Abstract
Background
Otd-related transcription factors are evolutionarily conserved to control anterior patterning and neurogenesis. In humans, two such factors, OTX2 and CRX, are expressed in all photoreceptors from early specification through adulthood and associate with several photoreceptor-specific retinopathies. It is not well understood how these factors function independently vs. redundantly, or how specific mutations lead to different disease outcomes. It is also unclear how OTX1 and OTX2 functionally overlap during other aspects of neurogenesis and ocular development. Drosophila encodes a single Otd factor that has multiple functions during eye development. Using the Drosophila eye as a model, we tested the ability of the human OTX1, OTX2, and CRX genes, as well as several disease-associated CRX alleles, to rescue the different functions of Otd.
Results
Our results indicate the following: OTX2 and CRX display overlapping, yet distinct subfunctions of Otd during photoreceptor differentiation; CRX disease alleles can be functionally distinguished based on their rescue properties; and all three factors are able to rescue rhabdomeric photoreceptor morphogenesis.
Conclusions
Our findings have important implications for understanding how Otx proteins have subfunctionalized during evolution, and cement Drosophila as an effective tool to unravel the molecular bases of photoreceptor pathogenesis.
Keywords: rhodopsin, orthodenticle, eye, sense organ, congenital disease, retina
Introduction
Irreversible photoreceptor loss in multiple retinopathies is a leading cause of blindness in the developed world. These inherited retinal degenerative diseases are clinically heterogeneous, differing by their time of onset and the photoreceptor population targeted. Leber’s Congenital Amaurosis (LCA), for instance, disrupts the development of rod and cone photoreceptors, causing blindness in early childhood, whereas Cone-Rod Dystrophy (CORD) and Retinitis Pigmentosa (RP) are later-onset neurodegenerative diseases primarily affecting mature cones and rods, respectively. Although more than 200 genes have now been associated with retinal diseases (www.retnet.org), the mechanisms of pathogenesis are often not understood. Two Otd-related homeobox (OTX) transcription factors, OTX2 and CRX, are expressed in all rods and cones from their early specification throughout adulthood, and are important for regulating a wide range of photoreceptor-specific genes (Chen et al., 1997; Furukawa et al., 1997; Nishida et al., 2003; Koike et al., 2007; Hennig et al., 2008; Corbo et al., 2010; Omori et al., 2011). Consistent with such functions, mutations in both OTX2 and CRX can lead to LCA (Freund et al., 1998; Jacobson et al., 1998; Sohocki et al., 1998; Swaroop et al., 1999; Rivolta et al., 2001; den Hollander et al., 2008; Henderson et al., 2009; Nichols et al., 2010). However, identical mutations in CRX associated with LCA are also linked to progressive vision loss in CORD and RP (Freund et al., 1997; Swain et al., 1997; Freund et al., 1998; Sohocki et al., 1998; Swaroop et al., 1999; Rivolta et al., 2001), whereas LCA-associated alleles of OTX2 are also associated with more severe ocular diseases (Henderson et al., 2009). Therefore, gaining a better understanding how OTX2 and CRX regulate normal and diseased photoreceptor form and function should help identify genetic modifiers associated with different retinal degenerative diseases, shed light on distinct molecular pathways disrupted in a variety of ocular disorders, and uncover disease-specific targets amenable to therapeutic intervention.
Many genes required for photoreceptor differentiation in humans have homologous gene products in Drosophila. Moreover, mutations in several of these highly conserved genes result in retinal degeneration, both in flies and humans (Cook and Zelhof, 2008; Cook et al., 2011). Thus Drosophila is becoming a powerful model for defining how retinal genes function in normal and pathologic photoreceptor physiology. Here, we dissect the role of the Otd/OTX family of transcription factors during retinogenesis. This family of proteins is important for anterior patterning, neural specification, and sensory organ development in animals ranging from Cnidaria to humans and is defined by a highly conserved 60 amino acid homeodomain. The single Drosophila orthodenticle (otd) gene is represented by three vertebrate OTX factors, OTX1, OTX2 and CRX (Fig. 1B). Otd and all three vertebrate Otd-related factors are critical regulators of ocular development (Vandendries et al., 1996; Chen et al., 1997; Furukawa et al., 1997; Martinez-Morales et al., 2001; Nishida et al., 2003; Tahayato et al., 2003; Koike et al., 2007). In addition, previous cross- and intra-species rescue experiments have shown that fly otd and mouse Otx1 and Otx2 can largely replace each other’s functions during early nervous system development (Acampora et al., 1998a; Leuzinger et al., 1998; Nagao et al., 1998; Acampora et al., 2001a; Adachi et al., 2001; Simeone et al., 2002), revealing that these distantly related family members have retained remarkably similar transcriptional regulatory properties. However, surprisingly little homology exists among Otd/OTX factors outside of the DNA-binding domain (Simeone et al., 1993; Swain et al., 1997; Liu et al., 2001; Plouhinec et al., 2003; Acampora et al., 2005; Browne et al., 2006) (Fig. 1B), and target genes for the developmental processes tested in cross-species experiments are still not known. Thus, very little progress has been made towards uncovering how these important regulatory factors regulate common developmental processes.
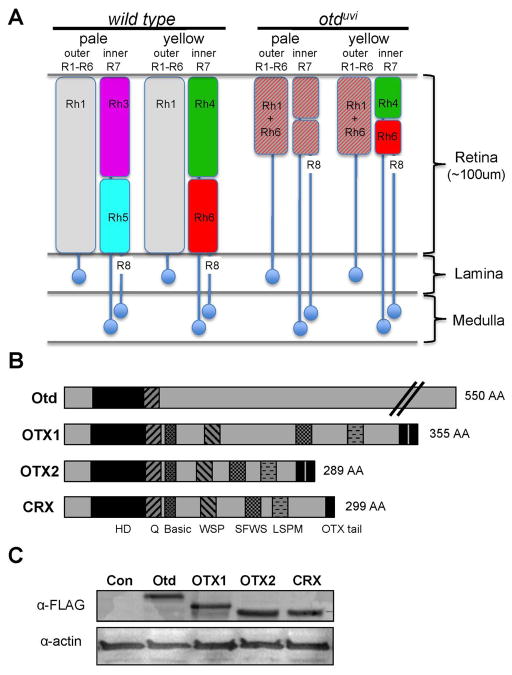
Figure 1.
Roles of Otd in photoreceptor development, and comparison of otd/OTX transcription factor proteins. A) Illustration of the known functions of Otd in the Drosophila eye: Rh3 activation in pale R7s, Rh5 activation in pale R8s, Rh6 repression in the outer photoreceptors, and rhabdomeric development and elongation. B) Schematic illustrating the regions of homology between the vertebrate OTX factors, including the homeodomain, the WSP motif, and the OTX tail(s). Drosophila Otd is longer that the vertebrate homologs, but only shares the well-conserved homeodomain followed by a downstream glutamine-rich (Q) region. C) Western blot analysis of Otd/OTX protein levels in rescue lines where each transgene was inserted at the same attB locus, 68E. Otd, OTX1, and CRX rescues were raised at 25°C, whereas OTX2 flies were raised at 18°C to achieve similar expression levels at this locus (see Experimental Procedures and quantification in Supp. Fig. 1A). α-actin serves as a loading control.
The role of Drosophila Otd in photoreceptor development is particularly well-defined. The Drosophila compound eye is comprised of ~750 individual eye units called ommatidia. Each ommatidium contains eight photoreceptor neurons: six outer photoreceptors (R1–R6), that like rod photoreceptors, function in motion detection and dim light conditions, and two inner photoreceptors (R7 and R8), that like cone photoreceptors, participate in color discrimination and bright light conditions (Hardie, 1985; Yamaguchi et al., 2008; Yamaguchi et al., 2010). Otd is essential for the formation of the rhabdomeres (functional equivalents to vertebrate outer segments) in all photoreceptors (Vandendries et al., 1996; Mishra et al., 2010) yet also regulates several photoreceptor subtype-specific functions (Tahayato et al., 2003; Xie et al., 2007; Mishra et al., 2010; Johnston et al., 2011). For example, Otd directly activates one of the two UV-sensitive rhodopsin-encoding genes, Rh3, in a subset of R7s, and activates the blue-sensitive rhodopsin, Rh5, in a subset of R8s (Fig. 1A) (Tahayato et al., 2003; Xie et al., 2007). Otd also prevents the expression of the green-sensitive rhodopsin, Rh6, in outer photoreceptors indirectly by activating the expression of a transcriptional repressor Dve (Tahayato et al., 2003; Johnston et al., 2011). Thus, Otd plays multiple roles in photoreceptor morphogenesis and opsin gene regulation during eye development.
Similar to Otd, OTX2 and CRX both play important roles in photoreceptor morphogenesis and gene expression (Chen et al., 1997; Furukawa et al., 1997; Nishida et al., 2003; Akagi et al., 2004; Koike et al., 2007; Hennig et al., 2008; Jomary and Jones, 2008; Corbo et al., 2010; Montana et al., 2011; Omori et al., 2011). While both OTX2 and CRX regulate many of the same genes, it remains unclear to what extent these factors play redundant vs. unique functions. Similar questions exist with relationship to OTX1 and OTX2 during the development of other regions of the nervous system. Thus, in the current study, we aimed to determine common and independent functions for human OTX1, OTX2 and CRX by testing their ability to rescue Otd-dependent functions during fly retinogenesis. We find that OTX1, OTX2 and CRX each mediate a defined subset of Otd-dependent functions in the fly eye, with OTX2 and CRX controlling unique cell-specific functions. We also examine several disease-associated CRX mutations and uncover specific functional deficits previously undetected by in vitro-based assays. These findings have important implications for understanding how Otx proteins have sub-functionalized during evolution, and cement Drosophila as an effective tool to unravel the molecular bases of photoreceptor pathogenesis.
Results
We previously developed a UAS/GAL4-based rescue system that allows us to test the ability of factors to rescue Otd-dependent functions during photoreceptor development (McDonald et al., 2010). This system relies on expressing UAS-driven transgenes in eye-specific otduvi mutants using a photoreceptor-restricted GAL4 driver. To test if the human OTX factors can functionally replace Otd during Drosophila eye development, we cloned the full-length OTX1, OTX2, and CRX cDNAs, with and without an N-terminus FLAG epitope tag, downstream of UAS sites. We created several independent transgenic lines for each construct with either random P-element insertions or site-specific phi31C/attB-dependent integrations (see Experimental Procedures). The latter allows each transgene to be expressed at the same genomic locus, minimizing position-based differences in expression levels (Bischof et al., 2007). Random and site-specific integration lines gave identical results, as did tagged and untagged versions of the proteins, in this assay system (data not shown, see Experimental Procedures). Here, for simplicity, we report data of lines that were specifically inserted at position 68E on chromosome III (Fig. 1C, Supp Fig. 1A).
Human OTX factors rescue rhabdomere morphogenesis similar to Otd
We first examined the ability of the human OTX factors to rescue photoreceptor morphogenesis. In wild-type eyes, the R1–R6 rhabdomeres are large rod-like structures that extend through the entire depth of the retina and form an asymmetric trapezoid around the central, smaller, and stacked R7 and R8 rhabdomeres (see Fig. 1A; Fig. 2A,G,M). In otduvi mutants, rhabdomeres are misshapen, often split, and only extend through ~30% of the retina (Fig. 2B,H,N). Similar to Otd rescue controls (Fig. 2C), all three vertebrate Otd-related factors rescue rhabdomere elongation (Fig. 2D–F). Also similar to Otd, all three factors generate recognizable, well-structured rhabdomeres in most photoreceptors at the distal region of the retina (Fig. 2I–L), Qualitatively, however, OTX1 and OTX2 rescues often have unorganized trapezoids and misshapen rhabdomeres (Figure 2P,Q), whereas almost all ommatidia in CRX rescues exhibit well-formed and organized trapezoids (Fig. 2R). We note, however, that not even CRX restores morphology to the same extent as Otd (Fig. 2C,I,O) (see Discussion). Together, these data indicate that OTX1, OTX2, and CRX are all largely capable of restoring rhabdomere formation in otduvi mutants, with CRX being most efficient.
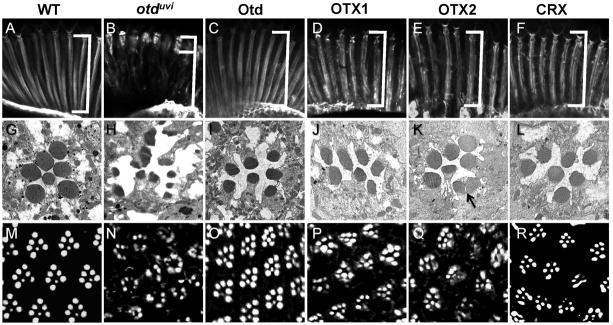
Figure 2.
All three human OTX factors rescue photoreceptor morphogenesis. A–F) Rhabdomere elongation was visualized in adult head cryosections stained with phalloidin to detect the actin-rich membranes of the rhabdomeres. Wild-type or control (O2-GAL4, pWIZ; TM2/TM6B) rhabdomeres extend the entire length of the retina (A), whereas otduvi retinas have rhabdomeres restricted to the distal-most 1/3 of the retina (B, brackets). Otd (C), OTX1 (D), OTX2 (E), and CRX (F) can each rescue rhabdomere elongation. G–L) Transmission electron microscopy of 1 μm distal sections of a control eye shows the trapezoidal arrangement of the six larger outer photoreceptor rhabdomeres surrounding the smaller, central R7 rhabdomere (G). otduvi rhabdomeres are often split and/or duplicated (H). Otd (I), OTX1 (J), OTX2 (K), and CRX (L) show round, and rarely duplicated rhabdomere (arrow, in K) in all rescues, although the trapezoidal arrangement and number of rhabdomeres in OTX1 rescues are less defined than in OTX2 or CRX rescues. M–R) Rhabdomere and trapezoid shape in multiple ommatidia was visualized by phalloidin staining of 8–10 μm cryosections from agarose-embedded adult heads. Controls have 6 large outer photoreceptors arranged in an asymmetric trapezoid, surrounding a smaller central R7 inner photoreceptor (M). otduvi flies show severely altered rhabdomeric structure (N), with some photoreceptors not detectable. Otd rescues (O) resemble wild-type eyes, while OTX1 (P) and OTX2 (Q) rescues show frequently disorganized ommatidia. CRX rescues (R) best resemble Otd rescues, although some outer photoreceptor rhabdomeres do not appear to reach wild-type sizes.
Vertebrate OTX factors exhibit differential activation potential on inner photoreceptor-specific opsins Rh3 and Rh5
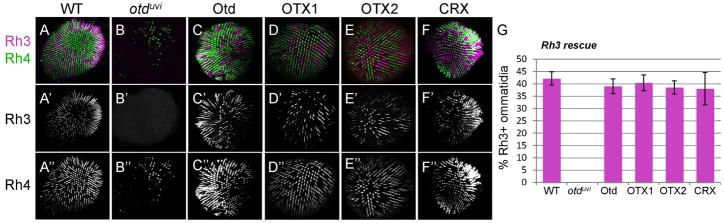
We next examined the ability of the vertebrate OTX factors to rescue Otd’s cell- and gene-specific functions. First, we analyzed Rh3 expression in R7 photoreceptors. In controls, Rh3 is expressed in approximately 40% of R7 cells (Fig. 3A, G), with the other UV-sensitive opsin, Rh4, being expressed in the remaining 60%. In otduvi mutants, Rh3 expression is lost (Fig. 3B,G), while Rh4 remains expressed (Tahayato et al., 2003). Like Otd, OTX1, OTX2 and CRX are each sufficient to restore Rh3 expression to 40% of R7s (Fig. 3C–G). These results indicate that, in contrast to morphogenesis, all three vertebrate factors function equivalently to Otd to rescue Rh3-expressing R7s.
Figure 3.
Rh3 expression is equally rescued in R7 cells by OTX1, OTX2, and CRX. Whole-mounted retinas from controls (O2-GAL4, pWIZ; TM2/TM6B (A), otduvi mutants (otduvi; O2-GAL4, pWIZ; TM2/TM6B) (B), and rescues with FLAG-tagged Otd (C), OTX1 (D), OTX2 (E), and CRX (F) were immunostained for Rh3 (magenta) and Rh4 (green) Representative retinas are shown at 200× magnification. G: Quantification of ommatidia expressing Rh3 in R7 cells demonstrates that Otd, OTX1, OTX2, and CRX each have equal abilities to restore Rh3 expression in ~40% ommatidia. Rh4 is not a direct target of Otd and serves as an internal control and counterstain for Rh3 quantifications. Error bars represent standard deviation.
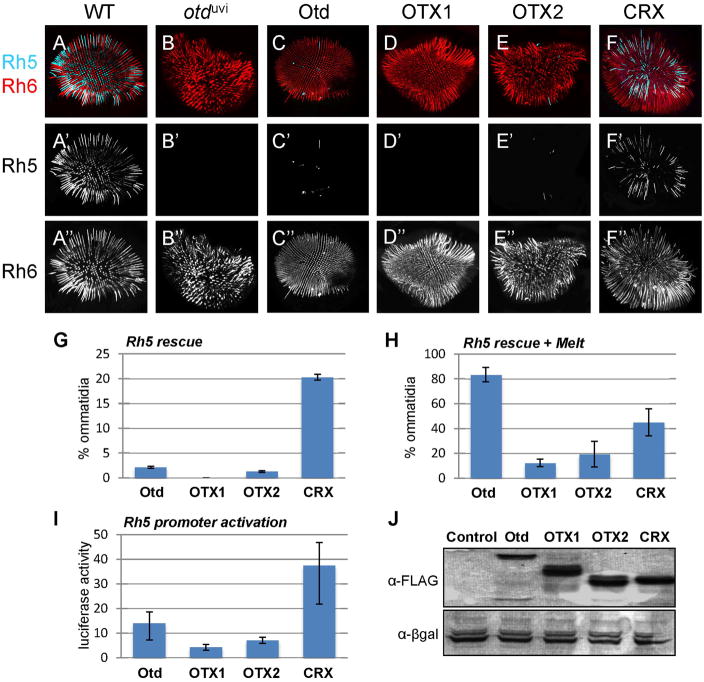
We next analyzed the ability of the vertebrate factors to activate Rh5 in R8 photoreceptors. In control flies, Rh5 is present in approximately 40% of R8 cells, while Rh6 is expressed in the remaining 60% (Fig. 4A). In otduvi flies, Rh5 expression is lost, and Rh6 expression is expanded (Fig. 4B) (Tahayato et al., 2003; McDonald et al., 2010). As previously reported, in this rescue paradigm, Otd only restores Rh5 expression in a small percentage of R8s (<3%, Fig. 4C,G). No Rh5-expressing cells are detected in OTX1 rescues, and OTX2 activates Rh5 in <2% of ommatidia (Fig. 4D,E,G). Surprisingly, CRX activates Rh5 in more than 20% of R8s (Fig. 4F,G). These data suggest that CRX is able to promote the development of Rh5-expressing R8s even more effectively than Otd, whereas OTX1 and OTX2 function less effectively than Otd in the process.
Figure 4.
Rh5 expression is only rescued in R8 cells by CRX. Whole-mounted retinas from controls (O2-GAL4, pWIZ; TM2/TM6B (A), otduvi mutants (otduvi; O2-GAL4, pWIZ; TM2/TM6B) (B), and rescues with FLAG-tagged Otd (C), OTX1 (D), OTX2 (E), and CRX (F) were immunostained for Rh5 (cyan) and Rh6 (red) and imaged with confocal microscopy. Representative images are shown at 200× magnification. G) Quantification of the percent of ommatidia expressing Rh5 in R8 cells reveals that CRX activates Rh5 in more cells than Otd, OTX2 can activate Rh5 in a small number of cells, and OTX1 has no ability to activate Rh5. H) Quantification of Rh5 expression when the OTX factors are coexpressed with Melted indicates that all factors are able to activate Rh5, but to a lesser degree than Otd. I) Relative luciferase activity of Rh5 promoter activation in S2 cells transfected with the vertebrate factors mirrors in vivo activation in the absence of Melt - CRX activates Rh5 more strongly than Otd, whereas OTX1 and OTX2 activate Rh5 weakly. J) Immunoblot analysis of OTX factors expressed in S2 cell lines indicate they are equally expressed in this system. Error bars in G–I represent standard deviation.
The low rescue efficiency by Otd is likely due to the fact that Otd not only directly activates Rh5 gene expression, but also controls the correct ratio of blue (Rh5+) vs. green (Rh6+) photoreceptors in the R8 layer (McDonald et al., 2010). To overcome this latter requirement of Otd and to sensitize the system to detect even weak Rh5 activation, we co-expressed Otd with Melted, a signaling protein that is sufficient to transform Rh6+ R8s into Rh5+R8s (Mikeladze-Dvali et al., 2005). In this genetic background, Otd activates Rh5 in >80% of R8 cells (Fig. 4H), whereas no Rh5 is present in otduvi mutants (data not shown) (McDonald et al., 2010), consistent with the requirement of Otd to activate Rh5. OTX1, OTX2, and CRX, with Melted co-expression, increase the percentage of Rh5+ R8 to ~15%, ~20%, ~40% R8s, respectively (Fig. 4H). These results show that introducing the Otd/OTX transgenes with Melted creates a more sensitive assay for Rh5 rescue, and indicate that all three factors are capable of activating Rh5, with CRX being the most effective of the three.
Since Otd can directly activate Rh5 promoter activity in vitro (Xie et al., 2007; McDonald et al., 2010), we also compared the activity of OTX1, OTX2, and CRX to function in this assay system. The results of these in vitro studies closely parallel our in vivo rescue experiments when the factors are expressed without Melted: OTX1 and OTX2 only weakly activate Rh5 (~5% and ~8%, respectively), whereas CRX activates Rh5 more than Otd (~38% vs. ~12%) (Fig. 4I). Western blot analysis confirms that these factors are expressed at similar levels to Otd in S2 cells (Fig. 4J).
Combined, these data suggest that all three Otd-related factors can equally activate Rh3, but activate Rh5 in the following increasing order of strength: OTX1, OTX2, and CRX. These data also reveal that CRX can function even more effectively than Otd to activate Rh5, but in a context-dependent manner.
OTX2 uniquely prevents Rh6 expression in outer photoreceptors
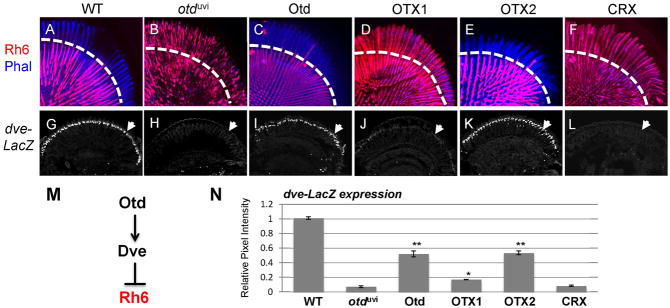
Besides directly activating the UV- and blue-sensitive Rh3- and Rh5-encoding genes in inner photoreceptors, Otd also indirectly represses the green-sensitive opsin-encoding gene, Rh6, in outer photoreceptors (Tahayato et al., 2003; Johnston et al., 2011). As shown in Fig. 5, Rh6 is normally restricted to a subset of R8 cells at the base of the retina (Fig. 5A); in contrast, in otduvi flies, Rh6 expression is expanded throughout the retina due to its de-repression into outer photoreceptors (Fig. 5B). Interestingly, no rescue of Rh6 repression is observed with OTX1 or CRX (Fig. 5D,F), whereas OTX2 is comparable with Otd to properly restrict Rh6 expression to R8 cells (Fig. 5C,E).
Figure 5.
Rh6 repression in outer photoreceptors is only rescued by OTX2. A–F) Whole-mounted retinas from controls (O2-GAL4, pWIZ; TM2/TM6B (A), otduvi mutants (otduvi; O2-GAL4, pWIZ; TM2/TM6B) (B), and rescues with FLAG-tagged Otd (C), OTX1 (D), OTX2 (E), and CRX (F) were immunostained for Rh6 (red) and actin-rich rhabdomeres were stained with phalloidin (blue). Dashed lines represent the boundary between the R7 and R8 layers. Rh6 is de-repressed into outer photoreceptors in otduvi flies (B), which is unchanged in OTX1 (D) and CRX (F) rescues. In contrast, Otd (C) and OTX2 (E) effectively prevent Rh6 expression in outer photoreceptors, thereby allowing proper restriction of Rh6 to the proximal R8 layer of the retina. G–H) Otd activates expression of the transcriptional repressor dve in outer photoreceptors, subsequently preventing Rh6 expression in these cells (M) (Johnston et al., 2011). Immunostaining of a dve-LacZ reporter (G–L) reveals that this line recapitulates wild-type Dve expression in adult outer photoreceptors (G), and like Dve, its expression is absent in otduvi mutants (H) (Johnston et al., 2011). Arrows indicate the outer photoreceptor nuclear layer, determined by co-staining with the pan-neural nuclear marker Elav (data not shown). I–L) Robust activation of dve expression is achieved in Otd (I) and OTX2 (K) rescues, very weak but consistent expression of dve is detected in OTX1 rescues (J), and no dve expression is observed in CRX rescues (L). N) Quantification of dve-LacZ expression in otduvi-dependent outer photoreceptor nuclei normalized to otduvi -independent brain neuron expression. * p < 0.002, ** p < 0.0001 compared to otduvi flies.
Otd represses Rh6 in outer photoreceptors by activating the expression of the transcriptional repressor Dve (Fig. 5M) (Johnston et al., 2011). To test whether OTX2 represses Rh6 through the same mechanism, we analyzed dve expression in our rescue flies, using a dve-LacZ enhancer trap line. This enhancer trap recapitulates previously reported Dve expression in outer photoreceptors (Fig. 5G) (Johnston et al., 2011) and is no longer detected in otduvi flies (Fig. 5H,N). Like Otd (Fig. 5I,N), OTX2 robustly restores dve expression in outer photoreceptors (Fig. 5K,N). OTX1 can also weakly activate dve expression (Fig. 5J,N). CRX, however, has no effect on dve expression (Fig. 5L,N), resembling otduvi mutants.
Together, these data indicate that OTX2 recapitulates Otd’s ability to restrict Rh6 expression to inner photoreceptors by activating dve, that OTX1 can weakly activate dve but this is insufficient to properly restrict Rh6 expression, and that Crx is unable to activate dve or repress Rh6 expression.
Differential functions of disease-associated alleles of CRX
In vertebrates, CRX is important for morphogenesis of the outer segment of photoreceptors and the activation of several photoreceptor-specific genes, including the rod and cone opsins. Similarly, our results indicate that CRX can rescue Otd-dependent photoreceptor morphology and activate opsin expression in the Drosophila retina. Previous studies aimed at understanding the molecular basis of disease-associated CRX mutations tested the ability of CRX mutants to activate a bovine rhodopsin reporter construct in cell culture-based assays (Swaroop et al., 1999; Chau et al., 2000; Mitton et al., 2000; Chen et al., 2002). Unfortunately, these studies were unable to identify obvious functional differences among different alleles or to establish genotype-phenotype correlations between mutations associated with distinct retinopathies - LCA, CORD, and RP. Thus, we sought to determine whether the fly eye could provide a useful model to more thoroughly analyze CRX mutant functions by testing a broader range of phenotypes in an in vivo context.
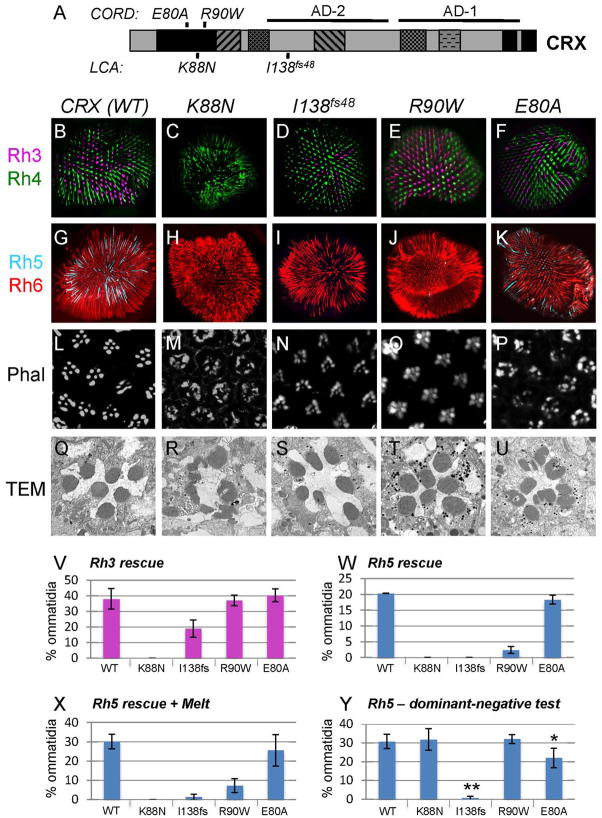
Here, we tested four different CRX alleles - K88N, I138fs48, R90W, and E80A (Fig. 6A). The K88N mutation, associated with LCA, changes a lysine residue present at position 50 of the homeodomain that is important for DNA-binding specificity (Wilson et al., 1993); this mutant was unstable in mammalian cells and stimulated bovine rhodopsin reporter expression ~10% of wild-type CRX in vitro (Nichols et al., 2010). Another LCA-associated mutation, I138fs48, truncates CRX’s C-terminus, deleting one and removing part of another of its activation domains (Chau et al., 2000; Nichols et al., 2010) (Fig. 6A). C-terminal truncations of CRX, including I138fs48, were shown to bind DNA similar to wild-type CRX but weakly activated bovine rhodopsin reporter expression (<30% of wild type) (Chen et al., 2002; Nichols et al., 2010). The third mutation, R90W, is associated with CORD when heterozygous and with LCA when homozygous (Swaroop et al., 1999). The R90W mutation, which is predicted to disrupt salt bridge formation between the third helix of the homeodomain with the major groove of DNA (Nichols et al., 2010), showed reduced DNA-binding activity in vitro, and activated bovine reporter expression only ~10% as wild-type CRX (Chen et al. 2002). Finally, the E80A mutation, linked with CORD, likely disrupts a critical electrostatic bond within the homeodomain, did not affect DNA-binding in vitro, and activated reporter expression two-fold higher than wild-type CRX (Chen et al., 2002).
Figure 6.
Differential properties of disease-associated CRX mutants in the Drosophila retina. A) Schematic illustrating the location of the mutations analyzed. AD-1 and AD-2 represent strong and weak transactivation domains, respectively, identified using cell culture-based luciferase reporter assays (Chau et al., 2000). Whole-mounted retinas (B–K) or agarose-embedded head cryosections (L–P) from otduvi rescues with wild-type CRX (B,G,L), CRX-K88N (C,H,M), CRX-I138fs48 (D,I,N), CRX-R90W (E,J,O), and CRX-E80A (F,K,P) were immunostained with Rh3 (magenta) and Rh4 (green) (B–F), Rh5 (cyan) and Rh6 (red) (G–K), or phalloidin (L–P). Q–U) TEM analysis of rhabdomere morphology in wild-type (Q), K88N (R), I138fs48 (S), R90W (T), and E80A (U) CRX rescues. I138fs48 and R90W mutant rescue rhabdomere morphology similarly to wild-type CRX, whereas K88N and E80A mutant rescues resemble otduvi mutants. V) Quantification of ommatidia expressing Rh3 in R7s, corresponding to experiments represented in B–F. W) Quantification of ommatidia expressing Rh5 in R8s, corresponding to experiments represented in G–K. X) Quantification of Rh5 rescue when CRX mutants are crossed with Melted. Y) Analysis of dominant negative function of CRX mutations assessed by percentage of Rh5-positive R8s present when CRX mutant alleles were expressed in a wild type background. Error bars represent standard deviation. * p < 0.005 and ** p < 0.0001 compared with wild-type CRX rescues.
When examined in our rescue assay, the four CRX mutations uncovered previously unrecognized functional differences among them (Table I). CRX-K88N is unable to activate Rh3 (Fig. 6C, V) or Rh5 (Fig. 6H,W), and cannot rescue photoreceptor morphogenesis (Fig. 6M,R). While this inactivity is similar to previous results with this mutant on bovine rhodopsin reporter expression, rather than being degraded as it is in mammalian ARPE 19 cells (Nichols 2010), CRX-K88N is expressed at similar levels as wild-type CRX in our rescue system (Supp. Fig. 1B). This supports the idea that the inactivity of this mutant in humans may not only be because of degradation, but may also reflect its inability to regulate gene expression. CRX-I138fs48 retains some ability to activate Rh3, albeit in fewer cells than wild-type CRX (Fig. 6D, V), has no capacity to activate Rh5 (Fig. 6I,W), yet largely restores rhabdomere and trapezoid formation (Fig. 6N,S). The R90W mutation activates Rh3 similar to wild-type (Fig. 6E, V), weakly activates Rh5 (Fig. 6J,W), and rescues morphology similar to wild-type CRX (Fig. 6O,T). Finally, the E80A mutation activates Rh3 (Fig. 6F,V) and Rh5 (Fig. 6K,W) similar to wild-type, but shows a reduced ability to rescue morphology (Figure 6P,U). Rh5 activation by all mutants in the presence of Melt largely mirrors that observed in the absence of Melt (Fig. 6W,X). In summary, K88N is non-functional, I138fs48 functions weaker than wild-type CRX in all assays, and R90W and E80A show differential activities: both activate Rh3 like wild-type CRX, but R90W largely loses its ability to activate Rh5 while E80A has a reduced ability to rescue morphology. These data suggest that these alleles may contribute to photoreceptor pathology through different mechanisms. In addition, at least in terms of their ability to activate Rh3 and Rh5, these results are consistent with K88N and I138fs48 being associated with a more severe retinal degeneration (LCA) than R90W and E80A (CORD).
Table I.
Comparison of vertebrate OTX factor function.
| Morphology | Activation | Repression Rh6 | Dominant Neg. Fxn | ||
|---|---|---|---|---|---|
| Rh3 | Rh5 | ||||
| Otd | ++++ | ++ | + | ++ | nd |
| OTX1 | ++ | ++ | − | − | nd |
| OTX2 | ++ | ++ | +/− | ++ | nd |
| CRX | +++ | ++ | ++ | − | − |
| K88N | − | − | − | − | − |
| I138fs48 | ++ | + | − | − | ++ |
| R90W | +++ | ++ | +/− | − | − |
| E80A | +/− | ++ | ++ | − | + |
n.d. = not determined
Because the majority of CRX mutations are linked to autosomal dominant diseases, we also assayed the ability of the four mutants tested above to function as dominant-negative proteins, using Rh5 as a readout. Expressing CRX- K88N or R90W mutants in otduvi heterozygotes (data not shown) or wild-type flies (Fig. 6Y) has no significant effect on the number of Rh5+ photoreceptors compared to controls, whereas E80A shows a slight but significant decrease and the I138fs48 truncation mutant almost eliminates all Rh5-expressing cells in both backgrounds (data not shown, and Fig. 6Y). It is worth noting that I138fs48 is considerably more stable in our in vivo system, similar ti previous in vitro studies. Together, these data suggest that the proteins encoded by the CRX- I138fs48 and -E80A alleles can compete with endogenous Otd to function as dominant-negative proteins, whereas the other homeodomain alleles do not.
Together with the rescue assays above, these findings indicate that we can detect subtle functional differences between CRX mutations associated with retinopathies, findings that may provide insight into the molecular bases of how these mutations exert their pathologic functions.
Discussion
Here, we show that the three human Otd-related transcription factors, OTX1, OTX2 and CRX, each have a distinct functional profile in the fly eye (Table I). All three factors can equally activate Rh3 in R7s, similarly to Otd. In addition, all three factors can largely rescue rhabdomere duplications and elongation defects in otduvi mutants. However, OTX1 has almost no capacity to activate Rh5 in R8s or repress Rh6 in outer photoreceptors, and does not fully rescue photoreceptor trapezoid formation; OTX2 rarely activates Rh5 in inner photoreceptors, only partially restores trapezoid formation, yet fully represses Rh6 in outer photoreceptors; and CRX activates Rh5, even better than Otd in some contexts, rescues trapezoid formation much like Otd, yet fails to repress Rh6 in outer photoreceptors. We also define differences in the ability of several CRX mutant alleles to rescue CRX-dependent functions in this system, which should be helpful for further understanding the disease mechanisms linked to these mutations. Below, we discuss the significance of these findings.
Functional conservation of Otd/OTX in photoreceptor morphogenesis
The discovery that Pax6 serves as a “master control gene” for visual system development, from the simple photoreceptive system in jellyfish, to the compound eye in Drosophila and the camera eye of humans, led to the now widely held model that all visual systems arose from a common precursor. However, one of the remaining controversies to this unifying theory of eye evolution relates to whether ciliary and rhabdomeric photoreceptor types arose as independent cell types or from a common precursor (Halder et al., 1995; Nilsson, 1996; Arendt and Wittbrodt, 2001; Arendt, 2003; Kozmik, 2005; Nilsson, 2005; Arendt, 2008; Erclik et al., 2009; Nilsson, 2009). Ciliary photoreceptors are primarily associated with vertebrate visual systems, and are defined by a cilia-based outer segment that houses a hyperpolarizing cGMP-based phototransduction pathway. In contrast, rhabdomeric photoreceptors are more commonly associated with invertebrate visual systems, which have an actin-based rhabdomere and a depolarizing cAMP-based pathway [reviewed by (Arendt and Wittbrodt, 2001; Wang and Montell, 2007; Sanes and Zipursky, 2010)]. These fundamental differences have led to the idea that these photoreceptors are not evolutionarily related and instead, arose through independent mechanisms. However, it is also clear that many of the same factors are commonly used for the morphogenesis and function of both photoreceptor cell types, including Crumbs/CRB1, Spacemaker/EYS, Prominin, and Rhodopsin (see review by Cook et al., 2011).
Given the importance of OTX2 and CRX in preventing ciliary photoreceptor degeneration, here we asked whether these factors shared Otd’s ability to rhabdomeric structures. Previous studies suggest that Otd/OTX transcription factors are central regulators of photoreceptor differentiation. For examples, these factors are expressed in photosensitive cells in a wide array of organisms and are critical to guide proper neural/eye specification and photoreceptor development (Finkelstein et al., 1990; Vandendries et al., 1996; Chen et al., 1997; Furukawa et al., 1997; Stornaiuolo et al., 1998; Umesono et al., 1999; Martinez-Morales et al., 2001; Salo et al., 2002; Lanjuin et al., 2003; Nishida et al., 2003; Plouhinec et al., 2003; Koike et al., 2007; Ward et al., 2008; Steinmetz et al., 2010; Passamaneck et al., 2011). High throughput techniques also show that Otd, OTX2 and CRX each regulate several common photoreceptor-specific genes (Ranade et al., 2008; Corbo et al., 2010; Mishra et al., 2010; Omori et al., 2011). Importantly, we find that all three Otd-related factors can largely rescue rhabdomeric morphogenesis. Since the homeodomain is the only highly conserved domain between the Otd and human OTX proteins, these data support the recent conclusion that Otd’s homeodomain is largely sufficient to mediate its role in photoreceptor morphogenesis (McDonald et al., 2010). Moreover, at least as it relates to OTX2 and CRX, our data reveal that OTX factors are able to build either ciliary or rhabdomeric photoreceptors. Interestingly, of the three, CRX qualitatively rescued photoreceptor morphogenesis more effectively that either OTX1 or OTX2, consistent with its strong association with diseases affecting the maintenance of photoreceptor integrity. Thus, it appears that CRX retained more “morphogenesis” functions than its other two counterparts during their evolutionary diversification. However, we do note that none of the factors individually rescue morphogenesis as effectively as Otd. Since we do not yet know the gene targets responsible for Otd, OTX2, or CRX-dependent photoreceptor morphogenesis, it is difficult to speculate what these partial rescues mean. Some possibilities are that the vertebrate factors do not effectively regulate all necessary targets due to differences in DNA affinity, post-translational modifications, cofactor recruitment, or ability to cooperate with other DNA-binding partners. Pph13/Hazy, for instance, was recently shown to cooperate with Otd to regulate rhabdomere formation, yet a Pph13 ortholog does not seem to be required for CRX function in vertebrates (Mishra et al., 2010); therefore, it is possible that the vertebrate OTX factors cannot cooperate with Pph13/Hazy in fly photoreceptors. Alternatively, it is possible that, similar to the complementary functions of OTX2 and CRX on Rh5 and Rh6 regulation in inner vs. outer photoreceptors (Fig. 4 and 5), the vertebrate factors may perform complementary functions during photoreceptor morphogenesis. Arguing against this latter possibility, we did not detect improvement in morphology in flies co-expressing OTX2 and CRX that were observed with either factor alone (data not shown). As we gain better insight into how Otd contributes to morphogenesis, we will be able to more directly test which aspects of this process the vertebrate factors can rescue and which ones they cannot. Nevertheless, the findings reported here contribute to a growing body of evidence that, despite major structural and biochemical differences, ciliary and rhabdomeric photoreceptors are built from a common, evolutionarily conserved molecular network, and suggest that this network relies on Otd-related factors.
Differential functions of human OTX factors in photoreceptor-specific gene regulation
Previous cross- and intra-species experiments, as well as the experiments described above, are limited in their ability to define common vs. unique properties used by OTX factors to regulate development, largely because of a lack of known target genes. In contrast, our ability to quantify the rescue of specific gene targets - Rh3, Rh5, and Rh6 -in distinct photoreceptor populations provided us with valuable tools to further explore these issues. Importantly, we found that OTX1, OTX2, and CRX are each equally able to rescue Rh3-expressing photoreceptors (Fig. 3), demonstrating that these factors are properly expressed and functional in the fly eye and can contribute to common processes. In contrast, only OTX2 can prevent Rh6 expansion into outer photoreceptors and only CRX signficantly activates Rh5 in R8 cells (Figs 4 and 5), revealing that these factors can also mediate distinct functions.
These data are in strong agreement with previous studies on Otx factors during mouse nervous system development, indicating that they exert similar and distinctive functions. Otx1 and Otx2, for example, are widely expressed and participate in overlapping processes during nervous system development and in the formation of ocular structures such as the lens, ciliary body, and retinal pigmented epithelia (Simeone et al., 1993; Frantz et al., 1994; Martinez-Morales et al., 2001), while Otx2 and Crx have overlapping patterns of expression in bipolar and photoreceptors and regulate many of the same genes in the retina (Liu et al., 2001; Wang et al., 2002; Peng and Chen, 2005; Koike et al., 2007; Glubrecht et al., 2009; Corbo et al., 2010; Omori et al., 2011). This, together with data indicating that Otx1 and Otx2 can largely replace each other’s functions in gene replacement studies (Acampora et al., 1998a; Acampora et al., 1998b; Acampora et al., 2001a; Acampora et al., 2001b), suggests that OTX factors can mediate common regulatory functions. However, it is also clear that Otx2 only partially restores many of Otx1-dependent functions, with no capacity to rescue Otx1-dependent regulation of inner ear development (Acampora et al., 1999); similarly, despite their similarities, Otx2 and Crx induce the formation of different cell types when overexpressed in Xenopus retinal explants (Viczian et al., 2003), and photoreceptor-specific knockout studies of Otx2 do not phenocopy Crx mutants (Nishida et al., 2003). Thus, OTX factors also appear have distinct functions as well. Our ability to quantify the extent of rescue of specific target genes now allows us to begin defining domains within the OTX proteins necessary to mediate their various functions and to dissect the molecular mechanisms underlying their overlapping and unique properties.
Our studies may also lend insight into previous studies suggesting that Otx1 and Otx2 function dose-dependently. For example, during ocular development, Otx1−/−; Otx2+/− mice exhibit strong defects in lens, retina, and retinal pigmented epithelium (RPE), which are not observe in Otx1−/− or Otx2+/− mice (Martinez-Morales et al., 2001). Here, we found that OTX2 can strongly induce dve expression and subsequently repress Rh6 expression in outer photoreceptors, whereas OTX1 can only weakly activate dve and this is not sufficient to repress Rh6. One could envision that similar events are underway on target genes in the mouse eye, with Otx2 primarily regulating the expression of a certain gene or set of genes required for some developmental events, and Otx1 contributing weakly to their expression. Hence, when both copies of Otx1 and a copy of Otx2 are removed, the target gene(s) are now no longer able to reach the appropriate threshold of expression necessary for function. Indeed, this may be the case with Otx2-dependent activation of the BEST-1 gene in the RPE (Esumi et al., 2009). Thus, we can now begin to identify domains between OTX1 and OTX2 that would explain how they can equally activate some targets (e.g. Rh3) but differentially regulate other targets (e.g. dve).
Finally, our studies expand our understanding of the cell-specific mechanisms that are integrated by Otd and co-opted by the vertebrate factors to differentially regulate gene expression in the eye. For instance, our previous molecular mapping of Otd suggested that it contains two activation domains: an N-terminal region that is sufficient for Rh3 activation in ~50% of its correct subtype but is insufficient to activate Rh5 expression, and a C-terminal region that also contributes to Rh3 activation, is essential for Rh5 activation, and controls Rh6 repression in outer photoreceptors (McDonald et al., 2010). Our findings here with OTX1, OTX2, and CRX suggest that: 1) the regulatory mechanism utilized by Otd’s N-terminus is commonly used by all three vertebrate factors, 2) Otd’s C-terminus may recruit up to three different regulatory complexes - one that contributes to Rh3 activation in R7s, one that contributes to Rh5 activation in R8s, and one that contributes to Rh6 repression in outer photoreceptors; and 3) the three human factors have each maintained distinct complements of these C-terminal activities -all three have maintained the Rh3 activation domain, whereas CRX maintained the Rh5 activation domain and OTX2 maintained the Rh6 repression (dve activation) domain. Importantly, as Fig. 1A illustrates, identifying such domains between fly and human OTX factors is not possible by primary sequence analysis. However, the rescue system described here will allow us to perform unbiased molecular mapping studies of the human OTX factors to identify structurally and/or functionally conserved regulatory motifs. In addition, we are actively pursuing the identification of Otd cofactors that allow it to perform cell-specific functions, which will also be tested for interactions with the vertebrate OTX factors as a means to test the hypothesis that they recruit common coregulatory factors.
Molecular bases of CRX subfunctions
By analyzing the abilities of naturally occurring CRX mutations to rescue retinogenesis in Drosophila otduvi mutants, we have already begun to gain insights into the molecular bases behind CRX subfunctions in the normal retina. For instance, by comparing homeodomain mutations, K88N, E80A, and R90W, with a C-terminal deletion that removes its activation domains, I138fs48, we further confirm that the homeodomain is important for morphogenesis (McDonald et al., 2010). Surprisingly, however, although the E80A mutant showed reduced morphogenesis rescue, it could still activate Rh3 or Rh5 expression similarly to wild-type CRX; R90W, on the other hand, could still rescue morphogenesis, but could not activate Rh5. These data suggest that the homeodomain is not just responsible for DNA-binding but also differentially contributes to subfunctions of CRX. One possible explanation for why R90W and E80A proteins behave differently is that they each lose the ability to recruit different homeodomain-interacting proteins. Indeed, several proteins have been shown to interact with CRX’s homeodomain (Mitton et al., 2000; Wang et al., 2002; Chen et al., 2004; Lerner et al., 2005; Palhan et al., 2005; Peng et al., 2005), but their precise modes of interactions have not been explored. However, at least with the case of NRL (neural retina leucine zipper protein) - a well-established CRX interactor linked to retinal degenerations (DeAngelis et al., 2002; Nishiguchi et al., 2004; Wright et al., 2004) - it has been shown to maintain its interactions with the R90W mutant but not the E80A mutant (Chen et al., 2002). Thus, it is possible that NRL and CRX together control genes important for photoreceptor morphogenesis, whereas another factor that can still interact with E80A mutants contribute to CRX’s ability to activate other targets. Therefore, our ability to detect subtle differences in a wide range of phenotypes enables us to characterize pathogenic mechanisms underlying CRX mutations in a more directed way than previously possible, highlighting the importance of conducting such studies in an in vivo genetic system.
Our findings also reveal that CRX’s C-terminus is essential for Rh5 expression, but is not critical for Rh3 activation. These results are identical to our findings with Otd (McDonald et al., 2010), supporting the model that a structurally-related region in Otd’s and CRX’s C-termini recruit a common coregulator necessary for activation of specific subsets of gene targets. In this respect, it is particularly interesting that the I138fs48 functions as a strong dominant-negative allele on Rh5 expression - it is a highly stable protein that is unable to directly activate Rh5 (Fig. 5W,Y), so it readily competes with endogenous Otd to prevent its ability to activate this particular target. However, I138fs48 does not appear to function as a dominant-negative protein with regards to Rh3 activation (data not shown), consistent with its ability to still activate this target by itself (Fig. 5V). This data suggests that I138fs48 can function selectively as a dominant-negative protein. Given that several CRX alleles lead to truncations, testing more alleles in this system may not only help define the domain within CRX that recruits specific coactivators, but may also help uncover mechanisms by which CRX mutants function as dominant alleles. Our continuing work aimed at identifying Otd/CRX coregulators may also lead to the discovery of other candidate factors that are disrupted distinct retinal degenerations.
Why E80A and other non-truncation mutants of CRX function as dominant-negative proteins remains to be determined. Nevertheless, this is, to our knowledge, the first in vivo analysis of dominant negative functions of CRX mutations. This has obvious significance for future therapies. For example, while loss-of-function alleles of CRX might potentially be treated by simple transgene replacement of wild type CRX, dominant-negative alleles of CRX will likely demand another therapeutic approach. Thus, our in vivo rescue should allow for a relatively inexpensive, rapid analysis of the many as of yet unanalyzed OTX/CRX disease-causing mutations that fall within this class.
Summary
The studies presented here indicate that the fly is a useful system for efficiently and effectively defining functionally conserved and protein-specific mechanisms that are used by OTX factors to control the development of different subsets of neurons. These studies also provide insight into what functions of CRX are disrupted in various disease alleles, information that should be helpful for developing more effective diagnostic and therapeutic tools for patients carrying such mutations.
Experimental Procedures
DNA constructs
Human OTX1 and OTX2 cDNAs were a gift from Shiming Chen (Washington University). 3XFLAG-pAc5.1 was generated by subcloning the 3XFLAG epitopes from CMV-3Tag (Agilent) into pAc5.1 (Invitrogen) using PCR primers containing the preferred Drosophila Kozak consensus site. OTX1 and OTX2 were PCR-amplified and subcloned into 3XFLAG-pAC5.1 using the following primers:
OTX1 5′: GCGAATTCTCTTACCTCAAACAACCCCCA,
OTX1 3′: AATGTCGACCTACAAGACCTGGAACCGCCA,
OTX2 5′: GCGGAATTCTCTTATCTTAAGCAACCGCCT,
OTX2 3′: GACTCGAGCTACAAAACCTGGAATTTCCACG,
FLAG-OTX1 and FLAG-OTX2 were then subcloned into pUAST (Brand and Perrimon, 1993) or pUAST-attB (Bischof et al., 2007). Full-length wild-type, K88N and I138fs48 mutant human CRX cDNAs were obtained from Brian Brooks (NEI). These were PCR-amplified using the following primers: CRX 5′: GAAGATCTTATGATGGCGTATATGAACCCG and CRX 3′: CCGCTCGAGCTACAAGATCTGAAACTTCCAGGC. The PCR products were digested with Bgl II and Xho I and ligated into an N-terminal-tagged FLAG pUAST vector (cloning details available upon request). R90W and E80A CRX mutants were created using the Stratagene Lightning site-directed mutagenesis kit to modify pUAST-attB FLAG-CRX using the primers:
R90W 5′-CAGGTTTCCTTCAAGAACTGGAGGGCTAAATGCAG
R90W 3′-CTGCATTTAGCCCTCCAGTTCTTGAACCAAACCTG
E80A 5′-GAAGATCAATCTGCCTGCGTCCAGGGTTCAGGTTT
E80A 3′-AAACCTGAACCCTGGACGCAGGCAGATTGATCTTC
Drosophila strains
Fly stocks were maintained on standard agar/cornmeal/molasses media at 25°C in 12 light/dark cycles. UAS-melt (III) was a gift from Claude Desplan (New York University), and the dve01738 LacZ enhancer trap line was obtained from the Bloomington Stock Center (stock #11073). The otd1.6-GAL4 driver, O2-GAL4 (McDonald et al., 2010) on the II chromosome was recombined with an RNAi directed against the white gene (Lee et al., 2003) to reduce autofluorescence generated by eye pigmentation. Transgenic lines were generated by random and attB site-specific insertions using standard methods (Rainbow Transgenics, Camarillo, CA). Similar results for each Otd/OTX factor were obtained, independent of insertion site. For example, N-terminal FLAG-tagged Otd was tested at four independent attB insertion sites: 51C (Bloomington Stock #24482), 58A (Bloomington Stock #24484), 68E (Bloomington Stock #24485), 86Fb (Bloomington Stock #24749), and each locus rescued the otduvi phenotype to the same degree. Two independent, randomly-inserted, untagged OTX1 and OTX2 lines (kindly provided by Ernst Wimmer, Georg-August-University, Goettingen) behaved similarly to FLAG-tagged versions of the same factors inserted at the 68E attB site, as did random and site-specific insertions of FLAG or untagged CRX constructs. All experiments reported here used FLAG-tagged transgenes integrated by phi C31/attB-mediated recombination at position 68E. At this site, OTX2 protein was detected at higher levels than the other proteins by western blot analysis when flies were raised at 25°C (data not shown). Therefore, to normalize OTX2 expression to similar levels as Otd, OTX1 and CRX, we raised OTX2-expressing flies at 18°C and the remaining flies at 25°C (see Fig. 1C, Supp. Fig. 1A).
Immunohistochemistry, morphologic analysis, and luciferase assays
Adult flies were decapitated and the heads were bisected. The heads were fixed in 3.2% paraformaldehyde, 1XPBS for 20 minutes at room temperature and washed three times for 10 minutes in PBX [PBS, 0.3%Triton]. The retinae were further dissected by removing the cuticle surrounding the eye and the corneal lens and blocked with BNT [0.5%BSA, 0.5mM NaCl, 0.1%Tween-20, and PBS] for 30 minutes. Primary antibodies were diluted to an experimentally determined concentration (see Antibodies section) in 300 μL BNT, and incubated with rocking overnight at 4°C. The tissue was then washed three times in PBX for 30 minutes, followed by addition of the appropriate secondary antibody for 2 hours at room temperature. The retinae were then washed three times for 30 minutes in PBX, mounted in Prolong™ Anti-Fade Reagent (Invitrogen), and cured for 4 hours prior to imaging. Samples were imaged using a Zeiss LSM-700 confocal microscope, and the images were processed using Zeiss Zen and Adobe Photoshop software. Morphologic analysis was carried out using agarose-embedded cryosections and transmission electron microscopy as previously described (McDonald, 2010; Mishra et al., 2010). Quantification of Rhodopsin ratios was determined by counting at least 500 ommatidia from a minimum of 6 retinas. When statistical values are presented, analysis was performed using a standard t-test. To quantify the relative level of dve-LacZ expression, the intensity of non-otduvi-dependent LacZ-expressing neurons in the brain and dve-LacZ expression in outer photoreceptors were quantified using Image J software. The intensity of at least 5 brain neurons and 5 regions of the outer photoreceptor nuclear layer were measured, averaged, and normalized for each section using at least 4 retinas per sample group. Statistical differences with otduvi levels was determined using a standard t-test. Luciferase assays were conducted in low passage S2 cells as previously published (McDonald, 2010).
Immunoblot analysis
Approximately 15 heads from each experimental group were frozen on dry ice, homogenized with a hand-held electric pestle, and resuspended in RIPA buffer [20 mM Tris-HCl (pH 7.5) 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate]. The tissue was incubated in RIPA for 30 minutes, and centrifuged at 12,000 rpm and 4° C for 10 minutes to remove the insoluble fraction of the cells. The resultant supernatant (total cell lysate) was quantitated using the BioRad protein assay (Biorad, Cat # 500-0006). S2 lysates were isolated as previously described (McDonald et al., 2010). Protein (10μg) was loaded onto 4–20% Biorad Ready Gels® and transferred to PVDF membrane overnight at 4°C in Tris-glycine buffer at a constant amperage of 40 mA. The membranes were washed using Tris-buffered saline (TBS), blocked in 5% milk/TBS, and incubated with primary antibody overnight at 4°C. Membranes were then washed three times in TBS and incubated with donkey α-mouse secondary antibodies (Santa Cruz), developed using ECL Plus Reagent (Amersham), and imaged on an 860 Storm scanner (Amersham). To confirm equal loading, membranes were subsequently stripped (2% SDS, 62.5mM Tris pH6.8, 100mM beta-mercaptoethanol) and immunoblotted with a pan-actin antibody (gift from James Lessard, CCHMC).
Antibodies
The antibodies and appropriate concentrations for indirect immunofluoresence were as follows: mouse α-Rh3 and α-Rh5 (1:50) (kindly provided by Steve Britt), rabbit α-Rh4 (1:100) (kindly provided by Charles Zuker and Nansi Colley), and rabbit α-Rh6 (1:1000) (kindly provided by Claude Desplan). Donkey anti-mouse AlexaFluor 647 and donkey α-rabbit AlexaFluor 488 were used as secondary antibodies (1:500) (Invitrogen). Polymerized actin was recognized using AlexaFluor (1:50) (488 or 555)-conjugated phalloidin (Invitrogen). The M2 mouse monoclonal FLAG antibody (Sigma, F3165) was used at 1:1000 to detect the expression of the vertebrate factors via immunofluorescence and immunoblot. Anti-CRX (Abnova, H00001406-M03) was used 1:500 for the western blot analysis in Supplemental Figure 1B, donkey anti-mouse HRP secondary antibodies (Santa Cruz, sc-2005) was used 1:2000, and the actin antibody used as a protein loading control (kindly provided by James Lessard, Cincinnati Children’s Hospital) was used at 1:1000.
Supplementary Material
A) Quantification of OTX expression shown in the western blot in Figure 1C. B) Western blot analysis of FLAG-tagged mutant CRX proteins from adult Drosophila head lysates. All mutant CRX proteins are expressed, albeit at different levels. Similarly to what was previously reported in vertebrate cell culture lines (Nichols et al., 2010), the CRX 138fs48 mutation increases the steady state levels of the protein. The R90W mutant has slightly reduced levels compared to wild-type CRX. Actin serves as a loading control.
Bullet points.
Human OTX factors - OTX1, OTX2 and CRX - perform overlapping, yet distinct subsets of Otd-dependent functions during Drosophila eye development, providing a system to study subfunctionalization of a family of related transcription factors.
Human OTX factors rescue the morphogenesis of rhabdomeric photoreceptors, suggesting that ciliary and rhabdomeric photoreceptors arose from a common ancestor requiring an Otd-related factor for its development.
Different CRX alleles associated with similar retinal degenerations can be functionally distinguished using the fly eye as a model system.
Acknowledgments
We thank Steve Britt, Brian Brooks, Richard Carthew, Shiming Chen, Claude Desplan, James Lessard, and Ernst Wimmer for kindly providing reagents, and Julie Kiefer and anonymous reviewers for their insightful comments to the manuscript. We also acknowledge the Drosophila Studies Hybridoma Bank, the Bloomington Stock Center, and the Drosophila Genome Resource Center for reagents. This work was supported by the Ziegler Foundation for the Blind, Research to Prevent Blindness, and NIH R01-EY017907 (TAC).
Abbreviations
- CORD
Cone-Rod Dystrophy
- LCA
Leber’s Congenital Amaurosis
- NRL
neural retina leucine zipper
- otd
Orthodenticle
- OTX
otd-related homeobox
- PR
photoreceptor
- RP
Retinitis Pigmentosa
- Rh
rhodopsin
References
- Acampora D, Annino A, Tuorto F, Puelles E, Lucchesi W, Papalia A, Simeone A. Otx genes in the evolution of the vertebrate brain. Brain Res Bull. 2005;66:410–420. doi: 10.1016/j.brainresbull.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Acampora D, Avantaggiato V, Tuorto F, Barone P, Perera M, Choo D, Wu D, Corte G, Simeone A. Differential transcriptional control as the major molecular event in generating Otx1−/− and Otx2−/− divergent phenotypes. Development. 1999;126:1417–1426. doi: 10.1242/dev.126.7.1417. [DOI] [PubMed] [Google Scholar]
- Acampora D, Avantaggiato V, Tuorto F, Barone P, Reichert H, Finkelstein R, Simeone A. Murine Otx1 and Drosophila otd genes share conserved genetic functions required in invertebrate and vertebrate brain development. Development. 1998a;125:1691–1702. doi: 10.1242/dev.125.9.1691. [DOI] [PubMed] [Google Scholar]
- Acampora D, Avantaggiato V, Tuorto F, Briata P, Corte G, Simeone A. Visceral endoderm-restricted translation of Otx1 mediates recovery of Otx2 requirements for specification of anterior neural plate and normal gastrulation. Development. 1998b;125:5091–5104. doi: 10.1242/dev.125.24.5091. [DOI] [PubMed] [Google Scholar]
- Acampora D, Boyl PP, Signore M, Martinez-Barbera JP, Ilengo C, Puelles E, Annino A, Reichert H, Corte G, Simeone A. OTD/OTX2 functional equivalence depends on 5′ and 3′ UTR-mediated control of Otx2 mRNA for nucleo-cytoplasmic export and epiblast-restricted translation. Development. 2001a;128:4801–4813. doi: 10.1242/dev.128.23.4801. [DOI] [PubMed] [Google Scholar]
- Acampora D, Gulisano M, Broccoli V, Simeone A. Otx genes in brain morphogenesis. Prog Neurobiol. 2001b;64:69–95. doi: 10.1016/s0301-0082(00)00042-3. [DOI] [PubMed] [Google Scholar]
- Adachi Y, Nagao T, Saiga H, Furukubo-Tokunaga K. Cross-phylum regulatory potential of the ascidian Otx gene in brain development in Drosophila melanogaster. Dev Genes Evol. 2001;211:269–280. doi: 10.1007/s004270100149. [DOI] [PubMed] [Google Scholar]
- Akagi T, Mandai M, Ooto S, Hirami Y, Osakada F, Kageyama R, Yoshimura N, Takahashi M. Otx2 homeobox gene induces photoreceptor-specific phenotypes in cells derived from adult iris and ciliary tissue. Invest Ophthalmol Vis Sci. 2004;45:4570–4575. doi: 10.1167/iovs.04-0697. [DOI] [PubMed] [Google Scholar]
- Arendt D. Evolution of eyes and photoreceptor cell types. Int J Dev Biol. 2003;47:563–571. [PubMed] [Google Scholar]
- Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet. 2008;9:868–882. doi: 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]
- Arendt D, Wittbrodt J. Reconstructing the eyes of Urbilateria. Philos Trans R Soc Lond B Biol Sci. 2001;356:1545–1563. doi: 10.1098/rstb.2001.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Browne WE, Schmid BG, Wimmer EA, Martindale MQ. Expression of otd orthologs in the amphipod crustacean, Parhyale hawaiensis. Dev Genes Evol. 2006;216:581–595. doi: 10.1007/s00427-006-0074-7. [DOI] [PubMed] [Google Scholar]
- Chau KY, Chen S, Zack DJ, Ono SJ. Functional domains of the cone-rod homeobox (CRX) transcription factor. J Biol Chem. 2000;275:37264–37270. doi: 10.1074/jbc.M002763200. [DOI] [PubMed] [Google Scholar]
- Chen S, Peng GH, Wang X, Smith AC, Grote SK, Sopher BL, La Spada AR. Interference of Crx-dependent transcription by ataxin-7 involves interaction between the glutamine regions and requires the ataxin-7 carboxy-terminal region for nuclear localization. Hum Mol Genet. 2004;13:53–67. doi: 10.1093/hmg/ddh005. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Xu S, Liu I, Li LY, Wang Y, Zack DJ. Functional analysis of cone-rod homeobox (CRX) mutations associated with retinal dystrophy. Hum Mol Genet. 2002;11:873–884. doi: 10.1093/hmg/11.8.873. [DOI] [PubMed] [Google Scholar]
- Cook B, Zelhof AC. Photoreceptors in evolution and disease. Nat Genet. 2008;40:1275–1276. doi: 10.1038/ng1108-1275. [DOI] [PubMed] [Google Scholar]
- Cook T, Zelhof A, Mishra M, Nie J. 800 facets of retinal degeneration. Prog Mol Biol Transl Sci. 2011;100:331–368. doi: 10.1016/B978-0-12-384878-9.00008-X. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Lawrence KA, Karlstetter M, Myers CA, Abdelaziz M, Dirkes W, Weigelt K, Seifert M, Benes V, Fritsche LG, Weber BH, Langmann T. CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 2010;20:1512–1525. doi: 10.1101/gr.109405.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis MM, Grimsby JL, Sandberg MA, Berson EL, Dryja TP. Novel mutations in the NRL gene and associated clinical findings in patients with dominant retinitis pigmentosa. Arch Ophthalmol. 2002;120:369–375. doi: 10.1001/archopht.120.3.369. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27:391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Erclik T, Hartenstein V, McInnes RR, Lipshitz HD. Eye evolution at high resolution: the neuron as a unit of homology. Dev Biol. 2009;332:70–79. doi: 10.1016/j.ydbio.2009.05.565. [DOI] [PubMed] [Google Scholar]
- Esumi N, Kachi S, Hackler L, Jr, Masuda T, Yang Z, Campochiaro PA, Zack DJ. BEST1 expression in the retinal pigment epithelium is modulated by OTX family members. Hum Mol Genet. 2009;18:128–141. doi: 10.1093/hmg/ddn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Smouse D, Capaci TM, Spradling AC, Perrimon N. The orthodenticle gene encodes a novel homeo domain protein involved in the development of the Drosophila nervous system and ocellar visual structures. Genes Dev. 1990;4:1516–1527. doi: 10.1101/gad.4.9.1516. [DOI] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis-Anagnostou A, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Freund CL, Wang QL, Chen S, Muskat BL, Wiles CD, Sheffield VC, Jacobson SG, McInnes RR, Zack DJ, Stone EM. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet. 1998;18:311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Glubrecht DD, Kim JH, Russell L, Bamforth JS, Godbout R. Differential CRX and OTX2 expression in human retina and retinoblastoma. J Neurochem. 2009;111:250–263. doi: 10.1111/j.1471-4159.2009.06322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. New perspectives on eye evolution. Curr Opin Genet Dev. 1995;5:602–609. doi: 10.1016/0959-437x(95)80029-8. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Footfre OD. Prog Sens Physiol. Vol. 5. Springer; Berlin, Heidelberg, New York, Toronto: 1985. pp. 1–79. [Google Scholar]; Ottoson D, editor. Prog Sens Physiol. Berlin, Heidelberg, New York, Toronto: Springer; 1985. Functional organization of the fly retina; pp. 1–79. [Google Scholar]
- Henderson RH, Williamson KA, Kennedy JS, Webster AR, Holder GE, Robson AG, FitzPatrick DR, van Heyningen V, Moore AT. A rare de novo nonsense mutation in OTX2 causes early onset retinal dystrophy and pituitary dysfunction. Mol Vis. 2009;15:2442–2447. [PMC free article] [PubMed] [Google Scholar]
- Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SG, Cideciyan AV, Huang Y, Hanna DB, Freund CL, Affatigato LM, Carr RE, Zack DJ, Stone EM, McInnes RR. Retinal degenerations with truncation mutations in the cone-rod homeobox (CRX) gene. Invest Ophthalmol Vis Sci. 1998;39:2417–2426. [PubMed] [Google Scholar]
- Johnston RJ, Jr, Otake Y, Sood P, Vogt N, Behnia R, Vasiliauskas D, McDonald E, Xie B, Koenig S, Wolf R, Cook T, Gebelein B, Kussell E, Nakagoshi H, Desplan C. Interlocked feedforward loops control cell-type-specific Rhodopsin expression in the Drosophila eye. Cell. 2011;145:956–968. doi: 10.1016/j.cell.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomary C, Jones SE. Induction of functional photoreceptor phenotype by exogenous Crx expression in mouse retinal stem cells. Invest Ophthalmol Vis Sci. 2008;49:429–437. doi: 10.1167/iovs.07-0812. [DOI] [PubMed] [Google Scholar]
- Koike C, Nishida A, Ueno S, Saito H, Sanuki R, Sato S, Furukawa A, Aizawa S, Matsuo I, Suzuki N, Kondo M, Furukawa T. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol. 2007;27:8318–8329. doi: 10.1128/MCB.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmik Z. Pax genes in eye development and evolution. Curr Opin Genet Dev. 2005;15:430–438. doi: 10.1016/j.gde.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev Cell. 2003;5:621–633. doi: 10.1016/s1534-5807(03)00293-4. [DOI] [PubMed] [Google Scholar]
- Lerner LE, Peng GH, Gribanova YE, Chen S, Farber DB. Sp4 is expressed in retinal neurons, activates transcription of photoreceptor-specific genes, and synergizes with Crx. J Biol Chem. 2005;280:20642–20650. doi: 10.1074/jbc.M500957200. [DOI] [PubMed] [Google Scholar]
- Leuzinger S, Hirth F, Gerlich D, Acampora D, Simeone A, Gehring WJ, Finkelstein R, Furukubo-Tokunaga K, Reichert H. Equivalence of the fly orthodenticle gene and the human OTX genes in embryonic brain development of Drosophila. Development. 1998;125:1703–1710. doi: 10.1242/dev.125.9.1703. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shen Y, Rest JS, Raymond PA, Zack DJ. Isolation and characterization of a zebrafish homologue of the cone rod homeobox gene. Invest Ophthalmol Vis Sci. 2001;42:481–487. [PubMed] [Google Scholar]
- Martinez-Morales JR, Signore M, Acampora D, Simeone A, Bovolenta P. Otx genes are required for tissue specification in the developing eye. Development. 2001;128:2019–2030. doi: 10.1242/dev.128.11.2019. [DOI] [PubMed] [Google Scholar]
- McDonald EC, Xie B, Workman M, Charlton-Perkins M, Terrell DA, Reischl J, Wimmer EA, Gebelein BA, Cook TA. Separable transcriptional regulatory domains within Otd control photoreceptor terminal differentiation events. Dev Biol. 2010 doi: 10.1016/j.ydbio.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T, Wernet MF, Pistillo D, Mazzoni EO, Teleman AA, Chen YW, Cohen S, Desplan C. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell. 2005;122:775–787. doi: 10.1016/j.cell.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Mishra M, Oke A, Lebel C, McDonald EC, Plummer Z, Cook TA, Zelhof AC. Pph13 and Orthodenticle define a dual regulatory pathway for photoreceptor cell morphogenesis and function. Development. 2010;137:2895–2904. doi: 10.1242/dev.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem. 2000;275:29794–29799. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- Montana CL, Lawrence KA, Williams NL, Tran NM, Peng GH, Chen S, Corbo JC. Transcriptional regulation of neural retina leucine zipper (Nrl), a photoreceptor cell fate determinant. J Biol Chem. 2011 doi: 10.1074/jbc.M111.279026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao T, Leuzinger S, Acampora D, Simeone A, Finkelstein R, Reichert H, Furukubo-Tokunaga K. Developmental rescue of Drosophila cephalic defects by the human Otx genes. Proc Natl Acad Sci U S A. 1998;95:3737–3742. doi: 10.1073/pnas.95.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols LL, 2nd, Alur RP, Boobalan E, Sergeev YV, Caruso RC, Stone EM, Swaroop A, Johnson MA, Brooks BP. Two novel CRX mutant proteins causing autosomal dominant Leber congenital amaurosis interact differently with NRL. Hum Mutat. 2010;31:E1472–1483. doi: 10.1002/humu.21268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson DE. Eye ancestry: old genes for new eyes. Curr Biol. 1996;6:39–42. doi: 10.1016/s0960-9822(02)00417-7. [DOI] [PubMed] [Google Scholar]
- Nilsson DE. Photoreceptor evolution: ancient siblings serve different tasks. Curr Biol. 2005;15:R94–96. doi: 10.1016/j.cub.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Nilsson DE. The evolution of eyes and visually guided behaviour. Philos Trans R Soc Lond B Biol Sci. 2009;364:2833–2847. doi: 10.1098/rstb.2009.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Nishiguchi KM, Friedman JS, Sandberg MA, Swaroop A, Berson EL, Dryja TP. Recessive NRL mutations in patients with clumped pigmentary retinal degeneration and relative preservation of blue cone function. Proc Natl Acad Sci U S A. 2004;101:17819–17824. doi: 10.1073/pnas.0408183101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori Y, Katoh K, Sato S, Muranishi Y, Chaya T, Onishi A, Minami T, Fujikado T, Furukawa T. Analysis of transcriptional regulatory pathways of photoreceptor genes by expression profiling of the Otx2-deficient retina. PLoS One. 2011;6:e19685. doi: 10.1371/journal.pone.0019685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamaneck YJ, Furchheim N, Hejnol A, Martindale MQ, Luter C. Ciliary photoreceptors in the cerebral eyes of a protostome larva. Evodevo. 2011;2:6. doi: 10.1186/2041-9139-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis Neurosci. 2005;22:575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- Plouhinec JL, Sauka-Spengler T, Germot A, Le Mentec C, Cabana T, Harrison G, Pieau C, Sire JY, Veron G, Mazan S. The mammalian Crx genes are highly divergent representatives of the Otx5 gene family, a gnathostome orthology class of orthodenticle-related homeogenes involved in the differentiation of retinal photoreceptors and circadian entrainment. Mol Biol Evol. 2003;20:513–521. doi: 10.1093/molbev/msg085. [DOI] [PubMed] [Google Scholar]
- Ranade SS, Yang-Zhou D, Kong SW, McDonald EC, Cook TA, Pignoni F. Analysis of the Otd-dependent transcriptome supports the evolutionary conservation of CRX/OTX/OTD functions in flies and vertebrates. Dev Biol. 2008;315:521–534. doi: 10.1016/j.ydbio.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta C, Berson EL, Dryja TP. Dominant Leber congenital amaurosis, cone-rod degeneration, and retinitis pigmentosa caused by mutant versions of the transcription factor CRX. Hum Mutat. 2001;18:488–498. doi: 10.1002/humu.1226. [DOI] [PubMed] [Google Scholar]
- Salo E, Pineda D, Marsal M, Gonzalez J, Gremigni V, Batistoni R. Genetic network of the eye in Platyhelminthes: expression and functional analysis of some players during planarian regeneration. Gene. 2002;287:67–74. doi: 10.1016/s0378-1119(01)00863-0. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D’Apice MR, Nigro V, Boncinelli E. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. Embo J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A, Puelles E, Acampora D. The Otx family. Curr Opin Genet Dev. 2002;12:409–415. doi: 10.1016/s0959-437x(02)00318-0. [DOI] [PubMed] [Google Scholar]
- Sohocki MM, Sullivan LS, Mintz-Hittner HA, Birch D, Heckenlively JR, Freund CL, McInnes RR, Daiger SP. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am J Hum Genet. 1998;63:1307–1315. doi: 10.1086/302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz PR, Urbach R, Posnien N, Eriksson J, Kostyuchenko RP, Brena C, Guy K, Akam M, Bucher G, Arendt D. Six3 demarcates the anterior-most developing brain region in bilaterian animals. Evodevo. 2010;1:14. doi: 10.1186/2041-9139-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stornaiuolo A, Bayascas JR, Salo E, Boncinelli E. A homeobox gene of the orthodenticle family is involved in antero-posterior patterning of regenerating planarians. Int J Dev Biol. 1998;42:1153–1158. [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, Sieving PA, Zack DJ. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Wang QL, Wu W, Cook J, Coats C, Xu S, Chen S, Zack DJ, Sieving PA. Leber congenital amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum Mol Genet. 1999;8:299–305. doi: 10.1093/hmg/8.2.299. [DOI] [PubMed] [Google Scholar]
- Tahayato A, Sonneville R, Pichaud F, Wernet MF, Papatsenko D, Beaufils P, Cook T, Desplan C. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev Cell. 2003;5:391–402. doi: 10.1016/s1534-5807(03)00239-9. [DOI] [PubMed] [Google Scholar]
- Umesono Y, Watanabe K, Agata K. Distinct structural domains in the planarian brain defined by the expression of evolutionarily conserved homeobox genes. Dev Genes Evol. 1999;209:31–39. doi: 10.1007/s004270050224. [DOI] [PubMed] [Google Scholar]
- Vandendries ER, Johnson D, Reinke R. orthodenticle is required for photoreceptor cell development in the Drosophila eye. Dev Biol. 1996;173:243–255. doi: 10.1006/dbio.1996.0020. [DOI] [PubMed] [Google Scholar]
- Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development. 2003;130:1281–1294. doi: 10.1242/dev.00343. [DOI] [PubMed] [Google Scholar]
- Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu S, Rivolta C, Li LY, Peng GH, Swain PK, Sung CH, Swaroop A, Berson EL, Dryja TP, Chen S. Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J Biol Chem. 2002;277:43288–43300. doi: 10.1074/jbc.M207952200. [DOI] [PubMed] [Google Scholar]
- Ward A, Liu J, Feng Z, Xu XZ. Light-sensitive neurons and channels mediate phototaxis in C. elegans. Nat Neurosci. 2008;11:916–922. doi: 10.1038/nn.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AF, Reddick AC, Schwartz SB, Ferguson JS, Aleman TS, Kellner U, Jurklies B, Schuster A, Zrenner E, Wissinger B, Lennon A, Shu X, Cideciyan AV, Stone EM, Jacobson SG, Swaroop A. Mutation analysis of NR2E3 and NRL genes in Enhanced S Cone Syndrome. Hum Mutat. 2004;24:439. doi: 10.1002/humu.9285. [DOI] [PubMed] [Google Scholar]
- Xie B, Charlton-Perkins M, McDonald E, Gebelein B, Cook T. Senseless functions as a molecular switch for color photoreceptor differentiation in Drosophila. Development. 2007;134:4243–4253. doi: 10.1242/dev.012781. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Desplan C, Heisenberg M. Contribution of photoreceptor subtypes to spectral wavelength preference in Drosophila. Proc Natl Acad Sci U S A. 2010;107:5634–5639. doi: 10.1073/pnas.0809398107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Wolf R, Desplan C, Heisenberg M. Motion vision is independent of color in Drosophila. Proc Natl Acad Sci U S A. 2008;105:4910–4915. doi: 10.1073/pnas.0711484105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Quantification of OTX expression shown in the western blot in Figure 1C. B) Western blot analysis of FLAG-tagged mutant CRX proteins from adult Drosophila head lysates. All mutant CRX proteins are expressed, albeit at different levels. Similarly to what was previously reported in vertebrate cell culture lines (Nichols et al., 2010), the CRX 138fs48 mutation increases the steady state levels of the protein. The R90W mutant has slightly reduced levels compared to wild-type CRX. Actin serves as a loading control.