Abstract
Fluorescent proteins photoswitchable with non-cytotoxic light irradiation and spectrally distinct from multiple available photoconvertible green-to-red probes are in high demand. We have developed a monomeric fluorescent protein, called PSmOrange2, which is photoswitchable with blue light from an orange (ex./em. at 546 nm/561 nm) to a far-red (ex./em. at 619 nm/651 nm) form. Compared to another orange-to-far-red photoconvertable variant, PSmOrange2 has blue-shifted photoswitching action spectrum, 9-fold higher photoconversion contrast, and up to 10-fold faster photoswitching kinetics. This results in the 4-fold more PSmOrange2 molecules being photoconverted in mammalian cells. Compared to common orange fluorescent proteins, such as mOrange, the orange form of PSmOrange has substantially higher photostability allowing its use in multicolor imaging applications to track dynamics of multiple populations of intracellular objects. The PSmOrange2 photochemical properties allow its efficient photoswitching with common two-photon lasers and, moreover, via Förster resonance energy transfer (FRET) from green fluorescent donors. We have termed the latter effect a FRET-facilitated photoswitching and demonstrated it using several sets of interacting proteins. The enhanced photoswitching properties of PSmOrange2 make it a superior photoconvertable protein tag for flow cytometry, conventional microscopy, and two-photon imaging of live cells.
Keywords: PSmOrange, mOrange, photoactivation, two-photon, far-red fluorescent protein
1. INTRODUCTION
A subclass of fluorescent proteins (FPs), termed irreversibly photoswitchable fluorescent proteins (PSFPs), is widely used for tracking intracellular proteins, organelles and individual cells 1,2. PSFPs are switched from one fluorescent color to another upon irradiation with light of a specific wavelength. PSFPs, together with so called photoactivatable FPs (PAFPs) (those which are initially dark but become fluorescent after irradiation with light), are important probes for super-resolution microscopy techniques 1. The majority of PSFPs, including Dendra2 3, mEos2 4, Kaede 5, mKikGR 6, mClavGR2 7 and their derivatives, change its fluorescent color from green to red upon irradiation with rather phototoxic violet light (390–410 nm). The blue-to-green proteins photoswitchable with violet light, PSCFP and PSCFP2, are also available 3.
Recently, a monomeric protein called PSmOrange has been developed from mOrange 8. PSmOrange is initially orange but becomes far-red after irradiation not with violet but with blue-green light (480–540 nm). PSmOrange is the first PSFP, which can be efficiently photoswitched with non-phototoxic visible light. The photoconverted far-red PSmOrange chromophore contains an N-acylimine substituent in the GFP-like chromophore core 8. The involvement of the N-acylimine functionality in the dihydrooxazole ring and the cleavage of the polypeptide chain results in a more efficient conjugation of the C=O group to the rest of the chromophore and in a larger red shift of the PSmOrange absorbance maximum and emission as compared to a DsRed-like chromophore. However, the photoconversion rate and photoconversion efficiency of PSmOrange are highly dependent on concentration of exogenous oxidants in the protein environment. When expressed in mammalian cells, PSmOrange could be only efficiently photoconverted using the high light intensities provided by 60–100x objective lens with a high numerical aperture. The photocoversion at the low light intensities was limited.
Two-photon (2P) microscopy with use of FPs is a powerful tool for imaging cells and tissues 9. Deeper tissue penetration, higher 3D resolution, reduced out-of-focus photobleaching and less autofluorescence are advantages of 2P microscopy. A 2P photoconversion of PSFPs is advantageous to the conventional one-photon (1P) photoconversion 10. With 1P photoconversion, the illuminated region within thick samples forms cones above and below the point of focus resulting in the loss of ability to perform selective photoswitching. With the 2P photoconversion, confinement of the light within all three spatial dimensions is possible. The 2P photoconversion of photoactivatable GFP (PAGFP) 11, EosFP 10 and KikGR 12 has been demonstrated but was not tested for PSmOrange that exhibit different photochemical behavior.
Förster resonance energy transfer (FRET) is a widely used technique to study protein-protein interactions in cells 13. FRET is a physical phenomenon whereby energy is transferred from an excited fluorophore, called the donor, to a nearby chromophore, the acceptor, via non-radiative dipole–dipole coupling. FRET occurs between two fluorophores only when the distance separating them is less than 10 nm 14,15. The FRET pairs most commonly used employ cyan and yellow FPs. It has been shown that irreversibly photoswitchable PSCFP 16 and reversibly photoswitchable rsTagRFP 17 can be used as the FRET donor and acceptor, respectively, in FRET pairs with yellow FPs. Previously, the FRET method called a FRET-sensitized acceptor photobleaching has been developed. It utilizes the measurement of photobleaching kinetics of a photolabile FRET acceptor dye excited by a donor dye via FRET 18,19.
In a similar way, when an acceptor of the FRET pair is PSFP, a non-radiative energy from the donor could be used to photoconvert PSFP. In standard so called sensitized-emission FRET imaging, great pains are taken to capture the transient FRET signal and isolate it from the donor signal that unavoidably bleeds through into the FRET imaging channel. The use of a PSFP as an acceptor would create a permanent and easily separable indicator of the presence of FRET. Despite these attractive qualities, a suitable FRET pair has not yet been reported in the literature. This is because all other existing PSFPs photoconvert by violet light for which there are no FP donors. Having a PSFP photoconvertible with a low-intensity blue-green light would enable this FRET scenario.
In this work, we have applied PSmOrange to mutagenesis to increase its rate of photoconversion, reduce photoswitching on oxidative environment, and improve other photoconversion properties. The resulted orange-to-far-red photoswitchable variant was characterized in purified form and in live mammalian cells, and further utilized as a FRET acceptor to demonstrate, for the first time, a FRET-facilitated acceptor photoswitching.
2. EXPERIMENTAL PROCEDURES
2.1. Mutagenesis and screening of libraries
A PSmOrange gene was inserted into a pBAD/His-B vector (Invitrogen) as a BglII-EcoRI fragment. For simultaneous mutagenesis at several positions, we applied an overlap-extension approach 20. Random mutagenesis was performed using either a GeneMorph II Random Mutagenesis Kit (Stratagene) or a Diversity PCR Random Mutagenesis Kit (Clontech) under conditions resulting in a mutation frequency of up to 16 mutations per 1,000 base pairs. After mutagenesis, a mixture of the mutated genes was electroporated into LMG194 bacterial host cells (Invitrogen).
Libraries of 106–108 independent clones of the PSmOrange mutants were photoswitched with a custom built 489 nm LED array (adjusted to 93 mW cm−2), screened using a MoFlo XDP FACS (Beckman Coulter) and followed by colony imaging with a Leica MZ16F fluorescence stereomicroscope, as previously described 21. After each round of FACS screening, typically 10–20 of the best photoswitchable candidate clones were sequenced, purified and characterized before the next round of mutagenesis.
2.2. Characterization of purified proteins
The PSmOrange and PSmOrange2 proteins with polyhistidine tags were expressed in LMG194 bacteria grown in an RM medium and supplemented with 0.002% arabinose for 24–48 h at 37°C and then purified using a Ni-NTA agarose (Qiagen). For spectroscopy, photoswitching of purified proteins was performed with the 489 nm LED array (adjusted to 280 mW cm−2) in 1.5 ml transparent Eppendorf tube on ice. Excitation and emission spectra of recombinant proteins were measured with a FluoroMax-3 spectrofluorometer (Jobin Yvon). For absorbance measurements, a Hitachi U-3010 spectrophotometer was used.
To determine the extinction coefficients of orange forms, we relied on measuring the mature chromophore concentration, as previously described for mOrange 22. For this, the purified PSmOrange and PSmOrange2 proteins were alkali-denatured. It is known that the extinction coefficient of the GFP-like chromophore is 44,000 M−1cm−1 at 447 nm in 1 M NaOH 23. Based on the absorbance of the native and denatured proteins, molar extinction coefficients for the native states were calculated. To determine extinction coefficients of far-red forms, we photoswitched the PSmOrange and PSmOrange2 purified proteins with the 489 nm LED array (adjusted to 280 mW cm−2) for 5 min in the presence of 5 mM or 0.25 mM K3Fe(CN)6, respectively. Under this oxidant concentration the maximally achievable far-red fluorescence was not dependent on the 489 nm light intensity above 40 mW. Extinction coefficients were calculated based on a comparison between the absorbance decrease of orange forms (at 548 and 546 nm for PSmOrange and PSmOrange2, respectively) and the absorbance increase of far-red forms (at 634 and 619 nm for PSmOrange and PSmOrange2, respectively). To determine quantum yields, we compared the fluorescence intensities of orange and far-red forms at pH 8.5 to the fluorescence intensities of equally absorbing amounts of mOrange in the orange form (quantum yield is 0.69 22) and mNeptune (quantum yield is 0.2 24), respectively.
Equilibrium pH titrations were performed using a series of buffers (100 mM NaOAc, 300 mM NaCl for pH 2.5–5.0, and 100 mM NaH2PO4, 300 mM NaCl for pH 4.5–10.0).
Photobleaching kinetics was measured using purified proteins in phosphate buffered saline (PBS) at 1 mg ml−1 in aqueous drops suspended in oil using an Olympus IX81 inverted microscope equipped with a 200 W metal halide arc lamp (Prior), a 100x 1.4 NA oil immersion objective lens (UPlanSApo, Olympus), 540/20 nm excitation and 575/30 nm emission filters for orange forms, and 605/40 nm excitation and 640LP nm emission filters for far-red forms. The microscope was operated with SlideBook 4.2 software (Intelligent Imaging Innovations). Light power densities were measured at a rear focal plane of the objective lens, and then a light power at the sample was estimated. Photoswitching kinetics was measured using the above conditions with a 480/40 nm filter for photoswitching. The data were normalized to a spectral output of the lamp, transmission profiles of the filters and dichroic mirror, absorbance spectra of the respective proteins, and their quantum yields.
To study protein maturation, LMG194 bacteria transformed with the PSmOrange or PSmOrange2 genes were grown in an RM medium supplemented with ampicillin at 37°C overnight. The next morning, we diluted bacterial cells to optical density 1.0 at 600 nm, and 0.2% arabinose was added. Upon induction of protein expression, bacterial cultures were grown at 37°C in 50 ml tubes filled to the brim and tightly sealed to restrict oxygen supply. After 2 hours, the cultures were centrifuged in the same tightly closed tubes. After opening the tubes, we purified the proteins using the Ni-NTA resin within 30 min, with all procedures and buffers at or below 4°C. Protein maturation occurred in PBS at 37°C. Orange fluorescence signal of the proteins was monitored using the FluoroMax-3 spectrofluorometer.
2.3. Mammalian plasmids and cell culture
To design pPSmOrange2-C1 and pPSmOrange2-α-Tubulin plasmids, the PSmOrange2 gene was swapped with the EGFP gene in the pEGFP-C1 and pEGFP-α-Tubulin vectors (Clontech), respectively. To design a pH2B-PSmOrange2 plasmid, the PSmOrange2 gene was swapped with the PAmCherry1 gene in the pH2B-PAmCherry1 plasmid 21. To design pVimentin-PSmOrange2, pKeratin-PSmOrange2 and pPaxillin-PSmOrange2 plasmids, the PSmOrange2 gene was swapped with the mTagBFP gene in the pVimentin-mTagBFP, pKeratin-mTagBFP and pPaxillin-mTagBFP vectors, respectively (provided by M. Davidson). To design a pPSmOrange2-N1 plasmid, the PSmOrange2 gene was PCR amplified as an AgeI-NotI fragment and swapped with the EGFP gene in a pEGFP-N1 vector (Clontech). To design a pT-Sapphire-L7-PSmOrange2-N1 plasmid (where L7 is the linker consisted of 7 amino acids: -GGSGGRS-), the T-sapphire and PSmOrange2 were amplified as an AgeI-BglII fragment and a BglII-NotI fragment, respectively and inserted into an AgeI-NotI fragment of a pEGFP-N1 plasmid instead of the EGFP gene. To design a pC4-RHE-PSmOrange2 plasmid encoding a PSmOrange2-FRB fusion protein, the PSmOrange2 gene was amplified as an XbaI fragment and inserted into the respective site of the pC4-RHE plasmid (Ariad Pharmaceuticals). To design a pGrb2-PSmOrange2 plasmid, the BamHI-HindIII fragment containing Grb2 gene was cut out of pGrb2-EYFP vector (provided by A. Sorkin) and inserted into the respective sites of the pPSmOrange2-N1 plasmid. To design a pC4M-F2E-T-Sapphire plasmid encoding a Myr-T-Sapphire-FKBP2 fusion protein (where Myr is the myristoilation signal that directs the protein to the cytoplasmic face of the membrane), the T-Sapphire gene was amplified as an XbaI fragment and inserted into the respective site of the pC4M-F2E plasmid (Ariad Pharmaceuticals). To design a pEGFR-T-Sapphire plasmid, a SacII-BsrGI fragment containing the T-Sapphire gene (provided by O. Griesbeck) was PCR amplified and swapped with the mRFP1 gene in the pEGFR-mRFP1 plasmid 25.
HeLa cells were grown in a Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum and 2 mM glutamine (all from Invitrogen). MTLn3 cells were grown in alpha-Modified Eagle Medium containing 5% fetal bovine serum and 2 mM glutamine (all from Invitrogen). Cells were grown in #1.5 glass bottom culture dishes (MatTek). Plasmid transfections were performed with an Effectene (Qiagen). Live HeLa cells were photoswitched and imaged in PBS or in a dye-free DMEM medium without serum (Invitrogen).
2.4. Imaging of mammalian cells
Wide-field epifluorescence imaging of live mammalian cells was performed 48–72 h after transfection. Cells were imaged using an Olympus IX81 inverted microscope described above. A 480/40 nm filter at 1,050 W cm−2 (here and below the light power densities are estimated at the sample) was used for photoswitching of PSmOrange2, and a 415/30 nm filter (900 W cm−2) was used for photoswitching of PSmOrange2 in the fusion construct T-Sapphire-L7-PSmOrange2. The 540/20 nm excitation and 575/30 nm emission filters (468 W cm−2) were used to image orange forms, and the 605/40 nm excitation in combination with 640LP nm emission filters (360 W cm−2) to image far-red forms. The 415/30 nm excitation and 480/40 nm emission filters (360 W cm−2) were used to image T-Sapphire. All filter sets were from Chroma. A power density of light was estimated in a focal plane of a 100x 1.4 NA oil immersion objective lens (UPlanSApo, Olympus).
For super-resolution imaging, we used an interferometric photoactivated localization microscopy (iPALM) method 26, which combines photoactivated localization microscopy with simultaneous multi-phase interferometry of photons from each fluorescent molecule. This approach allows imaging a high density of specific fluorescence-tagged molecules with three-dimensional nanoscale. 25×45 nm gold nanorods (NanoPartZ) immobilized to the coverglass were used as fiducials for calibration and drift correction. A paxillin-PSmOrange2 fusion protein was expressed in HeLa cells sparsely plated on fibronectin coated and fiducially marked cover glasses and fixed for imaging. The vertical coordinate calibration was performed before each cell was imaged. For each cell, 15,000–25,000 image triplets were acquired, with 50 ms per frame excitation/exposure time. The excitation was performed using a pulsed 637 nm laser (Opto Engine) with a power density of ~1,000 W cm−2. PSmOrange2 photoconversion was performed using a 488 nm pulsed laser (CrystaLaser). The photoconversion pulses varied from 3 to 20 ms; and the photoconversion power was ~100 W cm−2.
2.5. Two-photon characterization
2PE spectra of PSmOrange2 before and after photoswitching were measured using a custom built microscope system equipped with an optical parametric oscillator (OPO) (Opal, Spectra Physics), pumped with an automatically tunable Ti-Sapphire laser (Mai-Tai, Spectra Physics) 27. This system is capable of delivering over 100 mW of femtosecond pulsed light in the range of 780–1040 nm and 1,100–1,600 nm. Sample position is controlled by an automated XY stage (MS2000, ASI) driven by custom software written in LabVIEW. An Olympus 20x 0.95 NA water immersion objective lens with a working distance of ~3 mm is used throughout.
The process for collecting the two-photon excitation (2PE) spectrum consisted of the following steps. A pellet of bacteria expressing recombinant proteins was applied onto coverslips that were then placed on the microscope stage. For each excitation wavelength of interest, the fluorescence signal was split into four different spectral channels using Semrock dichroic and bandpass filters and detected with Hamamatsu PMTs. The source laser wavelength was then manually changed and the process repeated until the fluorescence signal at each wavelength was recorded.
To study 2PE induced photoswitching in both bacteria and mammalian MTLn3 cells, samples expressing recombinant proteins were mounted onto coverslips that were then placed on the microscope stage. Pre-photoswitching images for both the orange and the far-red form were acquired using 1,035 nm and 1,200 nm 2PE respectively. Photosignals were collected using a 580/60 nm emission filter for the orange channel and a 697/75 nm emission filter for the far-red channel. Photoswitching was then accomplished by tuning the femtosecond laser source to 900nm and scanning the beam at 1 frame per second from 30 s to 5 min. Finally, post-photoswitching images were acquired by tuning the laser source again to 1,035 nm and 1,200 nm to acquire the orange and far-red images, respectively. Care was taken to ensure that the fluorescence measurement was performed at the same laser intensity and PMT gain setting for each measurement. Laser intensity was measured at the input to the microscope and is typically ~2.7 times the intensity applied at the sample.
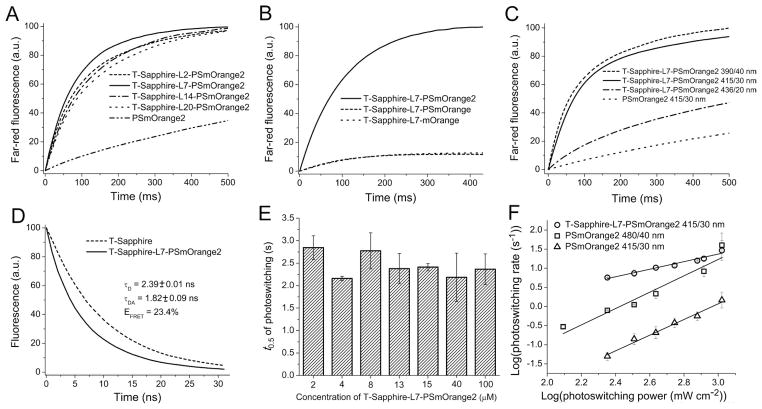
2.6. Characterization of PSmOrange2 in FRET
Streaks of bacteria expressing either PSmOrange2 alone, T-Sapphire-L2-PSmOrange2, T-Sapphire-L7-PSmOrange2, T-Sapphire-L14-PSmOrange2 and T-Sapphire-L20-PSmOrange2 (where L2, L7, L14 and L20 are the linkers consisted of 2, 7, 14 and 20 amino acids, respectively: -RS-, -GGSGGRS-, -EFGGSGSDEVDKGT-, and –EFGGSGSDEVDKLGGSGSGT-) or T-Sapphire-L7-PSmOrange2 purified protein in the presence of 0.25 mM K3Fe(CN)6 were photoconverted using Olympus IX81 inverted microscope equipped with a 200 W metal halide arc lamp (Prior), a 100x 1.4 NA oil immersion objective lens (UPlanSApo, Olympus), and 415/30 nm filter (900 W cm−2). To determine the light intensity dependence of the initial photoswitching rate for PSmOrange2 alone 480/40 nm and 415/30 nm bands were used. To determine the light intensity dependence of the initial photoswitching rate for T-Sapphire-L7-PSmOrange2 fusion 415/30 nm wavelength band was used. To characterize rapamycin-induced interaction between Myr-T-Sapphire-FKBP2 and PSmOrange2-FRB fusion proteins in the membrane of HeLa cells, 24 h after transfection 100 nM of rapamycin were added and incubated further for 24 h at 37°C. After that, images were acquired using the same settings described above for bacteria. To characterize EGF-induced interaction between EGFR-T-Sapphire and Grb2-PSmOrange2 in the membrane of HeLa cells, 48 h after transfection cells were incubated in DMEM without fetal bovine serum for 2 h at 37°C, and then 100 ng/ml of EGF were added. After that, dishes were placed for 20 min at 4°C and then for 15 min at 37°C and the images were acquired using the same settings described above for bacteria. Measurements of fluorescence lifetime, τ, were performed at room temperature on IX71 Olympus microscope using 10x dry objective lens equipped with a PicoStar (LaVision, Germany) time-gated camera and a 440 nm 5 mW picosecond pulsed laser (PicoQuant, Germany). Streaks of bacteria expressing either PSmOrange2 alone or T-SapphireL7-PSmOrange2 fusions were used. A beam width was adjusted using a 12.5x telescope to provide even illumination of the samples and intensity was regulated using ND filters. Laser scattering and background signal was removed from the emission signal with band pass filter and the remaining signal passed onto the time gated CCD intensifier of the PicoStar system. FRET efficiency, E, was calculated as E = (τD − τDA)/τD, where τD and τDA are lifetimes of the donor without and with acceptor, respectively.
3. RESULTS
3.1. Development of a fast photoswitchable orange fluorescent protein
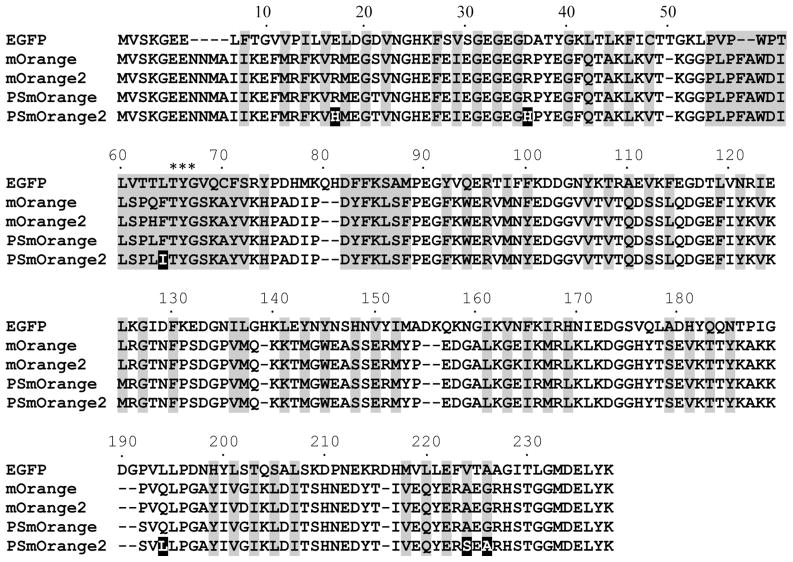
A PSmOrange’s gene was used as the template to develop a fast photoswitchable variant. First, PSmOrange was subjected to five rounds of random mutagenesis. Bacterial libraries of 106–108 clones were applied to high-throughput screening using a fluorescence-activated cell sorter (FACS). After each round, 10–20 clones with improved properties were sequenced, purified and spectroscopically characterized. A mixture of several of the best variants was then used as a template for the next round of mutagenesis. After five rounds of screening, three variants with R17H/R36H/F64I/A70P/Q194L, R17H/R36H/F64I/T183S/Q194L/G226A and R17H/R36H/F64I/Q194L/A224S substitutions in the PSmOrange sequence were selected. Next, we performed a site-specific saturated mutagenesis of these mutants at the identified positions, such as 63, 64, 70, 224 and 226 (see Figure 1 for the alignment), using an overlap extension mutagenesis technique 20. The subsequent screening has resulted in a PSmOrange/R17H/R36H/F64I/Q194L/A224S/G226A variant, which was named as PSmOrange2 (Figure 1).
Figure 1.
Alignment of the amino acid sequences for the EGFP, mOrange, mOrange2, PSmOrange and PSmOrange2 proteins. Alignment numbering follows that of EGFP. Residues buried in the protein β-barrel fold are shaded. Asterisks indicate residues that form the chromophore. Mutations in PSmOrange relative to PSmOrange are indicated in white on black background.
Based on an analysis of the variants obtained in several rounds of the random mutagenesis, we have found that an Ile64 amino acid residue played the critical role for the photoswitching rate and contrast, but decreased the brightness of the orange form. The Ser224 and Ala226 residues were responsible for the increase of quantum yield of the orange form in PSmOrange2.
Taking into account the important role of Ile64 for photoswitching properties, we thought that its surrounding residues could potentially affect photoswitching and brightness of the far-red form. To test this, the positions 16, 44, 46, 123, 125 and 220 were selected for site-specific mutagenesis, based on the crystal structure of mOrange 28. We have found that a Q220L substitution in PSmOrange2 had higher brightness of both orange and far-red forms, but the substantially lower photoswitching contrast. Mutations at the other surrounding positions did not result in better variants.
3.2. Properties of purified PSmOrange2
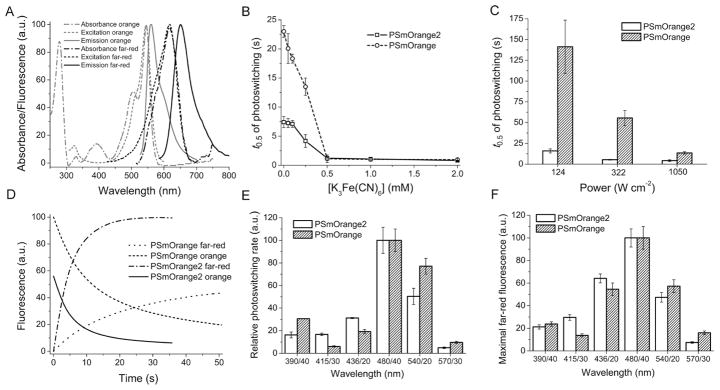
Before photoswitching, PSmOrange2 exhibited orange fluorescence with excitation (absorbance)/emission peaks at 546 nm/561 nm, similar to the parental PSmOrange (Figure 2A). After photoswitching with 489 nm light, far-red fluorescence with the excitation (absorbance)/emission peaks at 619 nm/652 nm developed (Figure 2A). These peaks were ~10 nm blue-shifted when compared to that of far-red form of the original protein (Table 1).
Figure 2.
Properties of the purified PSmOrange2 protein. (A) Absorbance, excitation and emission spectra of PSmOrange2 before and after photoswitching with 489 nm LED array. (B) Photoswitching half-times (t0.5) for PSmOrange2 and PSmOrange at indicated K3Fe(CN)6 concentrations and 1,050 mW cm−2 light power density. (C) Photoswitching half-times (t0.5) for PSmOrange2 and PSmOrange at indicated light power densities and 0.25 mM of K3Fe(CN)6. (D) Photoswitching kinetics for orange and far-red forms of PSmOrange and PSmOrange2. (E) Initial rates of photoswitching for PSmOrange and PSmOrange2 at indicated photoswitching wavelength bands normalized to 100% at 480/40 nm. (F) Maximal far-red fluorescence PSmOrange2 and PSmOrange achieved at indicated photoswitching wavelength bands normalized to 100% at 480/40 nm. In (B–D) the 480/40 nm wavelength band was used for photoswitching. In (D–F) the 0.25 mM of K3Fe(CN)6 and 1,050 mW cm−2 light power density were applied. The power densities were estimated at the sample. Error bars, SD.
Table 1.
Comparison of spectral and photochemical properties of the purified PSmOrange2, PSmOrange and mOrange proteins.
| Protein | mOrange | PSmOrange | PSmOrange2 | |||
|---|---|---|---|---|---|---|
| Orange form | Far-red form | Orange form | Far-red form | Orange form | Far-red form | |
| Absorbance (nm) | 546 | 631 | 548 | 634 | 546 | 619 |
| Emission (nm) | 562 | 662 | 565 | 662 | 561 | 651 |
| Extinction coefficient (M−1 cm−1) | 71,000 | 17,200 | 113,300 | 32,700 | 51,000 | 18,900 |
| Quantum yield | 0.69 | 0.19 | 0.51 | 0.28 | 0.61 | 0.38 |
| Brightness relative to common EGFP (%) | 148 | 10 | 176 | 28 | 95 | 23 |
| pKa | 6.5 a | ND | 6.2 ± 0.1 | 5.6 ± 0.1 | 6.6 ± 0.1 | 5.4 ± 0.1 |
| Photoswitching half- time, t0.5 (s) b | 12 ± 6 | 13.5 ± 1.5 | 4.2 ± 1.1 | |||
| Photoswitching contrast (fold) c | 2,800 ± 200 | 10,700 ± 500 | 96,000 ± 3,800 | |||
| Maturation half-time at 37°C (h) | 2.5 | 1.6 | 3.5 | |||
| Photobleaching t0.5(s) d | (0.65 ± 0.39) | (17 ± 4) | 103 ± 10 (15 ± 5) | 1302 ± 120 (49 ± 8) | 86 ± 2 (12± 4) | 667 ± 25 (20± 1) |
Data from 22.
Determined at 1,050 W cm−2 at the sample.
Determined as the product of the far-red fluorescence increase and the orange fluorescence decrease after photoswitching.
Determined at 133 (or 1,130 for the data in brackets) W cm−2 for the orange form and 103 (or 870 for the data in brackets) W cm−2 for the far-red form at a sample. The raw data were normalized to the spectral output of the lamp, transmission profile of the filter and dichroic mirror, absorbance spectra and quantum yields of the proteins, as described 45. ND, not determined. Errors, SD.
It has been shown that PSmOrange photoswitching in live mammalian cells occurred faster than for the purified protein without adding oxidants. This suggested that the intracellular redox environment affects the photoconversion process 8. The photoswitching half-time of PSmOrange in mammalian cells corresponded to that of the purified protein with 0.25 mM oxidant such as potassium ferricyanide (K3Fe(CN)6) was added 8. Therefore, we further compared effect of various K3Fe(CN)6 concentrations on the photoswitching rates of PSmOrange2 and PSmOrange. We found that at a high intensity of the photoconversion light PSmOrange2 photoswitches 3-fold faster than PSmOrange when incubated with low K3Fe(CN)6 concentrations of 0–0.25 mM (Figures 2B and S1). At the higher concentrations, the half-times of PSmOrange and PSmOrange2 photoconversion were the same and became independent of the oxidant concentration. In order to match the effect of intracellular redox agents on PSmOrange2 behavior, we further characterized purified PSmOrange2 in the presence of 0.25 mM K3Fe(CN)6.
Both 3-fold and 9-fold decrease in the intensity of photoswitching light resulted in ~10-fold faster photoswitching of PSmOrange2 compared to PSmOrange (Figure 2C). In other word, at the lower light intensities the larger differences in the photoswitching half-times were observed. A decrease in the intensity of photoswitching light also resulted in a decrease in the maximal far-red fluorescence (Figure S2).
A kinetics of the increase of the far-red forms and decrease of the orange forms for PSmOrange and PSmOrange2 at the highest tested light intensity is presented in Figure 2D. For both PSmOrange2 and PSmOrange the highest rate of photoswitching and the maximal far-red fluorescence intensity (also called as an action spectrum) were observed after irradiation with blue-green light (480/40 and 540/20 nm) (Figures 2E,F). Also, the photoswitching action spectrum for PSmOrange2 was blue-shifted compared to parental PSmOrange.
A molecular brightness (a product of extinction coefficient and quantum yield) of PSmOrange2 was 1.9-fold and 1.2-fold lower than that of PSmOrange before and after photoswitching, respectively (Table 1). However, a photoswitching orange-to-far-red contrast of PSmOrange2 was 9-fold higher than that for PSmOrange (Table 1). This indicates that the PSmOrange2 photoconversion is substantially more efficient than for the parental protein. PSmOrange2 has pKa values of 6.6 and 5.4 before and after photoswitching, respectively, that are similar to PSmOrange and mOrange (Table 1, Figure S3). Similarly to mOrange, purified PSmOrange2 exhibits monomeric behavior (Figure S4). The orange and far-red forms of PSmOrange2 were found to be less photostable than for PSmOrange but substantially more photostable than for commonly used mOrange (Table 1).
3.3. Behavior of PSmOrange2 in mammalian cells
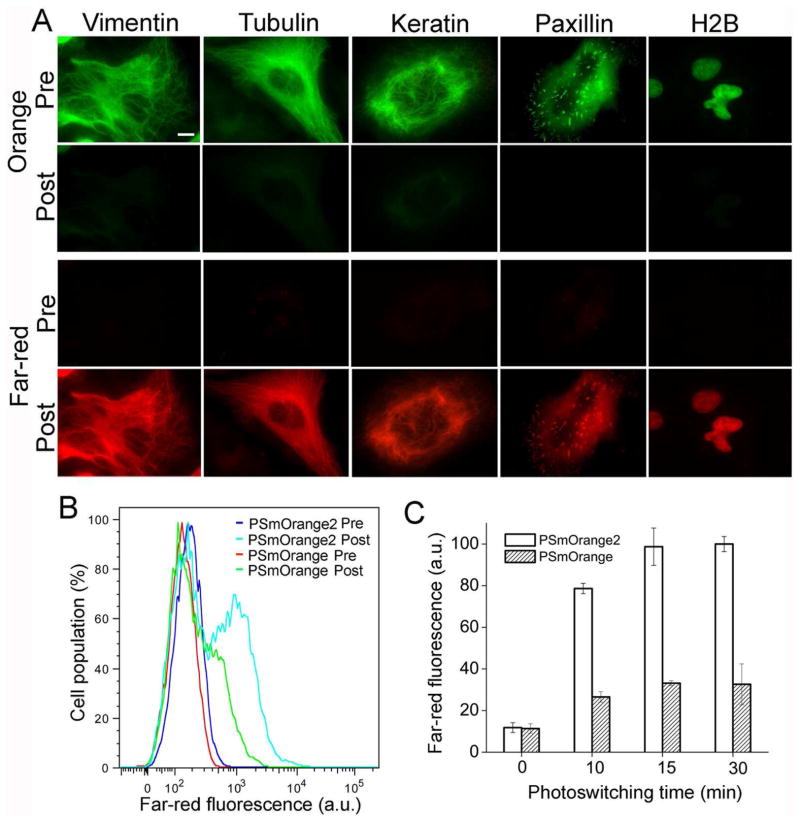
To study protein behavior in mammalian cells PSmOrange2 was fused to several cellular proteins. The PSmOrange2 fusions with vimentin, α-tubulin, keratin, paxillin and histone H2B properly localized in live HeLa cells and did not affect cell division (Figure 3A).
Figure 3.
PSmOrange2 in live HeLa cells. (A) Cells expressing PSmOrange2 fusions with indicated proteins before (Pre) and after (Post) photoswitching. Images in orange (green) and farred (red) channels are shown. Scale bar, 10 μm. (B) Flow cytometry analysis of cell populations expressing PSmOrange2 (blue and cyan lines) and PSmOrange (red and green lines) before (Pre) and after (Post) photoswitching. The 638 nm excitation laser and 660/20 nm emission filter were used. (C) Mean far-red fluorescence intensities of PSmOrange2 and PSmOrange expressing cells at indicated photoswitching times. The 647 nm excitation laser and 670/30 nm emission filter were used. Error bars, SD.
To compare brightness of the photoconverted far-red forms in mammalian cells PSmOrange2 and PSmOrange expressed in HeLa cells were photoconverted for various time periods with 489 nm light. The far-red fluorescence of PSmOrange2 was 3-fold brighter after photoconversion at all tested times (Figure 3B–C). Since the molecular brightness of the photoconverted PSmOrange2 is 78% of that of PSmOrange, approximately 4-fold more PSmOrange2 molecules were photoswitched in mammalian cells. Thus, the faster and more efficient photoconversion of PSmOrange2 resulted in substantially brighter live cells.
PSmOrange2 was fused to paxillin to characterize it for use in iPALM super-resolution imaging (Figure S5) 26. The paxillin-PSmOrange2 fusion protein was expressed in HeLa cells and imaged with TIRF illumination under 488 nm photoconversion and 637 nm excitation. As expected, paxillin formed a thin layer inside focal adhesion structure, confirming the biological viability of the paxillin-PSmOrange2 construct (Figure S5A and S5B). The lateral and axial localization accuracy was determined to be 10 nm and 15 nm, respectively, corresponding to 25 nm and 38 nm in full-width at half maximum (FWHM) (Figure S5C). Photon statistics of the photoconverted far-red PSmOrange2 molecules showed that they have properties sufficient for iPALM (Figure S5D).
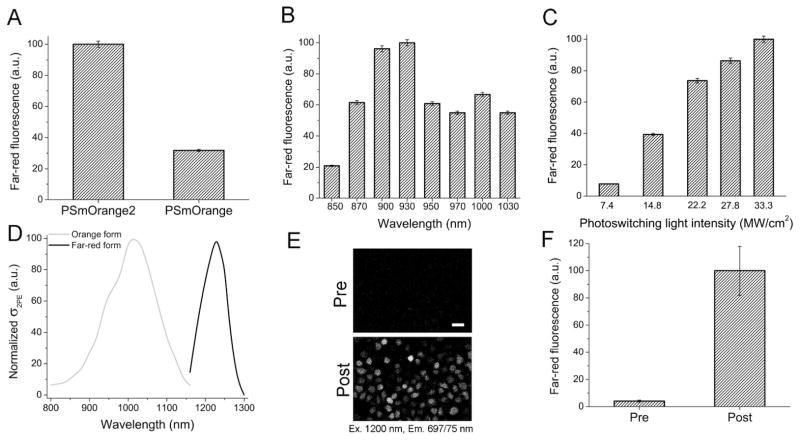
3.4. Two-photon photoconversion of PSmOrange2
To study effect of 2P irradiation on PSmOrange2 photoconversion we used a 2P microscope equipped with the standard Ti-Sapphire and OPO 2P light sources. After 5 min of irradiation at 900 nm, the PSmOrange2 sample showed 3-fold brighter far-red fluorescence detected with 1,200 nm 2PE (Figure 4A). To characterize dependence of 2P photoconversion of PSmOrange2 on wavelength, we scanned different wavelengths in the range from 850 nm to 1,030 nm (Figure 4B) while keeping the laser power and photodetector gains constant. The most efficient 2P photoconversion was observed using wavelengths from 900 to 930 nm. We then examined the dependence of the efficiency of 2P photoconversion on the laser power at 900 nm (Figure 4C). Increasing laser power from 20 to 90 mW resulted in a 13-fold increase in far-red fluorescence. We determined the spectral dependences of 2PE for the orange form of PSmOrange2 in the range of 800–1,160 nm and for the far-red form of PSmOrange2 in the range of 1,160–1,300 nm (Figure 4D). The maxima of 2PE were observed at 1,014 nm and 1,228 nm for the orange and far-red forms, respectively. Comparison of 2PE with 1P excitation spectra (plotted at the double wavelength) yielded blue shifts of 78 nm and 10 nm for the orange and far-red forms, respectively. It has been noted previously for FPs with the anionic chromophores such as mOrange, EGFP, Citrine, DsRed2, TagRFP and others that there is a distinct blue shift of the 2PE relative to the 1P band 9. This phenomenon was ascribed to an enhancement of certain vibronic transitions in 2P spectra 29.
Figure 4.
Properties of PSmOrange2 using a two-photon excitation (2PE). (A) Far-red fluorescence intensity of PSmOrange2 and PSmOrange achieved by 2P photoswitching using 40 mW of 900 nm light scanning at 1 frame per second for 5 min. (B) Far-red fluorescence intensity achieved at indicated wavelengths by 2P photoswitching using 90 mW of 900 nm light scanning at 1 frame per second for 30 s. (C) Far-red fluorescence intensity after 2P photoswitching with the indicated laser powers at 900 nm and scanning at 1 frame per second. (D) Relative 2PE spectra (σ2PE) of orange and photoswitched far-red PSmOrange2 forms. (E) 2P image s of MTLn3 live cells expressing PSmOrange2 before (Pre) and after (Post) photoswitching using 90 mW of 2PE at 900 nm for 40 s. 1,200 nm 2PE and a 697/75 nm emission filter were used to generate and collect the fluorescence signal. Scale bar, 50 μm. (F) Mean far-red fluorescence intensities of PSmOrange2 expressing cells shown in (E). Error bars, SD.
Using the chosen optimal conditions of 2P photoconversion of PSmOrange2, a stable MTLn3 cell line expressing PSmOrange2 was photoconverted with 900 nm using 90 mW light power for 40 s (Figure 4E). As a result, the far-red fluorescence of PSmOrange2 expressed in live mammalian cells excited at 1,200 nm increased by ~24-fold (Figure 4F). Thus, PSmOrange2 can be efficiently photoconverted and then imaged with 2P light sources providing a background to its use for deep-tissue imaging for which 2P microscopy is commonly used.
3.5. Evaluation of PSmOrange2 as a photoswitchable FRET acceptor
Irradiation of PSmOrange2 with violet light (390/40 nm and 415/30 nm) had a 6-fold lower rate of photoswitching (Figure 2E) and resulted in a 3.4–4.7-fold lower maximal far-red fluorescence than irradiation with blue-green light of the same intensity (Figure 2F). We therefore next tested if proximity of a FRET donor (one which efficiently absorbs in violet range) to PSmOrange2 could accelerate its photoconversion rate.
T-Sapphire is a green FP with a large Stokes shift with an excitation maximum at 399 nm and an emission maximum at 520 nm. Its emission spectrum has the substantial overlap with the photoswitching action spectrum of PSmOrange2 (Figure S6). We next fused T-Sapphire with PSmOrange2 using amino acid linkers of various length and studied kinetics of photoswitching of these fusion constructs with 415/30 nm light (Figure 5A). We compared the initial rates of the PSmOrange2 photoswitching in the fusions with that of non-fused PSmOrange2 (Table S1). The former ones were 10–15-fold higher than for the control. An optimal linker between two FPs consisted of seven amino acids (T-Sapphire-L7-PSmOrange2 construct in Figure 5A). The longer and shorter linkers resulted in 1.2–1.5-fold decrease of the photoswitching rate. These data suggested that FRET from T-Sapphire substantially accelerated PSmOrange2 photoswitching caused by 415/30 nm light. We have termed this phenomenon a FRET-facilitated photoswitching. The fusion constructs of T-Sapphire with mOrange and with parental PSmOrange resulted in 11–12-fold lower photoswitching rates (Figure 5B, Table S1). The accelerated photoconversion of PSmOrange2 in the fusion was only observed at the 390/40 nm and 415/30 nm wavelength bands, which were close to the excitation maximum of T-Sapphire (Figure 5C, Table S1).
Figure 5.
Properties of PSmOrange2 as a photoswitchable FRET acceptor. (A) Kinetics of farred fluorescence intensities for PSmOrange2 and for indicated T-Sapphire and PSmOrange2 fusions, in which proteins are connected with LX linkers of various lengths. X is a number of amino acid residues in a linker. (B) Kinetics of far-red fluorescence intensities for PSmOrange2, PSmOrange and mOrange linked to T-Sapphire with identical L7 linkers. (C) Kinetics of far-red fluorescence intensities for PSmOrange2 and for T-Sapphire and PSmOrange2 fusion at indicated photoswitching wavelength bands. (D) Fluorescence decay for T-Sapphire (dashed line) and T-Sapphire and PSmOrange2 fusion (solid line) at room temperature using 440 nm excitation light. (E) Photoswitching half-times (t0.5) for T-Sapphire and PSmOrange2 fusion at indicated concentrations. (F) Initial rates for PSmOrange2 photoswitching with 480/40 nm (squares) and 415/30 nm (triangles) light and T-Sapphire and PSmOrange2 fusion photoswitching with 415/30 nm light (circles) at indicated light power densities. The power densities were estimated at the sample. In (A, B, E) the 415/30 nm wavelength band was used for photoswitching. Error bars, SD.
We next determined the FRET efficiency in the T-Sapphire-L7-PSmOrange2 construct by fluorescence lifetime measurements using 440 nm light with ~20-fold lower intensity than in the photoswitching experiments described above. The lower light intensity avoids background photoconversion of the T-Sapphire-L7-PSmOrange2 construct. The lifetime of T-Sapphire fluorescence in the fusion was lower than that for non-fused T-Sapphire (Figure 5D). The FRET efficiency in the fusion construct was 23.4 %. The photoswitching half-time did not change over a wide range of the concentrations of the purified T-Sapphire-L7-PSmOrange2 (Figure 5E) suggesting that the accelerated photoswitching in the fusion construct occurred via intramolecular FRET.
The initial photoswitching rate depended linearly on the photoswitching light intensity when plotted on a logarithmic scale (Figure 5F). The slope of the dependency for the control non-fused PSmOrange2 was 2.14 ± 0.28 with 480/40 nm excitation and 2.05 ± 0.24 with 415/30 nm excitation suggesting photoswitching of a PSmOrange2 molecule required absorbance of two photons. The similar requirement of two photons for the photoconversion was earlier observed for parental PSmOrange 8. However, for the T-Sapphire-L7-PSmOrange2 construct the slope of the dependency was 0.99 ± 0.09 indicating that the photoswitching of a PSmOrange2 molecule in the fusion construct required absorbance of a single photon. It is likely that in the latter case the second portion of energy was delivered to PSmOrange2 non-radiatively via FRET from the T-Sapphire donor. This process was possibly substantially faster than a direct photon delivery to PSmOrange2 for photoswitching in the fusion construct. Thus, the direct photon delivery may be the rate limiting process. As a result, photoswitching in the fusion construct seems to only require absorbance of a single photon.
3.6. FRET-facilitated photoswitching in live mammalian cells
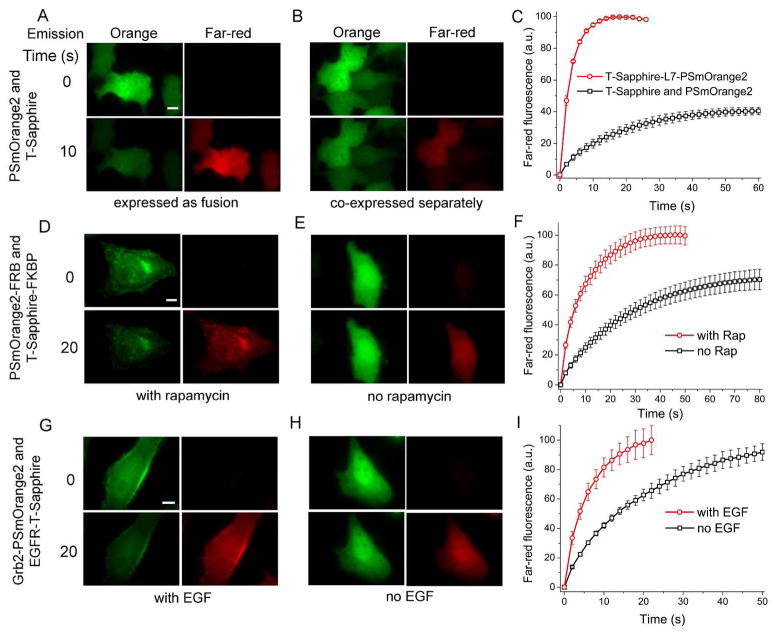
To evaluate FRET-facilitated photoswitching in mammalian cells, we compared the efficiency of PSmOrange2 photoswitching when it was in close proximity to T-Sapphire with that of PSmOrange2 expressed alone. For that we determined initial rates of PSmOrange2 photoswitching (Table S1). Three different sets of protein constructs were utilized.
In the first case, the same T-Sapphire-L7-PSmOrange2 fusion construct was expressed cytoplasmically in HeLa cells (Figure 6A). In the control experiment T-Sapphire and PSmOrange2 were co-expressed in non-fused states (Figure 6B). The initial rate of PSmOrange2 photoswitching in the fusion construct was 6.2-fold higher than in the control (Figure 6C, Table S1).
Figure 6.
FRET-facilitated photoswitching of PSmOrange2 in live HeLa cells. (A) Cytoplasmically expressed T-Sapphire and PSmOrange2 fusion construct and (B) co-expressed T-Sapphire and PSmOrange2 separately imaged in different fluorescence channels at indicated times during photoswitching. (C) Kinetics of far-red fluorescence intensities for cells in (A) and (B). Co-expressed membrane targeted T-Sapphire-FKBP and cytoplasmic PSmOrange2-FRB fusion proteins with (D) and without (E) supplemented 100 nM rapamycin imaged in different fluorescence channels at indicated times during photoswitching. (F) Kinetics of far-red fluorescence intensities for cells in (D) and (E). Co-expressed EGFR-T-Sapphire and Grb2-PSmOrange2 fusion proteins with (G) and without (H) supplemented 100 ng/ml EGF imaged in different fluorescence channels at indicated times during photoswitching. (I) Kinetics of far-red fluorescence intensities for cells in (G) and (H). The 415/30 nm wavelength band was used for photoswitching. Error bars, SD. Scale bars, 10 μm.
In two other sets, PSmOrange2 and T-Sapphire were brought into proximity when their fusion partners interacted under specific stimuli. In the second set, a human FK506-binding protein (FKBP), which binds to rapamycin and rapamycin analogs (rapalogs) with high affinity, was used. The FKBP–rapamycin complex is recognized by another protein, a 93 amino acid portion of FRAP, termed FKBP-rapamycin binding domain (FRB), thus allowing a rapamycin-induced dimerization of two target proteins 30. When FKBP and FRB were fused to other proteins, the rapamycin-induced dimerization of the fusion partners could be achieved. This system has been used to regulate protein expression 31,32, protein splicing 33 and glycosilation 34.
In our experiments FKBP and FRB were fused to T-Sapphire and PSmOrange2, respectively. In resting cells without rapamycin, T-Sapphire-FKBP containing the myristoilation signal was targeted to the plasma membrane, whereas PSmOrange2-FRB had cytoplasmic distribution (Figure 6E). When rapamycin was added, PSmOrange2-FRB fusion protein accumulated at the plasma membrane possibly dimerizing with the T-Sapphire-FKBP construct (Figure 6D). Photoconversion of the PSmOrange2-FRB fusion was found to be 3.3-fold faster when it was interacting with the T-Sapphire-FKBP fusion than when it was in the cytoplasm without rapamycin (Figure 6F, Table S1).
The third set of proteins was based on interaction between an epidermal growth factor (EGF) receptor (EGFR) and Grb2 adaptor protein induced upon stimulation with EGF. It has been shown that EGF-activated EGFR interacts with Grb2 at the plasma membrane and in endosomes 35,36. We transiently co-expressed EGFR-T-Sapphire and Grb2-PSmOrange2 fusion proteins in HeLa cells. The intracellular distributions of the fusions in resting cells were different. As expected, the EGFR-T-Sapphire signal mainly localized at the plasma membrane whereas the Grb2-PSmOrange2 fluorescence was evenly distributed throughout cell cytoplasm (Figure 6H). Addition of 100 ng/ml EGF led to a rapid accumulation of Grb2-PSmOrange2 at the membrane (Figure 6G). When bound to EGF-activated EGFR-T-Sapphire at the plasma membrane, Grb2-PSmOrange2 was photoconverted 2.3-fold faster than when it was distributed through the cytoplasm without EGF stimulation (Figure 6I, Table S1). Thus, in all sets of the intracellular constructs, the PSmOrange2 acceptor localized close to the T-Sapphire donor photoconverted substantially faster than in the control cases with the separated FRET donor and acceptor.
4. DISCUSSION
The PSmOrange2 protein has six amino acid substitutions compared to parental PSmOrange. The mechanism of the PSmOrange photocoversion, determined earlier by mass spectrometry, involves a break of the polypeptide chain between the main chain carbonyl and Cα of Phe64 8. Possibly, the PSmOrange2 photoconversion is similar and may consist of a break of the polypeptide chain between the main chain carbonyl and Cα of Ile64. This cleavage occurs faster in PSmOrange2 most likely because an electron donating Ile64 side chain has a positive inductive effect 37, that is not observed for Phe64. The shift of the electron density caused by this effect facilitates the polypeptide break and results in the faster PSmOrange2 photoswitching. Another PSmOrange2 variant which contains a different electron donating group in a side chain of the amino acid at position 64, Val64, also exhibited fast photoswitching kinetics giving support to this view (Figure S7).
PSmOrange2 has several advantages over parental PSmOrange. First, its photoconversion proceeds up to 10-fold faster, depending on the intensity of the photoswitching light. Second, the PSmOrange2 photoconversion is more efficient that results in the 9-fold higher photoswitching contrast. Third, its photoconversion is substantially faster at the oxidant concentrations lower or similar to that in live mammalian cells.
The PSmOrange2 photoconversion in mammalian cells leads to 3-fold brighter far-red fluorescence and correspondingly 4-fold more photoconverted molecules than with PSmOrange. PSmOrange2 is 3-fold more efficiently photoconverted with the 2P laser both in vitro and in live cells. This opens a possibility of using PSmOrange2 in 2P microscopy for selective optical highlighting of a cell population, an individual cell or part of a cell in deep-tissue imaging. Such selective photolabeling using 2P photoconversion has been used earlier with EosFP and KikGR proteins for highlighting mitochondrial network 10 and individual cells 38, respectively.
Using PSmOrange2 as a FRET donor we have observed, for the first time, an efficient FRET-facilitated photoswitching. The initial rates of the FRET-facilitated photoswitching differed in three different sets of the PSmOrange2 fusion constructs, from 10.5 to 16.6 s−1 (Table S1). The ratio between the initial rates of the FRET-facilitated photoswitching and photoswitching initial rates in the absence of FRET varied in the range of 2.3 to 6.2. The variation of these parameters is possibly caused by different localizations of the interacting partners and different FRET efficiencies between them.
According to the FRET-sensitized photobleaching approach 18,19, a FRET efficiency is proportional to a ratio of the acceptor photobleaching rate in the presence of the donor to the acceptor photobleaching rate in the absence of the donor. Similarly, we propose that a FRET efficiency in the FRET-facilitated photoswitching is proportional to a ratio of the initial rate of acceptor photoswitching in the presence of the donor to the initial rate of acceptor photoswitching without the donor. Based on this definition, the FRET efficiency between T-Sapphire and PSmOrange2 was 1.9-fold and 2.7-fold higher when the FRET fusion construct was in the cytoplasm than when PSmOrange2 and T-Sapphire chimeric proteins interacted at the plasma membrane in the rapamycin-induced and EGF-induced manner, respectively (Table S1).
FRET-facilitated photoswitching is a new approach for detection of protein-protein interactions. While there are several other FRET-based approaches to detect protein-protein interactions, such as sensitized emission, acceptor photobleaching, fluorescence lifetime imaging, and polarization anisotropy imaging 39 (Table S2), each has its own advantages and disadvantages. The sensitized emission method is suitable for observation of dynamic protein-protein interactions but requires a set of controls and is sensitive to donor-acceptor cross-talk 40. The acceptor photobleaching method is simple and does not require controls. However, it can be destructive for cells and, thus, is not suitable for dynamic interactions 41,42. Determination of FRET efficiency by this approach may also be incorrect because of incomplete photobleaching of an acceptor, or damage of a donor molecule. The fluorescence lifetime imaging approach is not sensitive to a donor-acceptor cross-talk and can be applied to non-fluorescent acceptors, however, it is slow and requires complex instrumentation 43. The polarization anisotropy method is based on measurements of fluorescence polarization of a donor 44. This approach is fast and has a high signal-to-noise ratio but is sensitive to cross-excitation of an acceptor and can be used for imaging with lenses of low numerical aperture only.
The FRET-facilitated photoswitching also has its strengths and weaknesses (Table S2). In contrast to other FRET methods this technique is not transient. Any PSmOrange2 acceptor interacting with a donor at the time of photoconversion will become photoconverted and remain a stable indicator of this interaction thereafter until degradation. Thus, this approach potentially allows observation and subsequent tracking of the photoconverted population of the acceptor molecules which have previously interacted with the donor molecules. Additionally, for systems involving only a few interacting molecules, or those with short molecular interaction times, this technique has the potential to allow signal to accumulate over time. That said, this approach does require controls and cannot be applied to multiple sequential protein-protein interactions because the PSmOrange2 acceptor loses its acceptor properties after photoswitching. At the same time, it is easy to detect FRET by the fast increase of far-red fluorescence intensity. Since photoconversion in the FRET-facilitated photoswitching requires relatively high light intensities, it can be combined with other FRET-approaches. For example, using a low light intensity for donor excitation, the sensitized emission, fluorescence lifetime imaging, or polarization anisotropy imaging can be first applied to detect dynamic protein-protein interactions. Next, the FRET-facilitated photoswitching can be used to determine intracellular localization of acceptor protein molecules that have interacted with the donor protein molecules.
5. CONCLUSIONS
The enhanced photoconversion properties of PSmOrange2 provide several advantages over parental PSmOrange for live cell imaging. These include substantially brighter far-red fluorescence of mammalian cells after the same photoconversion time or its similar far-red fluorescence intensity after the substantially shorter time during which cells are exposed to light. The former provides the higher signal-to-background ratio while the latter results in the lower photodamage of live cells. Furthermore, because of the faster photoswitching kinetics PSmOrange2 is more efficiently photoconverted with 2P lasers that will make it useful for deep-tissue imaging.
Similarly to PSmOrange, the red-shifted spectra of both PSmOrange2 forms will enable its simultaneous use with blue, cyan and green permanently fluorescent proteins or with cyan-to-green photoswitchable proteins in a live cell 8. Like the parental protein, PSmOrange2 can be used in a super-resolution PALM (photoactivated localization microscopy) microscopy with the common 488 nm blue lasers for photoactivation and 633–640 nm red lasers for readout. Because photoconverted PSmOrange2 has the far-red shifted excitation and emission spectra, similarly to PSmOrange it can be also applied to whole-body imaging of model animals 8.
Importantly, PSmOrange2 is the first FP whose photoswitching is substantially accelerated by a FRET process from the green emitting donor. Future engineering of PSFPs with the fast photoconversion after absorption of a single photon in blue-green spectral range will result in wide use of the FRET-facilitated photoswitching to visualize both ongoing and previously occurred protein-protein interactions in live cells.
Supplementary Material
Acknowledgments
We thank Gleb Shtengel and Herald Hess (both are from Janelia Farm, Howard Hughes Medical Institute, VA) for iPALM imaging of paxillin-PSmOrange2, Vladimir Malashkevich and Daria Shcherbakova (both are from Albert Einstein College of Medicine, NY) for the help with PSmOrange2 characterization. We thank Alexander Sorkin (University of Pittsburg, PA) and Michael Davidson (Florida State University, FL) for the plasmids with fusions of fluorescent proteins and Oliver Griesbeck (Max-Planck-Institute of Neurobiology, Germany) for the plasmid with the T-Sapphire gene. This work was supported by grants GM073913, CA163131 and CA164468 from the National Institutes of Health.
Footnotes
ACCESSION NUMBERS
The GenBank accession number for the PSmOrange2 sequence reported in this paper is JQ392581.
Seven supporting figures and two supporting tables are available. Photoswitching kinetics of PSmOrange and PSmOrange2 without adding oxidant. Dependence of maximal far-red fluorescence on the power of photoswitching light. The pH dependence of PSmOrange2. Seminative polyacrylamide gel with PSmOrange2. iPALM imaging of HeLa cells expressing the paxillin-PSmOrange2 fusion protein. Excitation and emission spectra of T-Sapphire overlaid with those of the orange form of PSmOrange2 and action spectrum of PSmOrange2. Photoswitching kinetics of the PSmOrange, PSmOrange2 and PSmOrange2/E32K/I64V/A70S proteins. Initial rates of PSmOrange2 photoswitching in different sets of fusion constructs in presence and absence of T-Sapphire FRET donor. Comparison of different FRET methods to detect protein-protein interactions. This material is available free of charge via Internet at http://pubs.acs.org.
References
- 1.Lippincott-Schwartz J, Patterson GH. Trends Cell Biol. 2009;19:555. doi: 10.1016/j.tcb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu B, Piatkevich KD, Lionnet T, Singer RH, Verkhusha VV. Curr Opin Cell Biol. 2011;23:310. doi: 10.1016/j.ceb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chudakov DM, Lukyanov S, Lukyanov KA. Nat Protoc. 2007;2:2024. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 4.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. Nat Methods. 2009;6:131. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. Proc Natl Acad Sci USA. 2002;99:12651. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Habuchi S, Tsutsui H, Kochaniak AB, Miyawaki A, van Oijen AM. PLoS One. 2008;3:e3944. doi: 10.1371/journal.pone.0003944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoi H, Shaner NC, Davidson MW, Cairo CW, Wang J, Campbell RE. J Mol Biol. 2010;401:776. doi: 10.1016/j.jmb.2010.06.056. [DOI] [PubMed] [Google Scholar]
- 8.Subach OM, Patterson GH, Ting LM, Wang Y, Condeelis JS, Verkhusha VV. Nat Methods. 2011;8:771. doi: 10.1038/nmeth.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drobizhev M, Makarov NS, Tillo SE, Hughes TE, Rebane A. Nat Methods. 2011;8:393. doi: 10.1038/nmeth.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanchenko S, Glaschick S, Rocker C, Oswald F, Wiedenmann J, Nienhaus GU. Biophys J. 2007;92:4451. doi: 10.1529/biophysj.106.103408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider M, Barozzi S, Testa I, Faretta M, Diaspro A. Biophys J. 2005;89:1346. doi: 10.1529/biophysj.104.054502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. EMBO Rep. 2005;6:233. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyawaki A. Nat Rev Mol Cell Biol. 2011;12:656. doi: 10.1038/nrm3199. [DOI] [PubMed] [Google Scholar]
- 14.Jares-Erijman EA, Jovin TM. Nat Biotechnol. 2003;21:1387. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 15.Miyawaki A. Annu Rev Biochem. 2011;80:357. doi: 10.1146/annurev-biochem-072909-094736. [DOI] [PubMed] [Google Scholar]
- 16.Souslova EA, Chudakov DM. Microsc Res Tech. 2006;69:207. doi: 10.1002/jemt.20278. [DOI] [PubMed] [Google Scholar]
- 17.Subach FV, Zhang L, Gadella TW, Gurskaya NG, Lukyanov KA, Verkhusha VV. Chem Biol. 2010;17:745. doi: 10.1016/j.chembiol.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mekler VM. Photochem Photobiol. 1994;59:615. [Google Scholar]
- 19.Mekler VM, Averbakh AZ, Sudarikov AB, Kharitonova OV. J Photochem Photobiol B. 1997;40:278. doi: 10.1016/s1011-1344(97)00070-5. [DOI] [PubMed] [Google Scholar]
- 20.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Gene. 1989;77:51. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 21.Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. Nat Methods. 2009;6:153. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Nat Biotechnol. 2004;22:1567. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 23.Chudakov DM, Verkhusha VV, Staroverov DB, Souslova EA, Lukyanov S, Lukyanov KA. Nat Biotechnol. 2004;22:1435. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- 24.Lin MZ, McKeown MR, Ng HL, Aguilera TA, Shaner NC, Campbell RE, Adams SR, Gross LA, Ma W, Alber T, Tsien RY. Chem Biol. 2009;16:1169. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verkhusha VV, Sorkin A. Chem Biol. 2005;12:279. doi: 10.1016/j.chembiol.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, Fetter RD, Hess HF. Proc Natl Acad Sci U S A. 2009;106:3125. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Entenberg D, et al. Nature Protocols. 2011;6:1500. doi: 10.1038/nprot.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Biochemistry. 2006;45:9639. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 29.Drobizhev M, Tillo S, Makarov NS, Hughes TE, Rebane A. J Phys Chem B. 2009;113:855. doi: 10.1021/jp8087379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Zheng XF, Brown EJ, Schreiber SL. Proc Natl Acad Sci U S A. 1995;92:4947. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho SN, Biggar SR, Spencer DM, Schreiber SL, Crabtree GR. Nature. 1996;382:822. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 32.Rivera VM, Clackson T, Natesan S, Pollock R, Amara JF, Keenan T, Magari SR, Phillips T, Courage NL, Cerasoli F, Jr, Holt DA, Gilman M. Nat Med. 1996;2:1028. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 33.Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. J Am Chem Soc. 2003;125:10561. doi: 10.1021/ja0362813. [DOI] [PubMed] [Google Scholar]
- 34.Kohler JJ, Bertozzi CR. Chem Biol. 2003;10:1303. doi: 10.1016/j.chembiol.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 35.Sorkin A, McClure M, Huang F, Carter R. Curr Biol. 2000;10:1395. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- 36.Yamazaki T, Zaal K, Hailey D, Presley J, Lippincott-Schwartz J, Samelson LE. J Cell Sci. 2002;115:1791. doi: 10.1242/jcs.115.9.1791. [DOI] [PubMed] [Google Scholar]
- 37.Smith MB, March J, editors. March’s advanced organic chemistry. Reactions, mechanisms, and structure. 6. Wiley-Interscience, John Willey & Sons, Inc; 2007. [Google Scholar]
- 38.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. EMBO Rep. 2005;6:233. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piston DW, Kremers GJ. Trends Biochem Sci. 2007;32:407. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Biophysical Journal. 1998;74:2702. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenworthy AK, Edidin M. Journal of Cell Biology. 1998;142:69. doi: 10.1083/jcb.142.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wouters FS, Bastiaens PI, Wirtz KW, Jovin TM. EMBO J. 1998;17:7179. doi: 10.1093/emboj/17.24.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gadella TWJ, Jovin TM, Clegg RM. Biophysical Chemistry. 1993;48:221. [Google Scholar]
- 44.Lidke DS, Nagy P, Barisas BG, Heintzmann R, Post JN, Lidke KA, Clayton AH, Arndt-Jovin DJ, Jovin TM. Biochem Soc Trans. 2003;31:1020. doi: 10.1042/bst0311020. [DOI] [PubMed] [Google Scholar]
- 45.Shaner NC, Steinbach PA, Tsien RY. Nat Methods. 2005;2:905. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.