by the uptake of peptides, which are cleaved in the cytoplasm to their respective amino acids (69, 70). In microorganisms, two different peptide uptake systems have been described (23, 41). The most common peptide transporters are binding-protein-dependent permeases, consisting of multiple components, belonging to the ATP-binding cassette (ABC) transporter superfamily (24). The process of peptide transport by ABC transporters involves the extracytoplasmic binding of the substrate by a substrate-binding protein, transfer of the substrate to two membrane-integrated permeases for translocation across the cytoplasmic membrane, and ATP hydrolysis by one or two proteins located on the cytoplasmic side of the membrane (24). In numerous bacteria, the uptake of di- and oligopeptides is mediated by these ABC transporters (41). In contrast, some microorganisms take up di- and/or tripeptides by single proteins with similarity to eukaryotic peptide transport proteins of the PRT family (62). These bacterial permeases use the proton motive force across the cytoplasmic membrane for the uptake of peptides (23, 37). They exhibit a broad substrate specificity, but size recognition is restricted to di- and tripeptides only (16). Examples of this kind of peptide symporter are the tripeptide permeases from Escherichia coli and Salmonella enterica serovar Typhimurium (17, 60), and the di- and tripeptide permeases (DtpT) of Lactococcus lactis and Lactobacillus helveticus (23, 37).
In L. lactis, whose peptide uptake systems have been the focus of intense research, di- and oligopeptides are taken up by two distinct ABC transporters, termed DppA-E and OppA-F, respectively (52, 64). Di- and tripeptide uptake is also mediated in L. lactis by the proton-driven permease DtpT (23). In contrast, in Streptococcus pyogenes, dipeptides are taken up exclusively by the ABC transporter system DppA-E (44).
In addition to nutrient uptake, peptide permeases of several organisms are involved in sensing pertinent environmental signals in the form of peptides, culminating in diverse responses such as sporulation (51), chemotaxis (1, 32), conjugation (29), development of competence (4, 5, 26), or expression of virulence determinants (11, 34, 44, 45). Signature-tagged mutagenesis identified the putative dipeptide ABC transporter DppA-E as a requirement for growth and survival of S. agalactiae in a rat sepsis model (28), suggesting an important role of peptides as nutrients or signaling molecules for the virulence of these bacteria.
S. agalactiae is the predominant cause of septicemia, pneumonia, and meningitis in neonates and poses a significant threat to parturient women (8, 54). A common cause of neonatal infection is the colonization of the human rectovaginal tract with S. agalactiae, ascending spread of the bacteria into the amniotic fluid, and aspiration of contaminated amniotic fluid by the infant (8). Although amniotic fluid contains only low amounts of free amino acids (35), it supports growth of S. agalactiae to high concentrations (14, 66), indicating that S. agalactiae satisfies its amino acid requirements in amniotic fluid by the uptake of peptides.
Recently, the genome sequences of the two S. agalactiae strains NEM316 and 2603 V/R were published, thereby allowing a genome-wide search for putative peptide permease genes in S. agalactiae.
The present report describes the characterization of four putative peptide permeases in S. agalactiae. Defined mutants in the putative peptide uptake systems were constructed and used to define the role of these permeases in the uptake of di- and oligopeptides and in the growth of S. agalactiae in various complex media. The transcriptional organization of the peptide permease genes was analyzed in S. agalactiae, and additionally, the expression of these genes was determined during growth in various media. Finally, we addressed the importance of the peptide permeases in the binding of S. agalactiae to host proteins, in the attachment to eukaryotic cells, and in the expression of a virulence gene in S. agalactiae.
MATERIALS AND METHODS
Bacterial strains, eukaryotic cells, and growth conditions.
The S. agalactiae strain O90R (ATCC 12386) is a capsule mutant of the serotype Ia clinical isolate O90. Strain O90R was used for the construction of insertion and deletion mutants in genes encoding putative peptide transporters in S. agalactiae. S. agalactiae strains belonging to the serotypes Ia, Ib, II, III, IV, and V, respectively, are clinical isolates that were described previously (53). S. agalactiae was generally grown in Todd-Hewitt broth supplemented with 1% yeast extract (THY). Recombinant S. agalactiae strains were selected in the presence of erythromycin (5 μg/ml). Growth experiments with S. agalactiae were performed in THY, in fetal calf serum (FCS), and in amniotic fluid. Amniotic fluid was obtained by amniocentesis from a patient suffering from polyhydramnios, and was kindly provided by the children's hospital in Ulm, Germany. Growth of S. agalactiae in THY or FCS was monitored by determining the optical density of the culture at 600 nm (OD600). Due to the turbid nature of amniotic fluid, growth of S. agalactiae in this medium was determined by measuring the viable cell counts of the culture. In addition to growth in complex media, S. agalactiae was also grown in chemically defined medium (CDM) (65). Depleted CDM (dCDM) was prepared without the essential amino acid isoleucine but containing isoleucine-containing di-, tri-, and oligopeptides at a final concentration of 0.1 mg ml−1. The isoleucine-carrying peptides were purchased from Bachem (Bubendorf, Switzerland). The amino acid requirements of different S. agalactiae strains were tested with dCDM lacking, in each case, 1 of the 20 proteinogenic amino acids. Amino acids in the growth medium were detected as their o-phthaldialdehyde derivatives via high-pressure liquid chromatography as described elsewhere (68).
E. coli DH5α was grown in Luria-Bertani medium, and pG+host6- or pTCV-lac-carrying clones were selected with ampicillin (100 μg/ml) or kanamycin (50 μg/ml).
The cell line A549 (ATCC CCL-185) was obtained from the American Type Culture Collection. A549 is a human lung carcinoma cell line which has many characteristics of type I alveolar pneumocytes. A549 cells were propagated in RPMI tissue culture medium (Gibco BRL) supplemented with 10% FCS. Tissue cultures were incubated in a humid atmosphere at 37°C with 5% CO2.
Construction of insertion and deletion mutants in S. agalactiae.
Primers used for the construction of insertion or deletion mutants of S. agalactiae O90R are listed in Table 1. Insertional inactivation of genes in S. agalactiae O90R was performed as described previously (48). Briefly, an internal fragment of the target gene was amplified by PCR and cloned into the thermosensitive plasmid pG+host6. The resulting plasmid was transformed into S. agalactiae according to the method described by Ricci et al. (49), and transformants were selected on erythromycin agar at 30°C. Cells carrying the plasmid within the chromosome were selected at 37°C under erythromycin pressure as described previously (31). The locations of the plasmid insertions were confirmed by Southern blot hybridization. The disruption of the oppA2 gene in S. agalactiae was confirmed by ClaI digestion of genomic DNA of the parental strains and of putative oppA2 integration mutants. Insertional inactivation of the gbs1577 gene was tested with HpaI-digested chromosomal DNA of the parental strains and of putative gbs1577 integration mutants. Digoxigenin-labeled probes of the genes oppA2 and gbs1577 were obtained with the primers 5′-GGATTGGAATGGTTCAAATGG and 5′-AGAGTCCGAATCAGCTGTG and with the oligonucleotides 5′-TTGAAGAGTCTAAAGGTGG and 5′-ACTTATTCCCTGCGAACTC, respectively. Integration mutants were constructed in the S. agalactiae O90R wild type and in S. agalactiae mutants that already carried deletions in the genes oppA1, oppB, dppB, and/or dpsA. The genes dppA, dppB, oppA1, oppB, and/or dpsA were deleted from S. agalactiae according to a procedure described by Schubert et al. (53). In brief, two DNA fragments flanking the target gene were amplified by PCR with the primers listed in Table 1. Due to 21 bases of cDNA in the two primer sets used for the amplification, the resulting PCR products possessed a stretch of 21 identical base pairs. This sequence identity was subsequently used to combine the two PCR products in a crossover PCR. The resultant PCR product consisted of a single DNA fragment that carried the flanking regions of the target gene. The crossover PCR product was cloned into the plasmid pG+host6, and the construct was used to transform S. agalactiae with subsequent erythromycin selection at 30°C. Cells in which the plasmid had integrated into the chromosome were selected at 37°C under erythromycin pressure. Six such integrant strains were serially passaged for 6 days in liquid medium at 30°C without erythromycin selection to facilitate the excision of the pG+host6 construct, leaving the desired target gene deletion in the chromosome. Dilutions of the serially passaged cultures were plated onto agar, and single colonies were tested for erythromycin sensitivity to identify pG+host6 excisants. Successful deletion of the genes oppA1, oppB, dppA, and/or dppB was confirmed by Southern blotting with EcoRI-digested chromosomal DNA of the S. agalactiae parental strains and of the putative deletion mutants. A digoxigenin-labeled probe for the detection of deletions in oppA1 and/or oppB was obtained by PCR with the primers 5′-GTATCTTGATAACAGACG and 5′-GAATACCTACGATGATGCGC. Deletions in the genes dppA and/or dppB were detected with a digoxigenin-labeled probe that had been obtained by PCR with the primers 5′-GTGATACACCGCTACTTC and 5′-CCTTGAGCACCTTTCCCAAC. The successful deletion of the dpsA gene was confirmed by Southern blot analysis with XbaI-digested chromosomal DNA from S. agalactiae and a digoxigenin-labeled probe obtained by PCR with the primers 5′-AACTTTTTGACTGCGTGAC and 5′-TAGGTGCTTTGACCGAGATG. Deletion mutants that had been confirmed by Southern blot analysis were subsequently used for the construction of double and triple mutants in genes encoding other putative peptide transporters.
TABLE 1.
Primers used for targeted deletion or disruption of S. agalactiae genes encoding putative peptide uptake systems
| Target gene | NEM316 gene designation | Type of inactivation | Primers (5′→3′)a | Cloning sites |
|---|---|---|---|---|
| oppA1 | gbs0144 | Deletion | CCGCGGATCCGTCAAGGAAGCAATAGCAGC | BamHI |
| CCCATCCACTAAACTTAAACAGCAACTGAAGCAAGTGTAAGG | ||||
| TGTTTAAGTTTAGTGGATGGGACTCTACTTCCGTAGTGG | ||||
| TGGCACAAGCTTATCATACGTGATACTGGCTG | HindIII | |||
| oppA2 | gbs0966 | Disruption | TGGCACAAGCTTTGAGTCAAAGAACTCTGG | HindIII |
| CCGCGGATCCGGACTTGTCAATTTAAAGG | BamHI | |||
| oppB | gbs0145 | Deletion | CCGCGGATCCAGATGCTGATGTTCCTGC | BamHI |
| CCCATCCACTAAACTTAAACAGCAAGAACTGGCAATTACC | ||||
| TGTTTAAGTTTAGTGGATGGGGCTATCTTGATAACAGACG | ||||
| TGGCACAAGCTTGAATACCTACGATGATGCGC | HindIII | |||
| dppA | gbs0187 | Deletion | GGGGGTACCTCGAGACAGCAATGGCGTGTATC | KpnI |
| CCCATCCACTAAACTTAAACACGCCAGAAACAAAGCAAC | ||||
| TGTTTAAGTTTAGTGGATGGGGACTTGGGATGAATCTGC | ||||
| CCGCGGATCCTCAATCAAGCAATTTCCC | BamHI | |||
| dppB | gbs0188 | Deletion | TGGCACAAGCTTTCAAGGCTAGTAACTGGG | HindIII |
| CCCATCCACTAAACTTAAACAGAGTCGAGGAAACCATTC | ||||
| TGTTTAAGTTTAGTGGATGGGGTGATACACCGCTACTTC | ||||
| CCGCGGATCCCCTTGAGCACCTTTCCCAAC | BamHI | |||
| dpsA | gbs1444 | Deletion | CCGCGGATCCCTACGAGCAAGTACACAGTC | BamHI |
| CCCATCCACTAAACTTAAACACGAAAATAGCACGCATACC | ||||
| TGTTTAAGTTTAGTGGATGGGCTCACCAGTTGGATTATCAG | ||||
| TGGCACAAGCTTCATTGGTACACAGATGG | HindIII | |||
| gbs1577 | gbs1577 | Disruption | GAGCGGGGTACCCTTACGCTTGCTTGGGGAG | KpnI |
| CCGCGGATCCCTTTTCACAACCCATTGAC | BamHI |
Restriction sites are underlined, and regions used for crossover PCR are in boldface type.
RNA preparation, RT-PCR, and LightCycler real-time PCR.
S. agalactiae O90R was grown to mid-log phase in 50 ml of THY, CDM, dCDM plus dipeptide (Ile-Val), or dCDM plus oligopeptide (Val-Tyr-Ile-His-Pro-Phe). Total RNA was prepared from the bacterial culture with the RNeasy midi extraction kit (Qiagen) and treated for 1 h with 150 U of RNase-free DNase (Promega). RNA samples were checked for DNA contamination by PCR without prior reverse transcription (RT). As no amplicons were obtained, DNA contamination during RNA preparation could be excluded. The transcriptional organization of the genes dppA-E, oppA1-F, and dpsA in S. agalactiae was elucidated by RT-PCR analysis with the primers listed in Table 2 by using 1-μg RNA samples prepared from THY-grown culture.
TABLE 2.
Primers used for RT-PCR and for quantitiative real-time PCR to analyze transcriptional organization and expression of genes dppA-E, oppA1-F, and dpsA in S. agalactiae
| Transcriptional region or gene tested | Gene(s) hybridized | First primer (5′→3′) | Second primer (5′→3′) |
|---|---|---|---|
| dpp region | gbs0183-dppA | ATGACTATGGACACCTTG | GCAGGATTAGGTCTAGGG |
| dppA-dppC | CCTTGAGCACCTTTCCCAAC | TAGCGGAGCAAGCTAAAGC | |
| dppB-dppE | CTCGACTTGGGTAATAAG | GTGATACACCGCTACTTC | |
| dppD-gbs0189 | GGTATCATTCTGTCAGAGGC | AAATCTCATGCGTGTGGCAC | |
| opp region | gbs0143-oppA1 | CAGTATAGCGCCTTTTTTC | GCAACTGAAGCAAGTGTAAGG |
| oppA1-oppB | ATCATACGTGATACTGGCTG | ACTCTACTTCCGTAGTGG | |
| oppB-oppC | ACGTTGACGTTCACTCTCC | GCTATCTTGATAACAGACG | |
| oppC-oppF | GATGGACTTCTATGTCTCG | ACGTTGACGTTCACTCTCC | |
| oppF-rDNA | AGCTATTGAGCAAGACGGAC | TCAGACTTCCGTCCATTGC | |
| dpsA region | gbs1512-dpsA | TGGACATTAGACAACTACG | CGAAAATAGCACGCATACC |
| dpsA-dpsA | GGTATGCGTGCTATTTTGC | CTGATAATCCAACTGGTGAG | |
| dpsA-gbs1514 | CTCACCAGTTGGATTATCAG | CATTGGTACACAGATGG | |
| dppA gene | dppA | TAGCGGAGCAGCTAAAGC | GCAGATTCATCCCAAGTC |
| dppB gene | dppB | GATCGAATTGTAAGGTGG | AGAAGTAGCGGTGTATCAC |
| dppE gene | dppE | CTCCTAGATCTGGTGAAG | CCATTGAAAGATACGATACG |
| oppA1 gene | oppA1 | TATGTTGCCCCAGGTTATG | GTTATAAGCGCCTTGCTC |
| oppB gene | oppB | CGCTAGGGATTTCTATGC | CGTCTGTTATCAAGATAGC |
| oppF gene | oppF | CGTGATATCGTAGCAGAAGG | ACGTTGACGTTCACTCTCC |
| dpsA gene | dpsA | GGTATGCGTGCTATTTTGC | CCAATAGCAGCAAGTGAG |
Quantitative real-time PCR experiments were performed after RT of RNA with random hexanucleotides and the RevertAid first-strand cDNA synthesis kit (MBI Fermentas) according to the instructions of the manufacturer. Expression analysis of the genes dppA, dppB, dppE, oppA1, oppB, oppF, and dpsA, respectively, was performed with the primers listed in Table 2 in a LightCycler as described previously (21). The quantity of cDNA for the investigated genes was normalized to the quantity of gyrA cDNA in each sample. Each experiment was performed at least four times with two independent RNA preparations.
Construction of the fbsA-lacZ transcriptional fusion plasmid pTfbsA.
Plasmid pTCV-lac (46) was used for expression analysis of the fbsA gene in S. agalactiae. A DNA fragment of 747 bp containing the fbsA promoter (unpublished data) was amplified by PCR from the genome of S. agalactiae with the primers 5′-CCGGAATTCAACTATTGCTCCCCTGC and 5′-GCCGGATCCTTTCCTGATTTCCAAGTTC. The EcoRI and BamHI sites used for cloning are underlined. After digestion of the PCR product and of plasmid pTCV-lac with EcoRI and BamHI, the fbsA promoter region was ligated into pTCV-lac, resulting in plasmid pTfbsA. The plasmids pTCV-lac and pTfbsA were subsequently transformed into S. agalactiae by electroporation (49), and recombinant clones were obtained by erythromycin selection.
Binding of fluorescein isothiocyanate-labeled group B streptococci to immobilized human proteins.
Terasaki microtiter plates were coated with the human proteins fibrinogen, fibronectin, and haptoglobin, respectively, and the binding of fluorescein isothiocyanate-labeled S. agalactiae to the immobilized fibrinogen was measured as described previously (21). Results were measured in triplicate, and each assay was repeated at least four times.
Adherence and internalization assays.
Adherence of S. agalactiae to A549 epithelial cells and internalization into this cell line were assayed as described previously (21). Briefly, A549 cells were transferred to 24-well tissue culture plates at approximately 4 × 105 cells per well and cultivated overnight in RPMI tissue culture medium supplemented with 10% FCS. After replacement of the medium with 1 ml of fresh medium, the cells were infected with S. agalactiae at a multiplicity of infection of 10:1 and incubated at 37°C for 2 h. The infected cells were subsequently washed three times with phosphate-buffered saline. The number of cell-adherent bacteria was determined by lysis of the eukaryotic cells with distilled water and subsequent determination of CFU by plating appropriate dilutions of the lysates on THY agar. Intracellular bacteria were determined after a further incubation of the infected cells for 2 h with RPMI medium containing penicillin G (10 U) and streptomycin (0.01 mg) to kill extracellular bacteria. After three washes with phosphate-buffered saline, the epithelial cells were lysed with distilled water and the amount of intracellular bacteria was quantitated by plating serial dilutions of the lysate onto THY agar plates. All samples were tested in triplicate, and experiments were repeated at least three times.
Determination of β-galactosidase activity.
Recombinant S. agalactiae strains harboring the plasmid pTCV-lac or pTfbsA were grown aerobically in THY broth at 37°C with shaking (100 rpm). At different time points, 1-ml samples were withdrawn to determine the OD600 and β-galactosidase activity of the culture. Prior to measuring β-galactosidase activity (36), cells were permeabilized with a 0.5% toluene-4.5% ethanol mixture as described previously (46). Enzymatic activity is expressed in Miller units, which were calculated as follows: (103 × OD420)/(time of the reaction [in minutes] × OD600 × dilution of the cells in the assay).
Nucleotide sequence accession number.
The genomic DNA sequence of S. agalactiae NEM316 can be accessed in the EMBL database under accession number AL732656. In the SwissProt TREMBL database, the polypeptides described in this report possess the following accession numbers: DppA, Q8E7H0; DppB, Q8E7G9; DppC, Q8E7G8; DppD, Q8E7G7; DppE, Q8G7G6; OppA1, Q8E7L0; OppA2, Q8E5L7; OppB, Q8E7K9; OppC, Q8E7K8; OppD, Q8E7K7; OppF, Q8E7K6; DpsA, Q8E489.
RESULTS
The genome of S. agalactiae possesses several putative peptide permease genes.
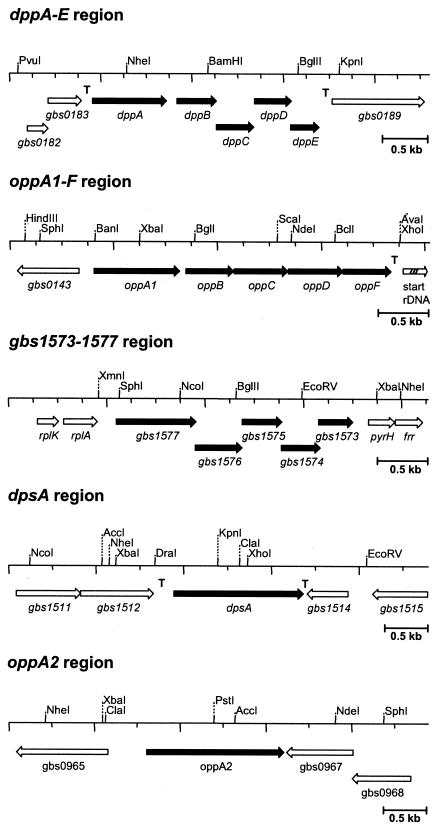
The annotated genome sequence of the S. agalactiae strains NEM316 (http://genolist.pasteur.fr/SagaList/index.html) and 2603 V/R (http://www.pnas.org/cgi/data/182380799/DC1/1) indicated the presence of three ABC transporters and one proton-driven symporter that are possibly involved in the import of peptides into the cell. In the genome of S. agalactiae NEM316, the putative ABC peptide transporters are encoded by the genes gbs0144 to gbs0148, gbs0184 to gbs0188, and gbs1573 to gbs1577. According to the functional analysis of these genes (see below), the S. agalactiae genes gbs0144 to gbs0148 were termed oppA1-F and the genes gbs0184 to gbs0188 were named dppA-E (Fig. 1). As the S. agalactiae genome carries two oppA homologs (63), one carried by gbs0144 and the other carried by gbs0966, the two genes were termed oppA1 and oppA2, respectively. The putative function of each of the gene products of the ABC transporters was inferred based on homology to functionally characterized ABC transporters (24). Thus, the proteins DppA, OppA1, OppA2, and Gbs1577 represent putative substrate-binding proteins, the polypeptides DppB,C, OppB,C, and Gbs1575,1576 represent hydrophobic integral membrane proteins, and DppD,E, OppD,F, and Gbs1573,1574 represent putative ATPases that couple ATP hydrolysis to substrate transport across the membrane.
FIG. 1.
Restriction maps of genomic regions in S. agalactiae encoding putative peptide permeases. According to the functional analysis of these regions, the genes dppA to dppE were shown to encode a dipeptide permease, the genes oppA1 to oppF code for di- and oligopeptide permeases, the dpsA gene encodes a dipeptide symporter, and the oppA2 gene represents a homolog of the oppA1 gene. The gene designations of the other open reading frames were adapted from the genome sequence of S. agalactiae NEM316. The arrow labeled “start rDNA” indicates the start of a ribosomal DNA operon in S. agalactiae. T indicates transcriptional terminators, as identified by RT-PCR analysis.
The genome of S. agalactiae also contains a gene, gbs1513, whose derived polypeptide sequence reveals 49% identity to the di- and tripeptide permease DtpT from L. lactis (23). Functional analysis of gbs1513 in S. agalactiae indicated that it encodes a dipeptide symporter (see below), and therefore, gbs1513 was termed dpsA.
Amino acid auxotrophies of S. agalactiae isolates.
Studying peptide uptake in S. agalactiae requires knowledge regarding amino acid auxotrophies of the strains in use. In a 1943 report, S. agalactiae strains were shown to be auxotrophic for the amino acids valine, leucine, isoleucine, phenylalanine, glutamic acid, arginine, lysine, histidine, and tryptophan (38). We tested 12 different S. agalactiae strains, belonging to six different serotypes, for growth in CDM lacking, in each case, one of the 20 proteinogenic amino acids. All of the S. agalactiae isolates investigated, including strain O90R, were auxotrophic for the amino acids valine, leucine, isoleucine, phenylalanine, tyrosine, arginine, lysine, histidine, and tryptophan. The tested S. agalactiae isolates therefore differ from the previously described strains (38) in that they require tyrosine but not glutamic acid for growth. Analysis of metabolic pathways that can be predicted from the genome sequence (http://www.genome.ad.jp/kegg/metabolism.html) of the S. agalactiae strains NEM316 and 2603 R/V confirmed for these strains the amino auxotrophies identified by the growth experiments. Peptide uptake was further investigated in S. agalactiae strain O90R by making use of its isoleucine auxotrophy.
Identification of the oligopeptide uptake system in S. agalactiae.
Oligopeptide import in S. agalactiae was studied by deleting the genes oppA1 and oppB and by insertionally inactivating the oppA2 gene in the genome of S. agalactiae O90R, resulting in the mutant strains ΔoppA1 ΔoppB and oppA2-int, respectively. Furthermore, a double mutant of the genes oppA1 and oppA2 was constructed and named ΔoppA1 oppA2-int. In THY complex medium, S. agalactiae O90R and its isogenic mutants revealed identical growth rates and final ODs (data not shown), suggesting that the genes oppA1, oppA2, and oppB are dispensable for growth in this medium. The role of these genes for the uptake of peptides was studied by growing strain O90R and its isogenic mutants in various formulations of CDM. All strains grew in complete CDM (Fig. 2) and, as determined by monitoring the OD of the cultures during growth, showed approximately identical growth rates and final ODs (data not shown). When CDM was depleted of the essential amino acid isoleucine (dCDM), there was no apparent growth of the strains (Fig. 2), confirming the isoleucine auxotrophy of S. agalactiae O90R. Addition of the isoleucine-containing dipeptide Ile-Val to dCDM restored growth of all the strains tested (Fig. 2), suggesting that the isoleucine auxotrophy can be supplemented by an isoleucine-containing dipeptide. This finding also indicates that the genes oppA1, oppA2, and oppB are not essential for the uptake of dipeptides. Also demonstrated in Fig. 2, S. agalactiae O90R revealed growth in dCDM with isoleucine-containing tri-, tetra-, and hexapeptides. However, no growth of the mutant ΔoppB was observed in these media (Fig. 2 and Table 3), indicating an essential function of oppB for the uptake of oligopeptides by S. agalactiae. Interestingly, the mutant strain ΔoppA1 and oppA2-int exhibited growth in dCDM with different tri-, tetra-, and hexapeptides while the double mutant ΔoppA1 oppA2-int did not grow on isoleucine-containing oligopeptides (Fig. 2). These data suggest that the substrate-binding proteins OppA1 and OppA2 are both involved in oligopeptide uptake and that they can substitute for each other functionally.
FIG. 2.
Growth of the S. agalactiae wild-type strain O90R and of different oligopeptide permease mutants in CDM or dCDM lacking the amino acid isoleucine. dCDM was supplemented with different isoleucine-containing peptides whose sequences are depicted in single letter code. Growth of the different strains was visually inspected.
TABLE 3.
Growth characteristics of different S. agalactiae strains in complete CDM or dCDMa lacking the amino acid isoleucine
| Strain (genotype) | Growth onb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CDM | dCDM | dCDM supplemented with:
|
|||||||
| Ile-Val | Ile-Glu | Ile-Pro | Ile-Phe | Val-Trp-Ile | Ile-Trp- Val-Asn | Val-Tyr-Ile- His-Pro-Phe | |||
| O90R (wild type) | + | − | + | + | + | + | + | + | + |
| ΔdppA | + | − | + | + | + | + | + | + | + |
| ΔdppB | + | − | + | + | + | + | + | + | + |
| ΔoppB | + | − | + | + | + | + | − | − | − |
| gbs1577-int | + | − | + | NDb | ND | + | + | + | + |
| ΔdpsA | + | − | + | + | + | + | + | + | + |
| ΔdppB gbs1577-int | + | − | + | ND | ND | + | + | + | + |
| ΔoppB gbs1577-int | + | − | + | ND | ND | + | − | − | − |
| ΔdppB ΔdpsA | + | − | + | + | + | + | + | + | + |
| ΔdppB ΔoppB | + | − | + | + | + | + | − | − | − |
| ΔoppB ΔdpsA | + | − | + | + | + | + | − | − | − |
| ΔdppB ΔoppB gbs1577-int | + | − | + | ND | ND | + | + | + | + |
| ΔdppB ΔoppB ΔdpsA | + | − | − | − | − | + | − | − | − |
dCDM was supplemented with the indicated peptides, and growth was visually inspected.
ND, not determined.
Identification of dipeptide import systems in S. agalactiae.
Dipeptide uptake was studied by deleting the genes dppA, dppB, and dpsA and insertionally disrupting the gene gbs1577 in the chromosome of S. agalactiae O90R, resulting in the mutant strains ΔdppA, ΔdppB, ΔdpsA, and gbs1577-int. In THY complex medium, the mutants revealed no difference in growth rate and final OD compared to S. agalactiae O90R (data not shown). All of the mutants, however, exhibited growth in dCDM that contained isoleucine-carrying dipeptides (Table 3), indicating that the import of dipeptides in S. agalactiae is mediated by different uptake systems with overlapping substrate specificities. This hypothesis was tested by constructing S. agalactiae double and triple mutants with different combinations of defects in the genes dppB, oppB, gbs1577, and dpsA. All double mutants revealed growth in dCDM with isoleucine-containing dipeptides (Table 3), suggesting that more than two uptake systems are involved in the import of dipeptides in S. agalactiae. Finally, a triple mutant with deletions in the genes dppB, oppB, and dpsA did not grow in dCDM with the isoleucine-carrying dipeptides Ile-Val, Ile-Glu, and Ile-Pro, respectively. This finding shows that dipeptide uptake is mediated by the oligopeptide permease OppA1-F, the dipeptide permease DppA-E, and the dipeptide symporter DpsA. Additionally, it shows that any of these permeases is sufficient for growth of S. agalactiae with dipeptides as an amino acid source. Our data also indicate that the putative ABC transporter genes gbs1573 to gbs1577 are not required for dipeptide import. However, the triple mutant ΔdppB ΔoppB ΔdpsA was still capable of growth on the dipeptide Ile-Phe, suggesting the presence of at least one more dipeptide uptake system in S. agalactiae. High-pressure liquid chromatography analysis did not detect free isoleucine during growth of S. agalactiae in Ile-Phe-containing dCDM (data not shown), indicating that this dipeptide is not externally hydrolyzed by S. agalactiae.
Transcriptional organization and expression control of dppA-E, oppA1-F, and dpsA.
The transcriptional organization of the genes dppA-E, oppA1-F, and dpsA was investigated by RT-PCR with total RNA from S. agalactiae O90R. As shown in Fig. 3A, RT-PCR amplified the regions dppA to dppC and dppB to dppE from S. agalactiae RNA, suggesting that the genes dppA-E comprise an operon. No amplification products were obtained with primer pairs specific to the genes gbs0183 to dppA and dppD to gbs0189, indicating transcriptional initiation upstream of dppA and transcriptional termination downstream of dppE, respectively. Similarly, RT-PCR amplified the regions oppA1 to oppB, oppB to oppC, and oppC to oppF from O90R RNA (Fig. 3A), suggesting transcriptional coupling of the genes oppA1-F. Here again, no amplification products were obtained with primer pairs specific to the genes gbs0143 to oppA1 and oppD to rDNA, indicating a promoter in front of oppA1 and a transcriptional terminator downstream of oppF. Analysis of the transcriptional organization of dpsA by RT-PCR revealed a dpsA-specific amplicon with dpsA-specific primers (Fig. 3A). No amplification products were obtained by RT-PCR with primers specific to the regions gbs1512 to dpsA and dpsA to gbs1514, indicating a monocistronic organization of the dpsA gene in S. agalactiae. All of the primer pairs used for RT-PCR analysis amplified products of the expected size from chromosomal S. agalactiae DNA (Fig. 3B). As no PCR product was amplified from total S. agalactiae RNA (data not shown), DNA contaminations in the RNA preparation could be excluded.
FIG. 3.
Transcriptional organization of the genes dppA-E, oppA1-F, and dpsA in S. agalactiae. The gene names on top indicate the genes to which the primer pairs annealed during RT-PCR with total RNA (A) or during PCR with chromosomal DNA (B) from S. agalactiae O90R.
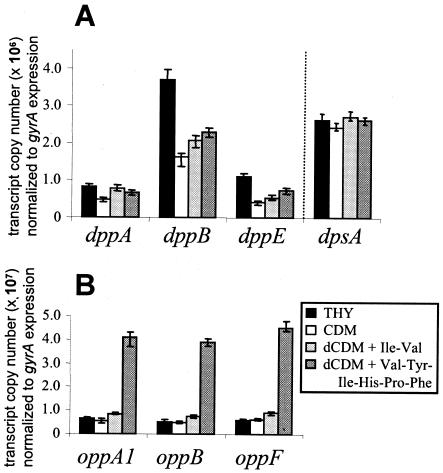
In several organisms, transcriptional attenuation behind dppA or oppA results in an increased amount of dppA- or oppA-specific mRNA compared to the polycistronic mRNA (2, 10, 44). Therefore, quantitative real-time PCR was performed in a LightCycler to determine the ratio of expression between dppA and the downstream genes, dppB and dppE, and between oppA1 and the downstream genes, oppB and oppF. The impact of medium components on the expression of dppA, dppB, dppE, oppA1, oppB, oppF, and dpsA was also studied by quantitative real-time PCR. Expression analysis was performed with total RNA from S. agalactiae O90R after growth in THY, CDM, or dCDM supplemented with the dipeptide Ile-Val or the oligopeptide Val-Tyr-Ile-His-Pro-Phe. Under different growth conditions, the expression of the dppA gene was similar to that of dppE and even weaker than that of dppB (Fig. 4A). Similarly, the amount of oppA1 mRNA was, under different growth conditions, nearly identical to the amount of transcript from the genes oppB and oppF (Fig. 4B). These findings suggest that in the operons dppA-E and oppA1-F in S. agalactiae, transcriptional attenuation does not take place behind the genes dppA and oppA1.
FIG. 4.
Expression analysis of the genes dppA, dppB, dppE, and dpsA (A) and of oppA1, oppB, and oppF (B) during growth of S. agalactiae O90R in THY complex medium, CDM, or dCDM containing the dipeptide Ile-Val or the hexapeptide Val-Tyr-Ile-His-Pro-Phe. The amount of mRNA of the different genes was measured by quantitative real-time PCR in a LightCycler and normalized to the expression of the gyrA gene. The expression of the different genes is presented as the transcript copy number per 1.7 × 106 copies of gyrA transcript. The dotted line indicates distinct transcription of the genes dppA, dppB, dppE, and dpsA. Note the different scales for the transcript copy number in panels A and B.
As depicted in Fig. 4, the expression of the oppA1-F operon significantly exceeded the expression of the dppA-E operon and that of the dpsA gene (note the different scale of the transcript copy number between the graphs), indicating that the oligopeptide permease might be required in higher amounts by the cell. Growth of S. agalactiae under different culture conditions revealed a moderately increased transcription of the genes dppA, dppB, and dppE in THY complex medium. In contrast, expression of the dpsA gene remained constant under the growth conditions tested (Fig. 4A). Interestingly, the transcription of the genes oppA1, oppB, and oppE was about fivefold induced in dCDM containing the hexapeptide Val-Tyr-Ile-His-Pro-Phe (Fig. 4B). However, growth of S. agalactiae in dipeptide-containing dCDM had no effect on the expression of either of the peptide permease-encoding genes. These findings indicate complex regulatory mechanisms that control the expression of peptide permease-encoding genes in S. agalactiae.
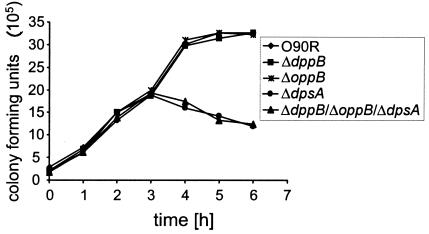
The dpsA gene plays an important role during growth in amniotic fluid.
S. agalactiae is known to infect the placenta and grow in amniotic fluid to high cell densities (15, 50, 66). The importance of peptide uptake for growth of S. agalactiae in amniotic fluid was studied with the strain O90R and the triple mutant ΔdppB ΔoppB ΔdpsA. As shown in Fig. 5, the mutant ΔdppB ΔoppB ΔdpsA grew to significantly lower cell numbers in amniotic fluid than S. agalactiae O90R, suggesting an important role of dipeptide and/or oligopeptide uptake during growth of S. agalactiae in amniotic fluid. To investigate this effect in more detail, growth in amniotic fluid was monitored for the S. agalactiae single mutants ΔdppB, ΔoppB, and ΔdpsA (Fig. 5). Interestingly, the mutant ΔdpsA revealed the same growth defect in amniotic fluid as observed for the triple mutant ΔdppB ΔoppB ΔdpsA, suggesting that the dpsA deficiency caused the growth impairment in amniotic fluid. The different peptide permease mutants and strain O90R were also tested for growth in various complex media. However, the tested strains did not reveal differences in growth rates and final ODs in either THY or FCS (data not shown). These findings suggest that S. agalactiae specifically requires the dipeptide permease DpsA to successfully proliferate in human amniotic fluid.
FIG. 5.
Growth of S. agalactiae O90R and isogenic peptide permease mutants in human amniotic fluid. The growth of the strains was monitored by plating serial dilutions onto THY agar plates and counting the CFU.
The oligopeptide permease is important for the binding of S. agalactiae to host proteins.
In Streptococcus gordonii and Streptococcus pneumoniae, the respective oligopeptide permeases control the interaction of the bacteria with host proteins (11, 27, 34). Therefore, the importance of the three peptide uptake systems for the binding of S. agalactiae to different host proteins was investigated. S. agalactiae strain O90R revealed significant binding to immobilized fibrinogen, fibronectin, and haptoglobin (Table 4). However, the fibrinogen binding of the S. agalactiae mutants ΔoppB and ΔdppB ΔoppB ΔdpsA was reduced by 42%. Similarly, both mutants revealed a 33% reduction in binding to human fibronectin. None of the tested mutants was impaired in its interaction with human haptoglobin. These findings indicate that the oligopeptide permease in S. agalactiae plays a role in the binding of the bacteria to human fibrinogen and fibronectin.
TABLE 4.
Binding of S. agalactiae O90R and isogenic peptide permease mutants to immobilized fibrinogen, fibronectin, and haptoglobina
| Strain | % Bacteria bound to immobilized protein
|
||
|---|---|---|---|
| Fibrinogen | Fibronectin | Haptoglobin | |
| O90R (wild type) | 52 ± 6 | 27 ± 7 | 24 ± 5 |
| ΔdppB | 44 ± 5 | 28 ± 6 | 25 ± 4 |
| ΔdpsA | 43 ± 7 | 25 ± 5 | 22 ± 5 |
| ΔoppA1 | 49 ± 7 | 26 ± 7 | 26 ± 5 |
| ΔoppB | 30 ± 5 | 18 ± 4 | 23 ± 4 |
| ΔdppB ΔdpsA | 42 ± 7 | 28 ± 5 | 27 ± 5 |
| ΔdppB ΔoppB ΔdpsA | 29 ± 4 | 19 ± 4 | 22 ± 5 |
Each assay was performed at least four times in triplicate.
The influence of peptide permeases on the bacterial adherence to and invasion of eukaryotic cells.
In S. pneumoniae, the oligopeptide permease was shown to be required for efficient adherence of the bacteria to human cells (11). The S. agalactiae wild-type strain O90R and the peptide permease mutants ΔdppB, ΔoppB, ΔdppB ΔdpsA, and ΔdppB ΔoppB ΔdpsA were therefore tested for their ability to adhere to and invade cells of the human lung epithelial cell line A549. Among the tested strains, the mutants ΔoppB and ΔdppB ΔoppB ΔdpsA revealed a 26% reduced adherence to host cells. However, none of the mutants exhibited altered invasion into host cells (data not shown), indicating that a deficiency in the oligopeptide permease only moderately reduces adherence of S. agalactiae to human cells.
The oligopeptide permease controls expression of the fbsA gene.
In S. agalactiae, the FbsA protein represents the major fibrinogen-binding protein (53), which also plays an important role in the adherence of the bacteria to host cells (unpublished data). As the previous result demonstrated an influence of the oligopeptide permease on fibrinogen binding and adherence of S. agalactiae to human cells, the expression of the fbsA gene was analyzed in S. agalactiae O90R and its isogenic mutant ΔoppB. Both strains were transformed with the vector pTCV-lac and with the plasmid pTfbsA, which carries the fbsA promoter in front of the promoterless lacZ gene. In pTfbsA-carrying strains, fbsA expression can be monitored by measuring the β-galactosidase activity. Strains that harbored the vector pTCV-lac did not possess β-galactosidase activity during growth (data not shown). However, both pTfbsA-carrying strains revealed a growth-phase-dependent expression of fbsA, peaking in the middle of the exponential growth phase with a moderate decline during late logarithmic and stationary growth (Fig. 6). The obtained results are identical to the expression analysis of fbsA in the S. agalactiae strain 6313-fbsA-luc (21), which carries in its chromosome a promoterless luciferase gene fused to the fbsA gene. This result indicates that plasmid-mediated expression of fbsA does not influence its growth-phase-dependent regulation. Growth experiments revealed identical growth of the S. agalactiae strains O90R(pTfbsA) and ΔoppB(pTfbsA) (Fig. 6). However, in strain ΔoppB(pTfbsA), the fbsA expression was approximately 35% reduced compared to the wild-type strain O90R(pTfbsA), indicating that the oligopeptide permease stimulates the expression of the fbsA gene in S. agalactiae.
FIG. 6.
Growth and expression profiles of an fbsA-lacZ transcriptional fusion in the S. agalactiae strains O90R and ΔoppB. The bacteria were grown aerobically in THY liquid medium, and at various time points, samples were withdrawn for the determination of β-galactosidase activity (A) and OD600 (B) of the culture. β-Galactosidase activity was expressed in Miller units.
DISCUSSION
S. agalactiae is a fastidious organism that requires 9 of the 20 proteinogenic amino acids for growth (reference 38 and the present report). To meet its amino acid requirements, S. agalactiae takes up amino acids in their free or peptide-bound form. Peptides are imported by the cell with the aid of specific peptide permeases and are subsequently cleaved by intracellular peptidases to their single amino acids (41). Four putative peptide permeases with homology to known peptide transport systems have been noted in the genomes of the two sequenced S. agalactiae strains (18, 63). However, none of these putative peptide permeases had been tested for functionality. In the present report, defined mutants in the four putative peptide permease systems were constructed and used to define the roles of these uptake systems in the overall process of peptide utilization and the expression of virulence traits in S. agalactiae.
Feeding experiments with defined S. agalactiae mutants demonstrated that peptide uptake in these bacteria is mediated by two ABC transporters and one putative proton-driven permease, as is also the case for L. lactis and E. coli (23, 41, 52, 60, 64). The ABC transporter Gbs1573-1577, however, appears not to be required for the uptake of peptides in S. agalactiae. As Gbs1573-1577 reveals similarity to peptide transporters but also to Ni2+ permeases (18), it is possibly involved in the uptake of Ni2+. Like other microorganisms (41), S. agalactiae requires an ABC transporter system (OppA1-F) for the import of oligopeptides of two to six residues. In the genome of S. agalactiae, two oppA homologs, termed by us oppA1 and oppA2, have been noted. The presence of two oppA genes readily explains the apparent dispensability of either for the transport of oligopeptides by the oligopeptide permease system. Indeed, the two proteins appear to have overlapping specificities for oligopeptides, as only the simultaneous inactivation of oppA1 and oppA2 abolished transport of oligopeptides. The presence of several oligopeptide binding proteins that serve the same membrane complex is rather common in microorganisms. S. pneumoniae, S. gordonii, and E. coli each possess three genes that encode oligopeptide binding proteins (3, 26, 40). In Borrelia burgdorferi, five oppA homologs have been identified (9). Finally, Enterococcus faecalis produces two peptide binding proteins with different affinities for the peptide sex pheromone cCf10, which is required for conjugative transfer of certain plasmids (29). It has been suggested that, as OppA1 and OppA2 from S. agalactiae are closely related to each other on the amino acid level, they presumably evolved from a common ancestor following gene duplication (63).
Dipeptide import by S. agalactiae appears to be a complex process, involving the two ABC transporters DppA-E and OppA1-F and the putative proton-driven symporter DpsA. DpsA and DppA-E homologs of several microorganisms have been shown to transport both dipeptides and tripeptides (6, 22, 37, 41, 67). The tripeptide Val-Trp-Ile was not transported in oligopeptide permease mutants of S. agalactiae, demonstrating that the dipeptide permeases DppA-E and DpsA are not involved in the transport of this tripeptide. However, this finding does not rule out other tripeptides as substrates for either of the two dipeptide permeases.
Unexpectedly, feeding experiments with S. agalactiae peptide permease mutants demonstrated the uptake of the dipeptide Ile-Phe even in the absence of functional DppA-E, OppA1-F, and DpsA permeases. Cleavage of the dipeptide Ile-Phe by extracellular peptidases is unlikely, as isoleucine was not detectable in Ile-Phe-containing dCDM and other isoleucine-containing dipeptides also were not cleaved by S. agalactiae. Our findings therefore indicate the presence of a further dipeptide permease that mediates the uptake of the dipeptide Ile-Phe in S. agalactiae. Interestingly, L. lactis triple mutants in the two ABC peptide transporters and the proton-driven peptide symporter also still grow on different dipeptides (52). The presence of four different dipeptide permeases is therefore discussed for L. lactis as well.
The presence of different dipeptide uptake systems in S. agalactiae indicates their involvement in the import of different types of dipeptides. However, the S. agalactiae mutants ΔdppB, ΔoppB, and ΔdpsA exhibited unaltered growth in dCDM with different isoleucine-containing dipeptides, suggesting overlapping substrate specificities of the three dipeptide uptake systems. In contrast, mutant ΔdpsA was significantly impaired for growth in human amniotic fluid, whereas the mutants ΔdppB and ΔoppB exhibited identical growth compared to the parental strain O90R. Amniotic fluid has a low concentration of free amino acids (55) but contains significant amounts of proteins and peptides (47, 56). It is therefore tempting to speculate that during growth of S. agalactiae in human amniotic fluid, the permease DpsA plays an essential role in the import of specific dipeptides to satisfy the bacterial amino acid requirements. Similarly, L. lactis also depends on the DpsA ortholog DtpT for growth in a mixture of caseins (59). A frequent cause of S. agalactiae infection is aspiration of infected amniotic fluid by the fetus. Amniotic fluid only has low titers of antibody against S. agalactiae (20) and supports growth of S. agalactiae to high cell densities (14, 66). As the present study demonstrates an important role of DpsA for growth of S. agalactiae in amniotic fluid, DpsA represents an interesting target for the prevention or treatment of in utero S. agalactiae infections.
In several organisms, the expression of genes encoding peptide permeases is induced by the presence of peptides (7, 25, 44) or repressed in the absence of substrate (39, 57). As shown by quantitative real-time PCR, the transcription of the S. agalactiae genes dppA to dppE was not affected in di- or hexapeptide-containing dCDM. However, the expression of the dppA-E operon increased about twofold during growth of the bacteria in THY medium, which is a complex mixture of free amino acids, dipeptides, and oligopeptides. This indicates that components in THY medium, putatively peptides, cause an induction of dppA-E gene expression in S. agalactiae. In contrast, the expression of the dppA gene in E. coli is repressed during growth of the bacteria in a peptide-rich complex medium (39). In S. pyogenes as well, the transcription of the dppA-E operon is down-regulated during growth of the bacteria in THY medium (44). This suggests different regulatory mechanisms in expression control of the dppA-E operon among these microorganisms. As demonstrated by our studies, the expression of the oppA1-F operon was induced about eightfold in dCDM containing the hexapeptide Val-Tyr-Ile-His-Pro-Phe. However, no induction of oppA1-F expression was observed during growth of S. agalactiae in THY complex medium, which also contains dipeptides and free amino acids in addition to oligopeptides. It can therefore be speculated that in S. agalactiae the transcription of the oppA1-F operon is repressed by the presence of free amino acids and/or dipeptides in the medium. In L. lactis, the expression of the oppA gene is also significantly repressed during growth of the bacteria in a complex medium that is rich in free amino acids and peptides (13), indicating a similar transcription control of the oppA-F operons in these bacteria. In our studies, the expression of the dpsA gene was not altered during growth of the bacteria in different media, whereas in L. lactis, transcription of the dpsA ortholog dtpT is regulated by the peptide content of the medium (22). Taken together, our findings suggest that the transcriptional regulation of genes encoding peptide permeases in S. agalactiae is complex and partially distinct from the regulatory mechanisms of peptide permease-encoding genes in other microorganisms.
In several gram-positive bacteria, peptide uptake systems are not only essential for nutrient accumulation but they are also involved in determining other cellular functions, including virulence mechanisms. In pneumococci and S. pyogenes, the oligopeptide permease modulates the adherence of the bacteria to human host cells (11, 12). As revealed by our studies, the oligopeptide permease from S. agalactiae also stimulates the adherence of the bacteria to human epithelial cells and modulates their binding to fibrinogen and fibronectin. In addition, the oligopeptide permease was shown to control the expression of the fbsA gene, which encodes a fibrinogen-binding adhesin. Similarly, the oligopeptide permease from S. gordonii modulates the expression of the cshA gene, encoding a 259-kDa protein that mediates the binding of the bacteria to immobilized fibronectin and to other microorganisms (33, 34). In S. pyogenes, the oligopeptide and the dipeptide permease both control the transcription of the speB gene, encoding a cysteine protease which is important for the virulence of the bacteria (30, 44, 45). Furthermore, in Bacillus cereus and Bacillus thuringiensis, the oligopeptide permease controls the expression of the plcR regulon, which is essential for the virulence of these bacteria (19, 58). As demonstrated by these examples, oligopeptide permeases are involved in the expression control of virulence genes in various organisms. At present, the molecular basis for the oligopeptide permease-dependent regulation of fbsA expression in S. agalactiae remains unknown. However, a putative quorum-sensing system was recently identified in S. agalactiae O90R (61). Interestingly, the inactivation of this quorum-sensing system also changed the fibrinogen-binding properties of the resultant mutant. As quorum sensing in gram-positive bacteria is mediated by small peptides, it is tempting to speculate that in S. agalactiae the oligopeptide permease is involved in the quorum-sensing-dependent regulation of fbsA expression. In fact, in several gram-positive bacteria, oligopeptide permeases are involved in quorum-sensing-dependent gene regulation and control cellular properties like sporulation in Bacillus subtilis (43, 51), development of competence in B. subtilis and S. pneumoniae (5, 42), and conjugation in E. faecalis (29). All these studies point to an involvement of peptide permeases in the transport of signaling molecules that in turn modulate gene expression by interacting with transcriptional regulators. The precise mechanism of oligopeptide permease-dependent gene regulation in S. agalactiae and the full complement of molecules that participate in this signaling process remain to be identified. Studies are therefore under way to unravel peptides with signaling function in S. agalactiae and to identify further genes revealing an oligopeptide permease-dependent expression.
REFERENCES
- 1.Abouhamad, W. N., M. Manson, M. M. Gibson, and C. F. Higgins. 1991. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol. Microbiol. 5:1035-1047. [DOI] [PubMed] [Google Scholar]
- 2.Abouhamad, W. N., and M. D. Manson. 1994. The dipeptide permease of Escherichia coli closely resembles other bacterial transport systems and shows growth-phase-dependent expression. Mol. Microbiol. 14:1077-1092. [DOI] [PubMed] [Google Scholar]
- 3.Alloing, G., P. de Philip, and J. P. Claverys. 1994. Three highly homologous membrane-bound lipoproteins participate in oligopeptide transport by the Ami system of the gram-positive Streptococcus pneumoniae. J. Mol. Biol. 241:44-58. [DOI] [PubMed] [Google Scholar]
- 4.Alloing, G., C. Granadel, D. A. Morrison, and J. P. Claverys. 1996. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol. Microbiol. 21:471-478. [DOI] [PubMed] [Google Scholar]
- 5.Alloing, G., B. Martin, C. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29:75-83. [DOI] [PubMed] [Google Scholar]
- 6.Alves, R. A., and J. W. Payne. 1980. The number and nature of the peptide-transport systems of Escherichia coli: characterization of specific transport mutants. Biochem. Soc. Trans. 8:704-705. [DOI] [PubMed] [Google Scholar]
- 7.Andrews, J. C., T. C. Blevins, and S. A. Short. 1986. Regulation of peptide transport in Escherichia coli: induction of the trp-linked operon encoding the oligopeptide permease. J. Bacteriol. 165:428-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker, C. J., and M. S. Edwards. 1995. Group B streptococcal infections, p. 980-1054. In J. S. Remington and J. O. Klein (ed.), Infectious disease of the fetus and newborn infant. W. B. Saunders Company, Philadelphia, Pa.
- 9.Bono, J. L., K. Tilly, B. Stevenson, D. Hogan, and P. Rosa. 1998. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology 144:1033-1044. [DOI] [PubMed] [Google Scholar]
- 10.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cundell, D. R., B. J. Pearce, J. Sandros, A. M. Naughton, and H. R. Masure. 1995. Peptide permeases from Streptococcus pneumoniae affect adherence to eucaryotic cells. Infect. Immun. 63:2493-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darmstadt, G. L., L. Mentele, A. Podbielski, and C. E. Rubens. 2000. Role of group A streptococcal virulence factors in adherence to keratinocytes. Infect. Immun. 68:1215-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Detmers, F. J., E. R. Kunji, F. C. Lanfermeijer, B. Poolman, and W. N. Konings. 1998. Kinetics and specificity of peptide uptake by the oligopeptide transport system of Lactococcus lactis. Biochemistry 37:16671-16679. [DOI] [PubMed] [Google Scholar]
- 14.Eickhoff, T. C., J. O. Klein, K. L. Daly, D. I. Ingall, and M. Finland. 1964. Neonatal sepsis and other infections due to group B beta-hemolytic streptococci. N. Engl. J. Med. 271:1221-1228. [DOI] [PubMed] [Google Scholar]
- 15.Eidelman, A. I., A. Nevet, B. Rudensky, R. Rabinowitz, C. Hammerman, D. Raveh, and M. S. Schimmel. 2002. The effect of meconium staining of amniotic fluid on the growth of Escherichia coli and group B streptococcus. J. Perinatol. 22:467-471. [DOI] [PubMed] [Google Scholar]
- 16.Fang, G., W. N. Konings, and B. Poolman. 2000. Kinetics and substrate specificity of membrane-reconstituted peptide transporter DtpT of Lactococcus lactis. J. Bacteriol. 182:2530-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson, M. M., M. Price, and C. F. Higgins. 1984. Genetic characterization and molecular cloning of the tripeptide permease (tpp) genes of Salmonella typhimurium. J. Bacteriol. 160:122-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 19.Gominet, M., L. Slamti, N. Gilois, M. Rose, and D. Lereclus. 2001. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 40:963-975. [DOI] [PubMed] [Google Scholar]
- 20.Gray, B. M., J. D. Springfield, and H. C. Dillon, Jr. 1987. Type-specific streptococcal antibodies in amniotic fluid. Am. J. Obstet. Gynecol. 156:666-669. [DOI] [PubMed] [Google Scholar]
- 21.Gutekunst, H., B. E. Eikmanns, and D. J. Reinscheid. 2003. Analysis of RogB-controlled virulence mechanisms and gene expression in Streptococcus agalactiae. Infect. Immun. 71:5056-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagting, A., J. Knol, B. Hasemeier, M. R. Streutker, G. Fang, B. Poolman, and W. N. Konings. 1997. Amplified expression, purification and functional reconstitution of the dipeptide and tripeptide transport protein of Lactococcus lactis. Eur. J. Biochem. 247:581-587. [DOI] [PubMed] [Google Scholar]
- 23.Hagting, A., E. R. Kunji, K. J. Leenhouts, B. Poolman, and W. N. Konings. 1994. The di- and tripeptide transport protein of Lactococcus lactis. A new type of bacterial peptide transporter. J. Biol. Chem. 269:11391-11399. [PubMed] [Google Scholar]
- 24.Higgins, C. F. 1992. ABC transporters: from microorganism to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 25.Jamieson, D. J., and C. F. Higgins. 1984. Anaerobic and leucine-dependent expression of a peptide transport gene in Salmonella typhimurium. J. Bacteriol. 160:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkinson, H. F., R. A. Baker, and G. W. Tannock. 1996. A binding-lipoprotein-dependent oligopeptide transport system in Streptococcus gordonii essential for uptake of hexa- and heptapeptides. J. Bacteriol. 178:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenkinson, H. F., and R. A. Easingwood. 1990. Insertional inactivation of the gene encoding a 76-kilodalton cell surface polypeptide in Streptococcus gordonii Challis has a pleiotropic effect on cell surface composition and properties. Infect. Immun. 58:3689-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, A. L., K. M. Knoll, and C. E. Rubens. 2000. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 37:1444-1455. [DOI] [PubMed] [Google Scholar]
- 29.Leonard, B. A., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukomski, S., C. A. Montgomery, J. Rurangirwa, R. S. Geske, J. P. Barrish, G. J. Adams, and J. M. Musser. 1999. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect. Immun. 67:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maguin, E., H. Prevost, S. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manson, M. D., V. Blank, G. Brade, and C. F. Higgins. 1986. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 321:253-256. [DOI] [PubMed] [Google Scholar]
- 33.McNab, R., H. Forbes, P. S. Handley, D. M. Loach, G. W. Tannock, and H. F. Jenkinson. 1999. Cell wall-anchored CshA polypeptide (259 kilodaltons) in Streptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J. Bacteriol. 181:3087-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNab, R., and H. F. Jenkinson. 1998. Altered adherence properties of a Streptococcus gordonii hppA (oligopeptide permease) mutant result from transcriptional effects on cshA adhesin gene expression. Microbiology 144:127-136. [DOI] [PubMed] [Google Scholar]
- 35.Mesavage, W. C., S. F. Suchy, D. L. Weiner, C. S. Nance, D. B. Flannery, and B. Wolf. 1985. Amino acids in amniotic fluid in the second trimester of gestation. Pediatr. Res. 19:1021-1024. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Nakajima, H., A. Hagting, E. R. Kunji, B. Poolman, and W. N. Konings. 1997. Cloning and functional expression in Escherichia coli of the gene encoding the di- and tripeptide transport protein of Lactobacillus helveticus. Appl. Environ. Microbiol. 63:2213-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niven, C. F. 1943. The nutrition of group B streptococcus. J. Bacteriol. 46:573-574. [Google Scholar]
- 39.Olson, E. R., D. S. Dunyak, L. M. Jurss, and R. A. Poorman. 1991. Identification and characterization of dppA, an Escherichia coli gene encoding a periplasmic dipeptide transport protein. J. Bacteriol. 173:234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, J. T., D. Raychaudhuri, H. Li, S. Normark, and D. Mengin-Lecreulx. 1998. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide l-alanyl-γ-d-glutamyl-meso-diaminopimelate. J. Bacteriol. 180:1215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Payne, J. W., and M. W. Smith. 1994. Peptide transport by micro-organisms. Adv. Microb. Physiol. 36:1-80. [DOI] [PubMed] [Google Scholar]
- 42.Pearce, B. J., A. M. Naughton, and H. R. Masure. 1994. Peptide permeases modulate transformation in Streptococcus pneumoniae. Mol. Microbiol. 12:881-892. [DOI] [PubMed] [Google Scholar]
- 43.Perego, M., C. F. Higgins, S. R. Pearce, M. P. Gallagher, and J. A. Hoch. 1991. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol. Microbiol. 5:173-185. [DOI] [PubMed] [Google Scholar]
- 44.Podbielski, A., and B. A. Leonard. 1998. The group A streptococcal dipeptide permease (Dpp) is involved in the uptake of essential amino acids and affects the expression of cysteine protease. Mol. Microbiol. 28:1323-1334. [DOI] [PubMed] [Google Scholar]
- 45.Podbielski, A., B. Pohl, M. Woischnik, C. Korner, K. H. Schmidt, E. Rozdzinski, and B. A. Leonard. 1996. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (opp) and its effect on cysteine protease production. Mol. Microbiol. 21:1087-1099. [DOI] [PubMed] [Google Scholar]
- 46.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193-198. [DOI] [PubMed] [Google Scholar]
- 47.Queenan, J. T. 1971. Amniotic fluid analysis. Clin. Obstet. Gynecol. 14:505-536. [DOI] [PubMed] [Google Scholar]
- 48.Reinscheid, D. J., B. Gottschalk, A. Schubert, B. J. Eikmanns, and G. S. Chhatwal. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J. Bacteriol. 183:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ricci, M. L., R. Manganelli, C. Berneri, G. Orefici, and G. Pozzi. 1994. Electrotransformation of Streptococcus agalactiae with plasmid DNA. FEMS Microbiol. Lett. 119:47-52. [DOI] [PubMed] [Google Scholar]
- 50.Rubens, C. E., H. V. Raff, J. C. Jackson, E. Y. Chi, J. T. Bielitzki, and S. L. Hillier. 1991. Pathophysiology and histopathology of group B streptococcal sepsis in Macaca nemestrina primates induced after intraamniotic inoculation: evidence for bacterial cellular invasion. J. Infect. Dis. 164:320-330. [DOI] [PubMed] [Google Scholar]
- 51.Rudner, D. Z., J. R. LeDeaux, K. Ireton, and A. D. Grossman. 1991. The spo0K locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J. Bacteriol. 173:1388-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanz, Y., F. C. Lanfermeijer, P. Renault, A. Bolotin, W. N. Konings, and B. Poolman. 2001. Genetic and functional characterization of dpp genes encoding a dipeptide transport system in Lactococcus lactis. Arch. Microbiol. 175:334-343. [DOI] [PubMed] [Google Scholar]
- 53.Schubert, A., K. Zakikhany, M. Schreiner, R. Frank, B. Spellerberg, B. E. Eikmanns, and D. J. Reinscheid. 2002. A fibrinogen receptor from group B streptococcus interacts with fibrinogen by repetitive units with novel ligand binding sites. Mol. Microbiol. 46:557-569. [DOI] [PubMed] [Google Scholar]
- 54.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schulman, J. D., J. T. Queenan, and L. Doores. 1972. Gas chromatographic analysis of concentrations of amino acids in amniotic fluid from early, middle and late periods of human gestation. Am. J. Obstet. Gynecol. 114:243-249. [DOI] [PubMed] [Google Scholar]
- 56.Schulman, J. D., J. T. Queenan, L. Doores, and B. Holland. 1973. Molecular weight distribution of amniotic fluid proteins during normal gestation. Obstet. Gynecol. 42:344-348. [PubMed] [Google Scholar]
- 57.Slack, F. J., J. P. Mueller, M. A. Strauch, C. Mathiopoulos, and A. L. Sonenshein. 1991. Transcriptional regulation of a Bacillus subtilis dipeptide transport operon. Mol. Microbiol. 5:1915-1925. [DOI] [PubMed] [Google Scholar]
- 58.Slamti, L., and D. Lereclus. 2002. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 21:4550-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smid, E. J., R. Plapp, and W. N. Konings. 1989. Peptide uptake is essential for growth of Lactococcus lactis on the milk protein casein. J. Bacteriol. 171:6135-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith, M. W., D. R. Tyreman, G. M. Payne, N. J. Marshall, and J. W. Payne. 1999. Substrate specificity of the periplasmic dipeptide-binding protein from Escherichia coli: experimental basis for the design of peptide prodrugs. Microbiology 145:2891-2901. [DOI] [PubMed] [Google Scholar]
- 61.Spellerberg, B., E. Rozdzinski, S. Martin, J. Weber-Heynemann, and R. Lutticken. 2002. rgf encodes a novel two-component signal transduction system of Streptococcus agalactiae. Infect. Immun. 70:2434-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steiner, H. Y., F. Naider, and J. M. Becker. 1995. The PTR family: a new group of peptide transporters. Mol. Microbiol. 16:825-834. [DOI] [PubMed] [Google Scholar]
- 63.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tynkkynen, S., G. Buist, E. Kunji, J. Kok, B. Poolman, G. Venema, and A. Haandrikman. 1993. Genetic and biochemical characterization of the oligopeptide transport system of Lactococcus lactis. J. Bacteriol. 175:7523-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van de Rijn, I., and R. E. Kessler. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27:444-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varner, M. W., J. W. Turner, C. R. Petzold, and R. P. Galask. 1985. Ultrastructural alterations of term human amniotic epithelium following incubation with group B beta-hemolytic streptococci. Am. J. Reprod. Immunol. Microbiol. 8:27-32. [DOI] [PubMed] [Google Scholar]
- 67.Verheul, A., A. Hagting, M. R. Amezaga, I. R. Booth, F. M. Rombouts, and T. Abee. 1995. A di- and tripeptide transport system can supply Listeria monocytogenes Scott A with amino acids essential for growth. Appl. Environ. Microbiol. 61:226-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vrljic, M., J. Garg, A. Bellmann, S. Wachi, R. Freudl, M. J. Malecki, H. Sahm, V. J. Kozina, L. Eggeling, M. H. Saier, Jr., L. Eggeling, and M. H. Saier, Jr. 1999. The LysE superfamily: topology of the lysine exporter LysE of Corynebacterium glutamicum, a paradyme for a novel superfamily of transmembrane solute translocators. J. Mol. Microbiol. Biotechnol. 1:327-336. [PubMed] [Google Scholar]
- 69.Willett, N. P., and G. E. Morse. 1966. Long-chain fatty acid inhibition of growth of Streptococcus agalactiae in a chemically defined medium. J. Bacteriol. 91:2245-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willett, N. P., G. E. Morse, and S. A. Carlisle. 1967. Requirements for growth of Streptococcus agalactiae in a chemically defined medium. J. Bacteriol. 94:1247-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]