Abstract
The renin gene has been previously reported to be associated with essential hypertension in a variety of ethnic groups. However, no studies have systematically evaluated the relationship between single nucleotide polymorphisms (SNPs) representing coverage of the entire renin gene and hypertension risk. To evaluate the association between renin gene variation and hypertension we investigated data on HyperPath cohort with 570 hypertensive and 222 normotensive Caucasian subjects. Six tagging SNPs and resultant haplotypes were tested for associations with hypertension risk, followed by mean arterial pressure (MAP), plasma renin activity (PRA) and the change in MAP in response to angiotensin II infusion (AngII ΔMAP). The A allele of SNP rs6693954 and the haplotype containing rs6696954A were significantly associated with higher risk for hypertension (OR=1.98, P=0.0001; OR=1.63 P=0.0005, respectively). The same haplotype block was also associated with altered PRA levels and blunted AngII ΔMAP (global P value=0.02, 0.047, respectively). Our results confirm that polymorphisms in the REN are associated with increased risk for hypertension in an independent cohort, and that the underlying mechanism may reside in the interaction of renin activity and vascular responsiveness to angiotensin II.
Keywords: Renin, SNP, haplotype, blood pressure and Hypertension
Introduction
As the first and rate-limiting enzyme of the renin-angiotensin system (RAS), renin plays an important role in the regulation of sodium homeostasis and blood pressure (BP). Alterations in renin activity have been associated with hypertension, salt-sensitivity of blood pressure and responsiveness to anti-hypertensive treatments1–3. Therefore, the renin gene (REN) is a rational candidate for unraveling the genetic basis of essential hypertension (EH).
REN maps to chromosome 1q32, spans 12.5kb in length and encodes 10 exons. Individual REN polymorphisms have been associated with BP 4,5, plasma renin activity (PRA) 6, and susceptibility to hypertension7–9 in a variety of ethnic groups, although often with inconsistent results 10,11. The inconsistency may in part be related to genetic heterogeneity across populations and studying different genetic variants by different groups, or more likely, failure to control environmental factors, such as dietary salt intake and body posture that have great impact on BP and PRA levels.
Therefore, to confirm the previous findings and further assess the contribution of REN genetic variants to hypertension, we have performed high-density genotyping with a tagging SNP approach, using the most recent single nucleotide polymorphisms (SNP) data available from the Haplotype Mapping Project (HapMap) to assess the association between individual SNPs and haplotypes of REN and hypertension risk in the Hypertension Pathotype (HyperPATH) cohort. Our study may help to explain previous inconsistencies by using SNPs representing coverage of the entire REN gene. Unlike general epidemiologic approaches, the HyperPath cohort 12–15 provides the unique opportunity to evaluate complex heritable disease such as hypertension due to its rigorous control of several known influencing conditions including dietary salt intake, body posture, hypertensive medication and evaluation in a controlled setting (Clinical Research Center). This enhances signal to noise ratios through cleaner phenotyping, reducing variability and increasing power while simultaneously providing a physiologic basis for associations.
The observed new associations between REN haplotypes and PRA level, BP and vascular response to exogenous AngII infusion provide useful mechanistic understanding involving gene function and risk. These data would be a critical first step in determining the utility of genetic markers of REN in a personalized medicine approach focused on prevention and/or treatment.
Methods
Objectives
To better understand the association between renin gene (REN) variation and hypertension, REN genotypes and haplotypes were first tested for association with hypertension risk and then for characteristic intermediate phenotypes following a strictly controlled protocol.
Participants and Protocol
For this study, data on 570 hypertensive and 222 normotensive white subjects in the HyperPATH cohort were used. The HyperPATH Cohort is an on-going multi-site international efforts aiming at investigating pathophysiological and genetic mechanisms underlying hypertension and cardiovascular diseases. The HyperPATH protocol is designed to minimize the known confounders of PRA (dietary sodium intake, body posture, diurnal variations and medications). Study participants were placed on a controlled high salt diet with sodium content of 200mmol/day approximating the average of Westernized diets. Details of this cohort have been previously described.12–15
Subjects were studied on the General Clinical Research Centers (GCRCs) of the Brigham and Women’s Hospital in Boston Massachusetts, USA (n=292), the University of Utah Medical Center in Salt Lake City, Utah, USA (n=254), Vanderbilt University in Nashville, Tennessee, USA (n=41), Hospital Broussais in Paris, France (n=105) and University La Sapienza in Rome, Italy (n=100). Of the 792 total subjects, there were 486 singletons, 254 from two-sibling families and 48 from three-sibling families. As per the original study design, hypertension was defined as a seated diastolic blood pressure ≥100 mmHg off antihypertensive medication, ≥90mmHg while taking at least one antihypertensive medication, or treatment with at least two antihypertensive medications. Eligibility was determined while on an ad lib diet. Hypertensive subjects requiring at least 4 medications were excluded. Normotensives were defined by three repeated blood pressure readings averaging less than 135/75mmHg and no first-degree relative diagnosed with hypertension before 60 years of age.
All subjects completed a screening visit consisting of a physical examination, a medical history, and laboratory assessment. Subjects with a history of diabetes mellitus, any form of secondary hypertension, overt renal insufficiency (serum creatinine >1.5mg/dL), or any significant medical or psychiatric illnesses were excluded. Subjects with severe obesity (body mass index>34kg/m2), current tobacco or illicit drug use, or alcohol intake >12oz per week were also excluded. Normal laboratory values for electrolytes, thyroid and liver function tests were required. Electrocardiographic evidence of heart block, ischemia, or previous coronary events led to exclusion. Subjects had to be between 18–65 years of age and race was self-defined. To minimize the interference of medication with our assessment of RAS activity, all angiotensin converting enzyme inhibitors (ACEi), angiotensin receptor blockers (ARB) or mineralocorticoid receptor antagonists were discontinued 3 months before the study. If needed, subjects were placed on amlodipine or hydrochlorothiazide to control BP. All antihypertensive medications were discontinued 3–4 weeks prior to hormonal and vascular assessment.
Subjects completed a 7-day caffeine and alcohol free isocaloric high salt diet (200mmol/day sodium, 100mmol/day potassium, 20mmol/day calcium). On the final day of high salt diet, subjects were admitted to the General Clinical Research Center where they remained fasting and supine overnight. Attainment of “high salt” balance was confirmed by measurement of sodium and creatinine excretion in a 24-hour urine collection and defined as ≥ 150 mmol/day sodium. All subjects had PRA measured from the baseline morning (8:00AM to 10:00AM) supine blood obtained on the first day of study using the Diasorin assay (Stillwater, MN). All blood samples were collected on ice and centrifuged immediately for 20 min and plasma separated and frozen without preservatives until the time of assay16. Brigham and Women’s Hospital served as the central laboratory for all lab processing. The detectable range of the assay is 0–50ng/mL/hr. The lowest detectable dose is 0.1ng/mL/hr. The intra-assay variation was 4.6–10% and the inter-assay variation was 5.6–7.6%. Mean arterial pressure (MAP) was measured supine at baseline and then again after an intravenous infusion of angiotensin II (AngII) (3ng/kg/min for 60 minutes) in order to assess the level of RAS responsiveness in the peripheral vasculature, ie. MAP (Dinamap; Critikon, Tampa, FL). Value was consisted of a mean of three consecutive readings at 2 min intervals. The protocols are standardized across all 5 study sites.
Ethics Statement
The Institutional Review Boards of Brigham and Women’s Hospital (Boston, MA), Vanderbilt University Medical Center (Nashville, TN), University of Utah Medical Center (Salt Lake City, UT), Hospital Broussais (Paris, France) and University La Sapienza (Rome, Italy) reviewed and approved the study protocol. Written informed consent was obtained from all participants at all sites prior to enrollment.
Tag SNP Selection and Genotyping
REN tagging SNPs were selected from HapMap CEU population (Phase II, November 2008) using the chromosomal coordinates, chr1:202,390,571–202,402,088 and including two 5 kb flanking regions (Figure 1A). Ten SNPs captured 100% of the variation in this region with a minor allele frequency >0.1 at an r2>0.9. Genotyping was performed using the Illumina Bead Station GoldenGate platform (Illumina Inc, San Diego, CA).
Figure.
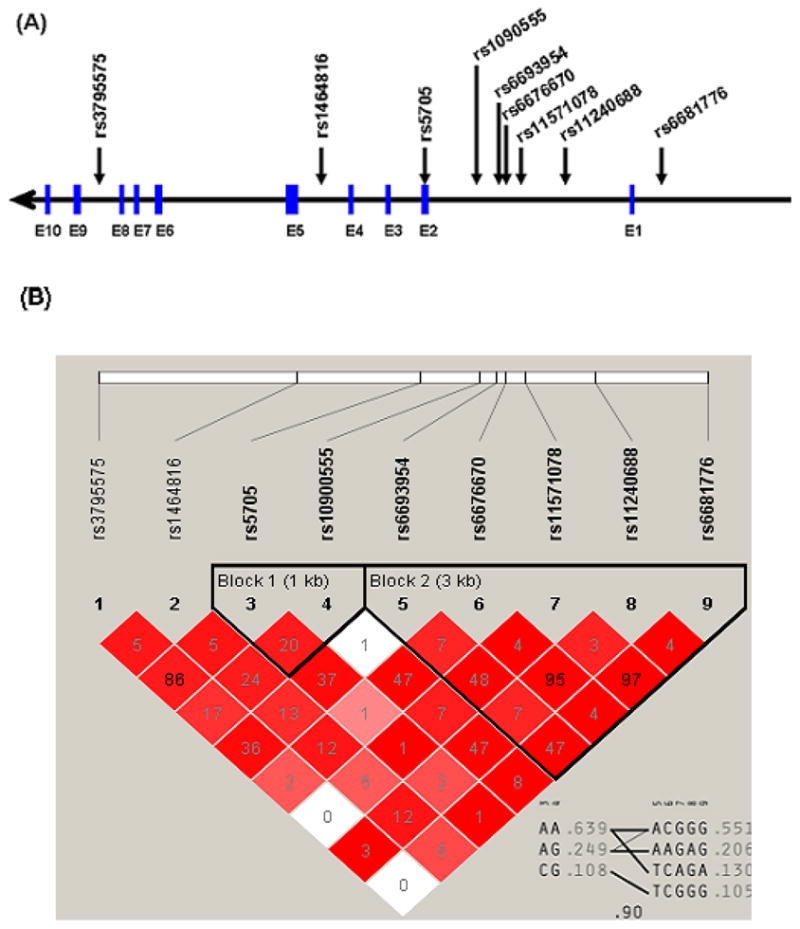
(A) Scheme of REN genomic structure and htSNPs included in the study. Exons are shown in blue boxes. (B) Haploview LD plot constructed using r2 statistic.
Phenotypes
The phenotypes analyzed included the primary outcome of risk for hypertension followed by a mechanistic analysis of MAP, PRA and the change of MAP in response to AngII infusion (AngII ΔMAP) within the hypertensive population.
Statistical Analysis
Hardy-Weinberg equilibrium (HWE) was assessed for each SNP using χ2 test. The D′ and r2 measures of linkage disequilibrium (LD) were calculated using Haploview 4.1 17.
All tests for single SNP associations were performed using SAS version 9.1 (SAS Institute, Cary, NC). A generalized-estimating-equation (GEE) logistic model with an exchangeable covariance matrix was used for binary data analyses; and a mixed effect linear regression model was used for all the continuous data analyses. All analyses were adjusted for age, gender, BMI, study site and familial relatedness, as some siblings were present in the study population. Logarithmic transformations were performed for PRA concentrations to normalize the distribution of data. An additive model was used to test associations between genotype and the outcome phenotype. Bonferroni correction was applied to account for multiple comparisons with a final significance level set at 0.05 divided by the number of independent SNP comparisons. Of the initial 10 SNPs genotyped, a total of 6 unique SNPs were identified in the study population based on LD plots (see Results). Thus, the final p-value for statistical significance was set at 0.05/6 or 0.008. Using a significance level of 0.008, a power of 80% and a disease allele frequency of 0.30–0.60, the minimum odds ratio for hypertension that we can detect in our model is 1.36–1.42.
Haplotypes were constructed and analyzed using Plink 1.06 (http://pngu.mgh.harvard.edu/purcell/plink/) 18. Plink estimates haplotype frequencies via the expectation-maximization (EM) algorithm, computing global and haplotype-specific score statistics for tests of association between a trait and haplotype weighted by their posterior possibility. Considering that 40% of the subjects were related as siblings, we compared the haplotype analyses with all subjects combined and then again separately with only one subject per family. Results were similar, thus we reported based on the output from all subjects. In the haplotype analyses, we only included haplotypes with represented frequencies of greater than or equal to 5%. Haplotype-specific odds ratios (OR) or regression coefficients were estimated using a logistic or a linear regression model adjusted for age, gender, and BMI, contrasting against all other haplotypes.
Results
General characteristics of the study population are shown in Table 1. Consistent with previous reports, hypertensive subjects were older and had characteristic differences in their biochemical and hemodynamic profiles compared to normotensives.
Table 1.
Characteristics of study subjects.
| Variable | Hypertensives | Normotensives | P value |
|---|---|---|---|
| Number | 567 | 222 | |
| Age, y | 48±9 | 40±11 | <.0001 |
| BMI, kg/m2 | 27.9±3.9 | 25.4±3.8 | <.0001 |
| Gender, female % | 222 (39%) | 111(50%) | 0.006 |
| MAP, mmHg | 108.7±14.0 | 80.5±8.2 | <.0001 |
| PRA, ng/mL/h | 0.6±0.7 | 0.5±0.4 | <.0001 |
| AngII ΔMAP* | 9.2±10.1 | 8.8±6.2 | <.0001 |
AngII ΔMAP = increase in mean arterial pressure in response to angiotensin II infusion (3ng/kg/min × 60 mins.). Data presented as the mean ± standard deviation.
REN SNP and Haplotype Report
Among the 10 SNPs that were genotyped, 2 SNPs were in the 5′ flanking region, 1 in the coding region, and 7 intronic (Figure 1A). The average genotyping success rate was >95% for all SNPs. In the normotensive control group, minor allele frequencies are shown in Table 2. All SNPs were in HWE. One SNP, rs16853062, was monomorphic and thus excluded from the analysis.
Table 2.
REN gene single nucleotide polymorphisms.
| SNP | Alleles * | Gene position | MAF(HTs/NTs)† | HWE P(HTs/NTs)‡ |
|---|---|---|---|---|
| rs16853062 | T/A | 5′ | 0/0.003 | NS/0.97 |
| rs6681776 | G/A | 5′ | 0.12/0.16 | 0.96/0.53 |
| rs11240688 | G/A | intron 1 | 0.21/0.22 | 0.17/0.93 |
| rs11571078 | G/A | intron 1 | 0.12/0.17 | 0.73/0.75 |
| rs6676670 | C/A | intron 1 | 0.21/0.22 | 0.08/0.81 |
| rs6693954 | A/T | intron 1 | 0.21/0.32 | 0.37/0.47 |
| rs10900555 | A/G | intron 1 | 0.34/0.40 | 0.85/0.15 |
| rs5705 | A/C | exon 2 | 0.10/0.16 | 0.23/0.58 |
| rs1464816 | C/A | intron 4 | 0.34/0.33 | 0.03/0.47 |
| rs3795575 | G/A | intron 8 | 0.09/0.16 | 0.71/0.22 |
indicates major/minor allele;
Minor allele frequencies of SNPs in hypertensives and normotensives;
P values for Hardy-Weinberg equilibrium testing in hypertensives and normotensives.
Of the remaining 9 SNPs, the LD map construct from this cohort reflected the LD pattern from HapMap, and revealed that 3 SNP pairs (rs6681776| rs11571078, rs11240688| rs6676670, and rs5705| rs3795575) were in strong LD (r2 >0.85). Thus, only 1 SNP from each pair was used for analysis (N=3). This left 3 additional independent SNPs from the original 10 genotyped. We limited our association analyses to these 6 haplotype tagging SNPs (htSNPs), which captured 100% of the common REN variation in this population with a mean r2 of 0.977. Two haplotype blocks were observed using these htSNPs. A multilocus r2 of 0.90 between two blocks showed strong LD between the blocks and minimal recombination (Figure 1B).
Single SNP Associations with Hypertension Risk
We first tested the association between the 6 individual SNPs with hypertension status (Table 3). Significant allelic associations with risk for hypertension were seen in 4 SNPs, with the strongest associations observed with rs6693954 and rs5705. The major allele A of either SNP demonstrated higher risk for hypertension (OR=1.98; 95% CI 1.40, 2.80; P=0.0001 for rs6693954 and OR=2.04; 95% CI 1.29, 3.21; P=0.002 for rs5705).
Table 3.
Single SNP association analyses with risk of hypertension.
| SNP | Major | Minor | MAF | OR | 95% CI | P value |
|---|---|---|---|---|---|---|
| rs11571078 | G | A | 0.17 | 1.58 | 1.07, 2.34 | 0.02 |
| rs6676670 | C | A | 0.22 | 1.06 | 0.74, 1.51 | 0.75 |
| rs6693954 | A | T | 0.32 | 1.98 | 1.40, 2.80 | 0.0001 |
| rs10900555 | A | G | 0.40 | 1.33 | 1.00, 1.78 | 0.048 |
| rs5705 | A | C | 0.16 | 2.04 | 1.29, 3.21 | 0.002 |
| rs1464816 | C | A | 0.33 | 0.91 | 0.66, 1.24 | 0.54 |
Significant associations with hypertension status by logistic regression with adjustment for covariates of age, sex, location and family relatedness are shown (bold font represents significance after Bonferroni correction). P values were obtained for the additive model with minor allele as reference.
Haplotype Variation in Relation to Hypertension Risk
Haplotype analysis was performed using 5 informative htSNPs necessary to build the haplotype. The SNP rs1464816 was not included in the haplotype analysis since it neither inferred any significance nor belonged to any haplotype block. There were 5 common haplotypes with frequencies >5% accounting for 94% of all haplotypes in this population.
We estimated haplotype frequencies first in LD block 1 and LD block 2, and then in block 1 and 2 combined. Significant associations with hypertensive status were observed in all 3 constructs (Table 4). In the overall analysis, haplotype H1A|H2A combination demonstrated the greatest risk for hypertension, accounting for 33% of alleles in the study population. This was in agreement with single SNP analysis, as the common allele tended to have an increased risk for hypertension compared with the minor allele. The most significant effect was noted in LD block 2, with the most common haplotype, H2A (ACG), resulting in a 1.63-fold odds for hypertension (P=0.0005). In LD block 1, haplotype H1A (AA) also demonstrated an increased risk for hypertension (OR=1.31; P=0.046). These 2 haplotypes (AA and ACG) correlated and appeared to cosegregate as shown in the combined block 1 and 2 analysis, and thus, the observed individual bock associations do not appear to be independent. Interestingly, a single allele change at rs6693954 as noted in haplotype H2D, resulted in a decreased risk for hypertension (OR=0.56; P=0.005). Likewise, H1C (CG) was associated with decreased risk for hypertension.
Table 4.
Haplotype analyses with risk for hypertension.
| Haplotype | HT% | NT% | OR | P value* |
|---|---|---|---|---|
| Block 1: rs5705|rs10900555 | 0.01 | |||
| H1A: AA | 65.8 | 59.3 | 1.31 | 0.046 |
| H1B: AG | 25.0 | 25.1 | 0.99 | 0.98 |
| H1C: CG | 9.2 | 15.6 | 0.55 | 0.002 |
| Block 2: rs6693954|rs6676670|rs11571078 | 0.001 | |||
| H2A: ACG | 58.8 | 47.5 | 1.63 | 0.0005 |
| H2B: AAG | 20.3 | 21.2 | 0.95 | 0.75 |
| H2C: TCA | 12.1 | 16.7 | 0.69 | 0.05 |
| H2D: TCG | 8.7 | 14.5 | 0.56 | 0.005 |
| Overall: rs5705|rs10900555|rs6693954|rs6676670|rs11571078 | 0.001 | |||
| H1A|H2A: AA-ACG | 33.9 | 31.6 | 1.55 | 0.001 |
| H1B|H2B: AG-AAG | 20.6 | 22.3 | 0.93 | 0.68 |
| H1A|H2C: AA-TCA | 11.8 | 15.3 | 0.72 | 0.08 |
| H1C|H2D: CG-TCG | 9.1 | 15.4 | 0.57 | 0.005 |
| H1B|H2A: AG-ACG | 4.0 | 2.6 | 1.61 | 0.25 |
Bolded P-values in the first rows represent the global P-value; other P-values respresent haplotype-specific P-values for each versus all others in the block.
Analysis of Intermediate Phenotypes
In order to understand the possible mechanisms underlying the associated increased risk for hypertension, we limited our analysis to only the hypertensive group using LD block 1 and 2. Associations were tested between haplotypes and blood pressure characteristics (baseline MAP and AngII ΔMAP) and PRA levels (Table 5). Despite a reduction in statistical power from a smaller sample size, we nonetheless observed significant associations. LD block 2 was significantly associated with PRA level and AngII ΔMAP with global P values both at 0.02. LD block1 was also significantly associated with baseline MAP and AngII ΔMAP (global P value=0.01 and 0.04, respectively). At the individual haplotype level, MAP for haplotype H2D (rs6693954T|rs6676670C|rs11571078G) was 3.59 mmHg higher than for all other haplotypes (P=0.04). Although there was no statistical difference of MAP among the 4 haplotypes in block 2 as a whole (global P value=0.07), the adjacent block 1 exhibited a stronger signal. It has been suggested that the heightened RAS activity at the tissue level (i.e. kidney, peripheral vasculature) may contribute directly to the development of hypertension, and that this dysregulation may present as a blunted BP response to exogenously infused AngII12,19–21. As expected, the haplotype H2D (rs6693954T|rs6676670C|rs11571078G) that showed relatively higher MAP also displayed 3.46mmHg lower BP response to AngII infusion compared with all others (P=0.01). As shown in Figure 2B, the haplotype H1C(rs5705C|rs10900555G) in the block 1 and H2D(rs6693954T|rs6676670C|rs11571078G) in the block 2 are co-transmitted, this was reflected with a similar associated heightened MAP and blunted AngII ΔMAP observed with H1C(rs5705C|rs10900555G).
Table 5.
Associations of REN LD blocks and quantitative traits.
| Haplotype | MAP mmHg (P value) | PRA ng/mL/h (P value) | AngII ΔMAP mmHg (P value) |
|---|---|---|---|
| Block 1: rs5705|rs10900555 | |||
| Global P value | 0.01 | 0.46 | 0.04 |
| H1A: AA | −2.97(0.005) | −0.01(0.88) | 0.63(0.47) |
| H1B: AG | 2.08(0.08) | −0.05(0.54) | 0.92(0.35) |
| H1C: CG | 3.24(0.06) | 0.11(0.39) | −3.16(0.02) |
|
| |||
| Block 2: rs6693954|rs6676670|rs11571078 | |||
| Global P value | 0.08 | 0.02 | 0.02 |
| H2A: ACG | −1.23 (0.24) | 0.16 (0.03) | 0.44 (0.59) |
| H2B: AAG | 1.36 (0.29) | −0.08 (0.38) | 1.92 (0.06) |
| H2C: TCA | −1.92 (0.19) | −0.29 (0.006) | −0.83 (0.46) |
| H2D: TCG | 3.59 (0.04) | 0.13 (0.34) | −3.46 (0.01) |
Significant associations with MAP, PRA and MAP response to AngII infusion estimated by linear regression model with adjustment for covariates of age, gender, BMI and study site. Global P values are obtained by omnibus tests jointly estimating all haplotype effects. Values in the cell represent the effect size for each haplotype and the haplotype-specific P values when comparing against all others in the block.
Moreover, it is hypothesized that altered REN gene function and expression might change the PRA level, further lead to elevated BP. In our study, the PRA level for the hypertension risk haplotype H2A were 0.16 ng/mL/h higher than other haplotypes (P=0.03). However, Haplotype H2C was associated with lower PRA (−0.29 ng/mL/h, P=0.006).
Discussion
This study used a comprehensive htSNP set capturing all known variants of REN in HapMap in order to redefine the association between REN gene and hypertension. In this strictly salt-controlled, medication-washout study population, we found multiple REN htSNPs significantly associated with risk for hypertension. Haplotype analyses provided consistent evidence that haplotype H1A|H2A (AA-ACG) is a risk locus for hypertension and that this relationship is mainly explained by haplotype H2A (ACG).
In the single SNP analysis, the strongest association with hypertension was seen with rs6693954, which is in high LD with rs2368564 (r2=0.96) located in intron 9. The latter has been previously recognized as a REN MboI restriction fragment length polymorphism (RFLP) and found to be associated with risk for EH in a range of ethnic groups7,8. Thus, the replication of the findings previously reported in other populations suggests that the observed association is unlikely to be spurious. It is uncertain why one study failed to detect a significant association between rs2368564 and hypertension10. This lack of association could result from genetic heterogeneity across populations, small sample sizes, or failure to control environmental factors. Lending further support to our findings, rs2368564 has recently been shown to be associated with a reduced risk of edema caused by peroxisome proliferator activated receptor agonist 22, with the proposed mechanism being modification of the levels of REN expression.
Our haplotype results are consistent with the findings from the single SNP analyses and suggest the most frequent haplotype H1A|H2A is a susceptibility locus for hypertension. This association is primarily driven by the haplotype H2A in LD block 2 that contains the allele A of rs6693954. Interestingly, a one allele difference at this SNP locus, as observed in haplotype H2D, results in a decreased risk for hypertension, underscoring the important role of rs6693954 in EH. LD block 2 is located in intron 1 in which it is highly conserved across species and contains important regulatory elements 23–25. Thus, it is possible that this region is important in identifying a regulatory element that influences REN expression and altered risk for hypertension.
Our haplotype results suggested that two primary haplotypes were associated with hypertension risk; H2A predisposes higher risk and H2D is protective. Within hypertensives, we also observed an association between haplotype H2A and higher PRA level. Few recent randomized clinical trials have shown that BP-lowering response to high-dose antirenin (aliskiren) monotherapy were greatest in those with the highest baseline PRA level3,26. Therefore, people who carry haplotype H2A, or more specifically those who are rs6693954A carriers, could potentially exhibit preferential response to Aliskiren relative to the other haplotypes. It is well recognized that circulating RAS plays a crucial role in regulating arterial pressure and sodium homeostasis. However, recent studies also provided the notion that RAS activities at tissue level, particularly in the kidney and peripheral vasculature, are associated with the development of hypertension and the increased susceptibility of nephropathy among diabetics, although these may be considered speculative12,19–21,27. Moreover, responsiveness to exogenous AngII infusion can provide more sensitive information regarding target organ sensitivity, and the insensitivity state is reversible by using ACEi. In our study, the observed blunted response of haplotype H2D suggested that REN could be involved in the vascular RAS sensitivity, and people who carry this haplotype might more likely benefit from ACEi by downregulating the heighted tissue RAS.
The present study provides additional evidence linking REN and hypertension. However, since this study was designed to sample representative regions of the whole gene, fine mapping would be required to locate possible causal variants. It is important to interpret findings in genetic associations within the context of the study population and environment. Although we adjusted study site as a confounding factor in our analyses and did not observe significant differences in genotype distributions in the renin gene among the study sites; population heterogeneity across different study centers still could be a concern to take into consideration, and merits replication in similar populations as well as mechanistic or causal confirmation.
Our study confirms the association between risk for hypertension and variants in the REN. Furthermore the above results provide clues to the possible mechanistic underpinnings resulting in the hypertension, including altered REN expression and/or vascular reactivity. This underscores the importance and advantage of paying attention to known confounders in deciphering the genetic underpinnings of complex diseases such as hypertension. This knowledge may assist in identifying individuals at specific risk for developing hypertension while also supporting possible gene-oriented targeted therapy, i.e., a personalized medicine approach. Future studies should explore whether these relatively common variants may respond preferentially to pharmacologic manipulation of the RAS.
Footnotes
Disclosure: The authors have nothing to disclose.
References
- 1.Lambers Heerspink HJ, Perkovic V, de Zeeuw D. Renal and cardio-protective effects of direct renin inhibition: a systematic literature review. J Hypertens. 2009;27:2321–2331. doi: 10.1097/HJH.0b013e3283310f92. [DOI] [PubMed] [Google Scholar]
- 2.Williams GH, Moore TJ, Hollenberg NK. Dysregulation of aldosterone secretion and its relationship to the pathogenesis of essential hypertension. Endocrinol Metab Clin North Am. 1991;20:423–447. [PubMed] [Google Scholar]
- 3.Stanton AV, Dicker P, O’Brien ET. Aliskiren monotherapy results in the greatest and the least blood pressure lowering in patients with high- and low-baseline PRA levels, respectively. Am J Hypertens. 2009;22:954–957. doi: 10.1038/ajh.2009.114. [DOI] [PubMed] [Google Scholar]
- 4.Moore N, Dicker P, O’Brien JK, Stojanovic M, Conroy RM, Treumann A, et al. Renin gene polymorphisms and haplotypes, blood pressure, and responses to renin-angiotensin system inhibition. Hypertension. 2007;50:340–347. doi: 10.1161/HYPERTENSIONAHA.106.085563. [DOI] [PubMed] [Google Scholar]
- 5.Mansego ML, Redon J, Marin R, Gonzalez-Albert V, Martin-Escudero JC, Fabia MJ, et al. Renin polymorphisms and haplotypes are associated with blood pressure levels and hypertension risk in postmenopausal women. J Hypertens. 2008;26:230–237. doi: 10.1097/HJH.0b013e3282f29865. [DOI] [PubMed] [Google Scholar]
- 6.Hasimu B, Nakayama T, Mizutani Y, Izumi Y, Asai S, Soma M, et al. Haplotype analysis of the human renin gene and essential hypertension. Hypertension. 2003;41:308–312. doi: 10.1161/01.hyp.0000049762.77830.89. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad U, Saleheen D, Bokhari A, Frossard PM. Strong association of a renin intronic dimorphism with essential hypertension. Hypertens Res. 2005;28:339–344. doi: 10.1291/hypres.28.339. [DOI] [PubMed] [Google Scholar]
- 8.Frossard PM, Malloy MJ, Lestringant GG, Kane JP. Haplotypes of the human renin gene associated with essential hypertension and stroke. J Hum Hypertens. 2001;15:49–55. doi: 10.1038/sj.jhh.1001107. [DOI] [PubMed] [Google Scholar]
- 9.Chiang FT, Hsu KL, Tseng CD, Lo HM, Chern TH, Tseng YZ. Association of the renin gene polymorphism with essential hypertension in a Chinese population. Clin Genet. 1997;51:370–374. doi: 10.1111/j.1399-0004.1997.tb02493.x. [DOI] [PubMed] [Google Scholar]
- 10.Fu Y, Katsuya T, Asai T, Fukuda M, Inamoto N, Iwashima Y, et al. Lack of correlation between Mbo I restriction fragment length polymorphism of renin gene and essential hypertension in Japanese. Hypertens Res. 2001;24:295–298. doi: 10.1291/hypres.24.295. [DOI] [PubMed] [Google Scholar]
- 11.Berge KE, Berg K. No effect of a BglI polymorphism at the renin (REN) locus on blood pressure level or variability. Clin Genet. 1994;46:436–438. doi: 10.1111/j.1399-0004.1994.tb04413.x. [DOI] [PubMed] [Google Scholar]
- 12.Chamarthi B, Williams JS, Williams GH. A mechanism for salt-sensitive hypertension: abnormal dietary sodium-mediated vascular response to angiotensin-II. J Hypertens. 2010;28:1020–1026. doi: 10.1097/HJH.0b013e3283375974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams GH, Tuck ML, Sullivan JM, Dluhy RG, Hollenberg NK. Parallel adrenal and renal abnormalities in young patients with essential hypertension. Am J Med. 1982;72:907–914. doi: 10.1016/0002-9343(82)90851-8. [DOI] [PubMed] [Google Scholar]
- 14.Fisher ND, Hurwitz S, Ferri C, Jeunemaitre X, Hollenberg NK, Williams GH. Altered adrenal sensitivity to angiotensin II in low-renin essential hypertension. Hypertension. 1999;34:388–394. doi: 10.1161/01.hyp.34.3.388. [DOI] [PubMed] [Google Scholar]
- 15.Pojoga L, Kolatkar NS, Williams JS, Perlstein TS, Jeunemaitre X, Brown NJ, et al. Beta-2 adrenergic receptor diplotype defines a subset of salt-sensitive hypertension. Hypertension. 2006;48:892–900. doi: 10.1161/01.HYP.0000244688.45472.95. [DOI] [PubMed] [Google Scholar]
- 16.Emanuel RL, Cain JP, Williams GH. Double antibody radioimmunoassay of renin activity and angiotensin II in human peripheral plasma. J Lab Clin Med. 1973;81:632–640. [PubMed] [Google Scholar]
- 17.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velez JC. The importance of the intrarenal renin-angiotensin system. Nat Clin Pract Nephrol. 2009;5:89–100. doi: 10.1038/ncpneph1015. [DOI] [PubMed] [Google Scholar]
- 20.Hollenberg NK, Price DA, Fisher ND, Lansang MC, Perkins B, Gordon MS, et al. Glomerular hemodynamics and the renin-angiotensin system in patients with type 1 diabetes mellitus. Kidney Int. 2003;63:172–178. doi: 10.1046/j.1523-1755.2003.00701.x. [DOI] [PubMed] [Google Scholar]
- 21.Kobori H, Nangaku M, Navar LG, Nishiyama A. The intrarenal renin-angiotensin system: from physiology to the pathobiology of hypertension and kidney disease. Pharmacol Rev. 2007;59:251–287. doi: 10.1124/pr.59.3.3. [DOI] [PubMed] [Google Scholar]
- 22.Geese WJ, Achanzar W, Rubin C, Hariharan N, Cheng P, Tomlinson L, et al. Genetic and gene expression studies implicate renin and endothelin-1 in edema caused by peroxisome proliferator-activated receptor gamma agonists. Pharmacogenet Genomics. 2008;18:903–910. doi: 10.1097/FPC.0b013e32830a6ea0. [DOI] [PubMed] [Google Scholar]
- 23.Germain S, Bonnet F, Fuchs S, Philippe J, Corvol P, Pinet F. Dissection of silencer elements in first intron controlling the human renin gene. J Hypertens. 1999;17:899–905. doi: 10.1097/00004872-199917070-00005. [DOI] [PubMed] [Google Scholar]
- 24.Voigtlander T, Ganten D, Bader M. Transcriptional regulation of the rat renin gene by regulatory elements in intron I. Hypertension. 1999;33:303–311. doi: 10.1161/01.hyp.33.1.303. [DOI] [PubMed] [Google Scholar]
- 25.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanton A, Jensen C, Nussberger J, O’Brien E. Blood pressure lowering in essential hypertension with an oral renin inhibitor, aliskiren. Hypertension. 2003;42:1137–1143. doi: 10.1161/01.HYP.0000101688.17370.87. [DOI] [PubMed] [Google Scholar]
- 27.Hopkins PN, Hunt SC, Jeunemaitre X, Smith B, Solorio D, Fisher ND, et al. Angiotensinogen genotype affects renal and adrenal responses to angiotensin II in essential hypertension. Circulation. 2002;105:1921–1927. doi: 10.1161/01.cir.0000014684.75359.68. [DOI] [PubMed] [Google Scholar]


