Abstract
F-box proteins play diverse roles in regulating numerous physiological processes in plants. This study isolated a gene (OsFbx352) from rice encoding an F-box domain protein and characterized its role in seed germination. Expression of OsFbx352 was upregulated by abscisic acid (ABA). The transcripts of OsFbx352 were increased upon imbibition of rice seeds and the increase was markedly suppressed by glucose. Germination of seeds with overexpression of OsFbx352 was less suppressed by glucose than that of wild-type seeds, while glucose had greater inhibition for germination of seeds with knockdown of OsFbx352 by RNA interference (RNAi) than that of wild-type seeds. The differential response of germination of the transgenic and wild-type seeds to glucose may be accounted for by differences in ABA content among overexpressing, RNAi, and wild-type seeds such that overexpression of OsFbx352 and knockdown of OsFbx352 led to lower and higher ABA contents, respectively, than that of wild-type seeds in the presence of glucose. Overexpression of OsFbx352 led to a reduction in expression of genes responsible for ABA synthesis (OsNced2, OsNced3) and an increase in expression of genes encoding ABA catabolism (OsAba-ox2, OsAba-ox3) in the presence of glucose. These findings indicate that OsFbx352 plays a regulatory role in the regulation of glucose-induced suppression of seed germination by targeting ABA metabolism.
Key words: ABA, F-box protein, glucose, OsFbx352, rice (Oryza sativa), seed germination
Introduction
The ubiquitin (Ub)-26S proteasome pathway is involved in the mediation of rapid and precise degradation of cellular proteins and plays a critical role in multiple cellular processes such as differentiation, cell division, hormonal responses, protein trafficking, and responses to environmental stress (Smalle and Vierstra, 2004; Dreher and Callis, 2007; Vierstra, 2009; Santner and Estelle, 2010; Lee and Kim, 2011). The ubiquitination pathway involves the covalent attachment of Ub to target proteins through a cascade catalysed by the three known enzymes of E1, E2, and E3. In most cases, E3 proteins that are responsible for identifying specific target substrate can be classified into two groups based on their composition of subunit (Smalle and Vierstra, 2004). The HECT and RING/U-box E3 families are comprised of single polypeptide, whereas the SCF and APC E3 ligases consist of multiple subunits (Moon et al., 2004). The SCF complex, which is composed of four primary subunits: Cullin1, Rbx1, Skp1, and a member of a large family of proteins known as F-box proteins, is one of the best characterized and most important E3 types (Smalle and Vierstra, 2004). In the SCF complex, the F-box protein interacts with Skp1 via an N-terminal conserved F-box motif and with the substrate via a C-terminal variable protein–protein interaction domain. The bipartite function allows the F-box proteins to play a crucial role in the SCF complex by identifying specificity of target substrate (Ho et al., 2008).
There are more than 692 and 779 F-box genes in the Arabidopsis thaliana and rice genomes, respectively (Xu et al., 2009). Several F-box proteins have been characterized to regulate diverse physiological processes ranging from hormonal signalling cascades to stress responses (Santner and Estelle, 2010; Lee and Kim, 2011). For instance, an F-box protein TIR1/AFB that acts as an auxin receptor, can bind to the transcriptional repressor protein Aux/IAA and promote their proteasomal degradation, activating transcription of related genes (Dharmasiri et al., 2005; Tan et al., 2007). Another F-box protein, ZTL, has been shown to play a role in control of circadian period and early photomorphogenesis by targeting PRR5 for proteasome-dependent degradation (Kiba et al., 2007). In addition, Zhang et al. (2008) reported that the F-box protein DOR acts as a negative regulator in the mediation of responses to abscisic acid (ABA) and drought stress. ABA is closely related to sugar signalling in regulation of seed germination (Rook et al., 2006; Dekkers et al. 2008). However, there has been no study to evaluate the role of F-box proteins in sugar-mediated seed germination in the literature.
Seed germination, a key process in plant life cycle, is regulated by many phytohormones and sugar signalling cascades (Koornneef et al., 2002; Leo’n and Sheen, 2003; Rolland et al., 2006). Glucose has been well documented to play a regulatory role in seed germination and post-germination development (Smeekens, 2003; Gibson, 2004; Rolland et al., 2006). Recent studies reveal that the effect of glucose on seed germination is closely related to ABA levels and ABA signalling cascades (Gibson 2004; Rook et al., 2006; Yuan and Wysocka-Diller, 2006; Dekkers et al., 2008). The endogenous ABA level in plant cells is determined by the balance between its biosynthesis and catabolism. Glucose-induced suppression of seed germination in Arabidopsis has been shown to be associated with elevation of endogenous ABA contents in seeds by upregulation of genes encoding ABA biosynthesis (Cheng et al., 2002; Chen et al., 2006) and suppression of ABA catabolism at the transcriptional level (Zhu et al., 2011). Similarly to Arabidopsis, suppression of seed germination in rice by glucose has been reported to result from increased ABA level due to inhibition of ABA catabolism (Zhu et al., 2009). The observations that seed germination of ABA-deficient and ABA-insensitive Arabidopsis mutants display similar phenotypes to that of sugar-insensitive mutants confirm the crosstalk between glucose and ABA in regulation of seed germination (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rolland et al., 2006; Dekkers et al., 2008). However, the signalling networks between ABA and glucose in modulation of seed germination remain to be fully dissected.
This study identified an F-box gene, designed OsFbx352, from rice, and characterized its function during seed germination by generating transgenic rice plants with overexpression and knockdown of OsFbx352. More specifically, this study evaluated the role of OsFbx352 in seed germination with emphasis on the interactions among ABA, OsFbx352, and glucose. The results reveal that OsFbx352 was involved in the regulation of glucose-induced delay of seed germination by modulating ABA metabolism.
Materials and methods
Plant growth and treatments
Rice (Oryza sativa L. ssp. japonica cv. Zhonghua 10) seedlings were grown in the greenhouse at 27/23 °C with a 14/10 light/dark cycle. The rice seeds were supplied by the China National Rice Research Institute (Hangzhou, Zhejiang, China). To determine OsFbx352 transcript levels,2-week-old wild-type (WT) seedlings were exposed to incubation solution containing 0.1mM ABA (Sigma-Aldrich) and leaves were sampled after 2 and 5h. For determination of seed germination, seeds were exposed to solutions containing water (control), 0.1mM ABA, 3% mannitol, or 3% glucose. Germination rate was scored after varying periods (0, 2, 6, 12h). A similar treatment regime was used to study the gene expression of WT and transgenic rice seeds.
Plasmid construction and plant transformation
The full-length cDNA of OsFbx352 was amplified from rice plants with the primers OsFbx352-FL-F (5'-CGCGGATCCATGCCA- CCGCGCGCTCTGCC-3'; BamHI site in bold) and OsFbx352-FL-R (5'-CGGGGTACCTCATATCGACCGAATAAATG-3'; KpnI site in bold). The sequencing confirmed that PCR fragment was directionally cloned into the BamHI and KpnI sites of a pUN1301 vector to create the pUN1301-OsFbx352 construct. To construct the RNA interference (RNAi) vector, the 490-bp coding sequence was amplified with the primers OsFbx352-Ri-F (5'-GGGGTACCACTA- GTTCTCCTCCCATACTCCC-3'; KpnI and SpeI sites in bold) and OsFbx352-Ri-R (5'-CGGGATCCGAGCTCATGCCACTGCTAA- CAAAG-3′; BamHI and SacI sites in bold), digested by KpnI and BamHI and then SpeI and SacI, and ligated to the pTCK303 vector. The protocols used for transformation of rice plants followed those described by Yang et al. (2004). Briefly, rice seeds were sterilized and cultured on MS medium plus 4mg 2,4-dichlorophenoxyacetic acid l–1 (Sigma) for 4 weeks in the dark at 28 °C to induce embryogenic calluses, and the above constructs were introduced into the calluses by Agrobacterium tumefaciens EHA105-mediated transformation.
Seed germination assays
To determine seed germination, 40 seeds of T2 generation of different transgenic lines and WT were sterilized and spread on half-strength MS medium containing different concentrations of glucose and mannitol (pH 5.8). The MS medium itself contained no sugar. Seeds were placed in a growth chamber with 12/12 light/dark cycle at 25 °C. Germination is defined as the emergence of the radicles through the seed coat. Every experiment was repeated three times.
Determination of endogenous ABA content in seeds
Seeds were treated with water, mannitol, or glucose for 0 and 6h and immediately frozen in liquid nitrogen after determination of their freshweight, and were subsequently stored at –80 °C for measurement of endogenous ABA content. Samples were ground in liquid nitrogen using a mortar and pestle, extracted with 4ml of 80% (v/v) methanol containing 1mM butylated hydroxytoluene as an antioxidant. The extract was incubated at 4 °C for 24h and centrifuged at 13,400 g for 15min at 4 °C. After the residues were suspended in the same extraction solution and stored at 4 °C for 1h, they were centrifuged again at 13,400 g for 15min at the same temperature. The two supernatants were combined and passed through a C18 Sep-Pak cartridge (Waters, Milford, MA, USA).The efflux was collected and dried in N2 and then dissolved in 0.5ml phosphate-buffered saline containing 0.1% (v/v) Tween 20 and 0.1% (w/v) gelatin (pH 7.5) for analysis of ABA. ABA concentration was determined by enzyme-linked immunosorbent assay (ELISA) following methods described elsewhere (Yang et al., 2001; Zhu et al., 2005; Chen et al., 2006). The results are mean ± SE of at least three replicates.
RNA isolation and quantitative real-time PCR
RNA isolation and quantitative real-time PCR (qRT-PCR) was carried out as described by Song et al. (2012). Total RNA was extracted with Trizol reagent (Invitrogen) and treated with RNase-free DNase I (Promega). The total RNAs were reverse transcribed into first-strand cDNA in a 20-µl volume with M-MLV reverse transcriptase (Promega). The samples were diluted to 100 µl with water and 5 µl of each sample (~8ng RNA equivalent) was amplified using SYBR GreenER qPCR SuperMix Universal (Invitrogen) in a 25 µl reaction, containing 5 µl diluted cDNA, 12.5 µl SYBR GreenER qPCR SuperMix Universal, 0.5 µl Rox Reference Dye, 1 µl 10 µM forward primer, 1 µl of 10 µM reverse primer, and 5 µl water. The thermal cycle program was 95 °C for 3min; 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. The corresponding specific primers were: for OsNced2, 5'-GGTATGGAAACGAGGATAGTGGTT-3' and 5'-TGCTTATTGTTGTGCGAGAAGTTC-3'; for OsNced3, 5'-CCCCTCCCAAACCATCCAAACCGA-3' and 5'-TGTGAGCATAT- CCTGGCGTCGTGA-3'; for OsAba-ox2, 5'-CTACTGCTGATGG- TGGCTGA-3' and 5'-CCCATGGCCTTTGCTTTAT-3'; for OsAba-ox3, 5'-AGTACAGCCCATTCCCTGTG-3' and 5'-ACGCCTAATCAAAC- CATTGC-3'; for OsFbx352, 5'-GGATTCAAACTTCTATGCCCTAA-3' and 5'-CATGTAAACGATATTTCCGTAAGC-3'. Rice Actin1 (accession no. AB047313) was used as internal control with primers 5'-ACCACAGGT- ATTGTGTTGGACTC-3' and 5'-AGAGCATATCCTTCATAGATG- GG-3'. The relative expression levels were determined as described by Livak and Schmittgen (2001).
Statistics
All data were analysed by analysis of variance using the SAS statistics program. Statistical differences were referred to as significant when P < 0.05 or P < 0.01.
Results
OsFbx352 encodes a F-box domain protein and is suppressed by glucose
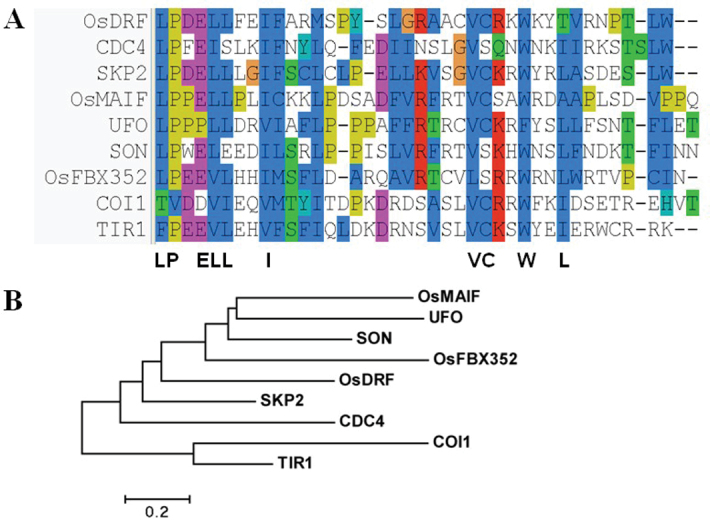
A OsFbx352 gene was isolated from a salt-stress rice DNA microarray. The gene corresponding to locus Os10g03850 consisted of three exons and three introns with a 1275-bp open reading frame and encoded a putative protein of 425 amino acids. A blastp search revealed that OsFBX352 was an F-box protein containing an F-box domain in its N-terminal region, which is a hallmark of F-box proteins from Arabidopsis, rice, yeast, and humans. A database search with the predicted polypeptide sequence showed that the F-box domain of OsFBX352 was related to those of the previously identified F-box proteins, including Arabidopsis SON (70% similarity), yeast CDC4 (65% similarity), rice OsDRF (67.5% similarity), and human SKP2 (57.5% similarity) (Fig. 1A). Phylogenetic analysis also revealed that OsFBX352 was closely related to Arabidopsis SON1 and UFO and to rice MAIF (Fig. 1B). These results suggest that OsFBX352 may be involved in the formation of E3 ubiquitin ligase complex.
Fig. 1.
OsFbx352 contains a conserved F-box domain.(A) Alignment of the conserved F-box domain in OsFbx352 with other representative F-box-containing proteins: blue indicates residues identical to the OsFbx352 sequence; other colours indicate similar amino acids. The sequence at the bottom is the consensus sequence. (B) Phylogenetic tree analysis of the conserved F-box domain in OsFbx352 with other known F-box proteins from diverse organisms. Alignment and phylogenetic analysis were performed using clustal x 2 and mega 5, respectively. Approximately 40 amino acids that constitute the F-box motif in OsFBX352 were aligned with comparable regions from reported F-box proteins in humans (SKP2, BAB87202.1), yeast (CDC4, NP_116585.1), Arabidopsis (COI1, NP_565919; SON1, NP_179323; UFO, NP_564368.1; TIR1, NP_567135.1), and rice (OsMAIF, NP_001047698.1; OsDRF, NP_001052823.1).
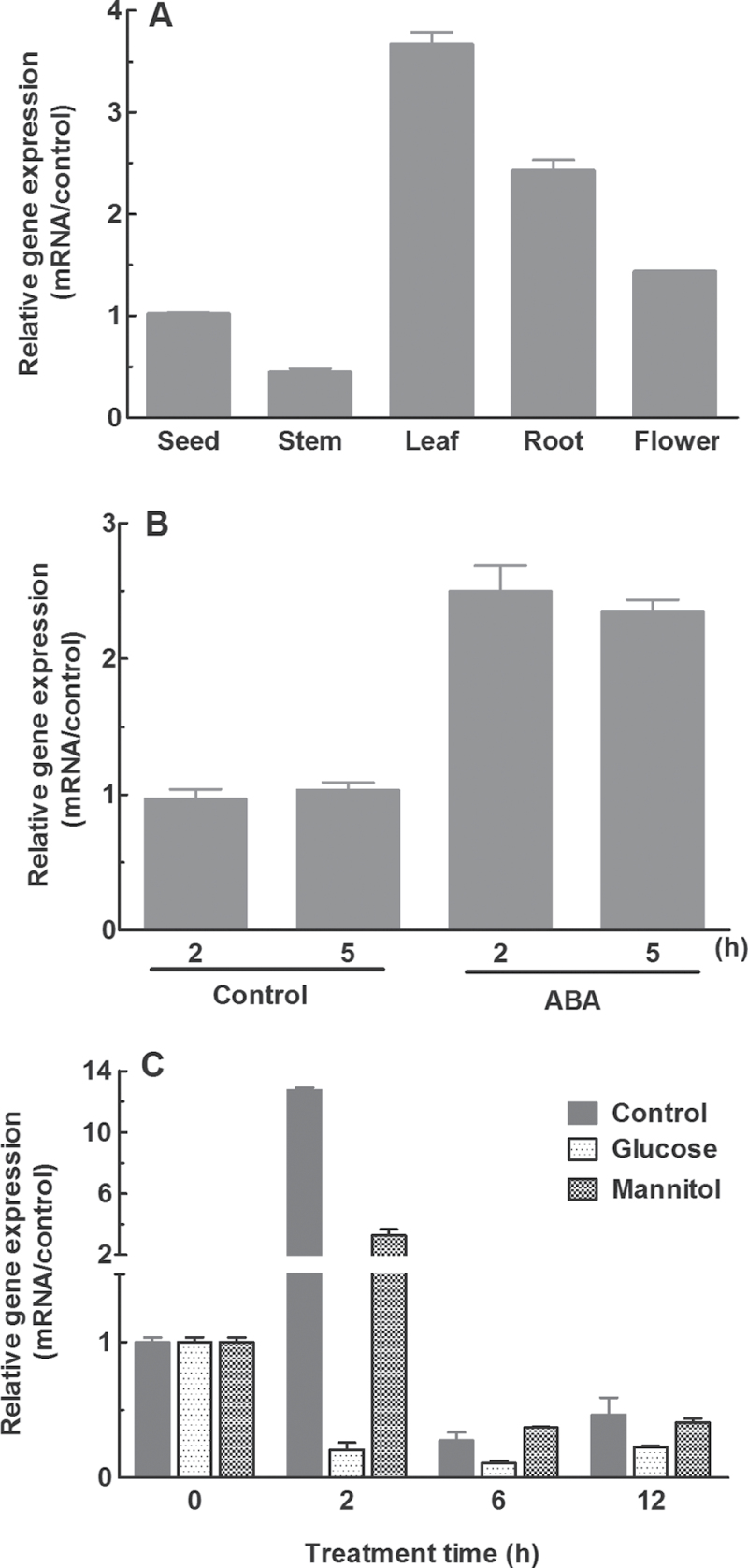
The expression patterns of OsFbx352 were analysed by qRT-PCR (Fig. 2). As shown in Fig. 2A, the expression of OsFbx352 was detected in the organs examined with the expression being highest in leaves and lowest in stems (Fig. 2A). The transcripts of OsFbx352 were increased by treatment with ABA (Fig. 2B). ABA has been shown to be involved in glucose-induced delay of seed germination (Gibson, 2004; Rook et al., 2006; Yuan and Wysocka-Diller, 2006; Zhu et al., 2009). To test whether OsFbx352 is associated with glucose-delayed seed germination, the response of OsFbx352 to glucose at transcriptional level was investigated. There was a rapid and transient increase in the transcripts of OsFbx352 upon imbibition of rice seeds such that the abundance of OsFbx352 transcripts peaked after 2h of imbibition and declined thereafter (Fig. 2C). The increase in OsFbx352 transcripts was markedly suppressed by glucose (Fig. 2C). The expression of OsFbx352 was much less increased when an identical concentration of mannitol was used to treat the seeds (Fig, 2C), suggesting that the inhibitory effect of glucose on OsFbx352 expression could not be accounted for by its osmotic effect.
Fig. 2.
Quantitative real-time PCR analysis of the expression patterns of OsFbx352. (A) Expression of OsFbx352 in different organs. (B) Effect of abscisic acid (ABA, 100 µM) on the expression of OsFbx352. (C) Response of OsFbx352 expression to water, mannitol, and glucose treatments in seed germination. Data are mean ± SE for three replicates.
Generation of transgenic rice plants with altered expression of OsFbx352
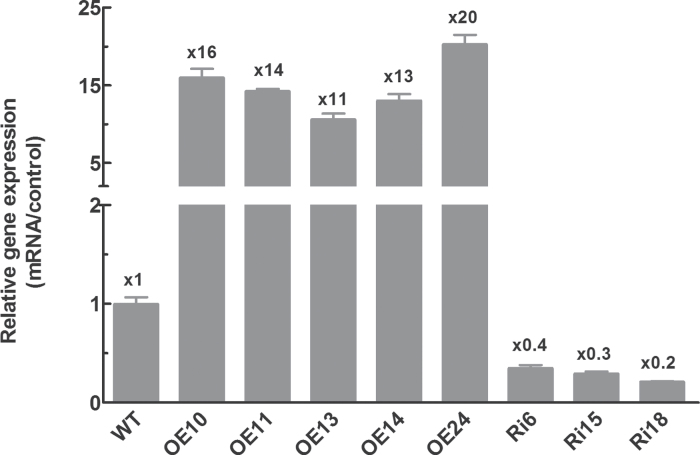
To characterize the function of OsFbx352, transgenic rice plants were generated by constructing a RNAi construct with part of the coding sequence fragment to knock down OsFbx352 expression, as well as an overexpression construct with the OsFbx352 full-length open reading frame driven by a maize ubiquitin promoter. Both constructs were introduced into rice calluses via Agrobacterium-mediated transformation. Several transgenic lines were obtained and subsequently confirmed by qRT-PCR. The expression of OsFbx352 in the overexpression seeds was at least 11-fold higher than that in WT seeds, while OsFbx352 transcripts in seeds of the knockdown lines were reduced by approximately 70% compared with WT seeds (Fig. 3). Among the transgenic lines, two overexpression lines (OE10, OE24) and two knockdown lines (Ri15, Ri18) were selected to study the physiological function of OsFbx352 during seed germination.
Fig. 3.
Quantitative real-time PCR analysis of OsFbx352 expression in wild-type (WT), overexpression (OE10, OE11, OE13, OE14, and OE24), and RNAi (Ri6, Ri15, and Ri18) plants. Data are mean ± SE for three replicates. Values above columns are the changes in gene expression relative to the WT.
Germination of transgenic seeds displayed different sensitivity to ABA
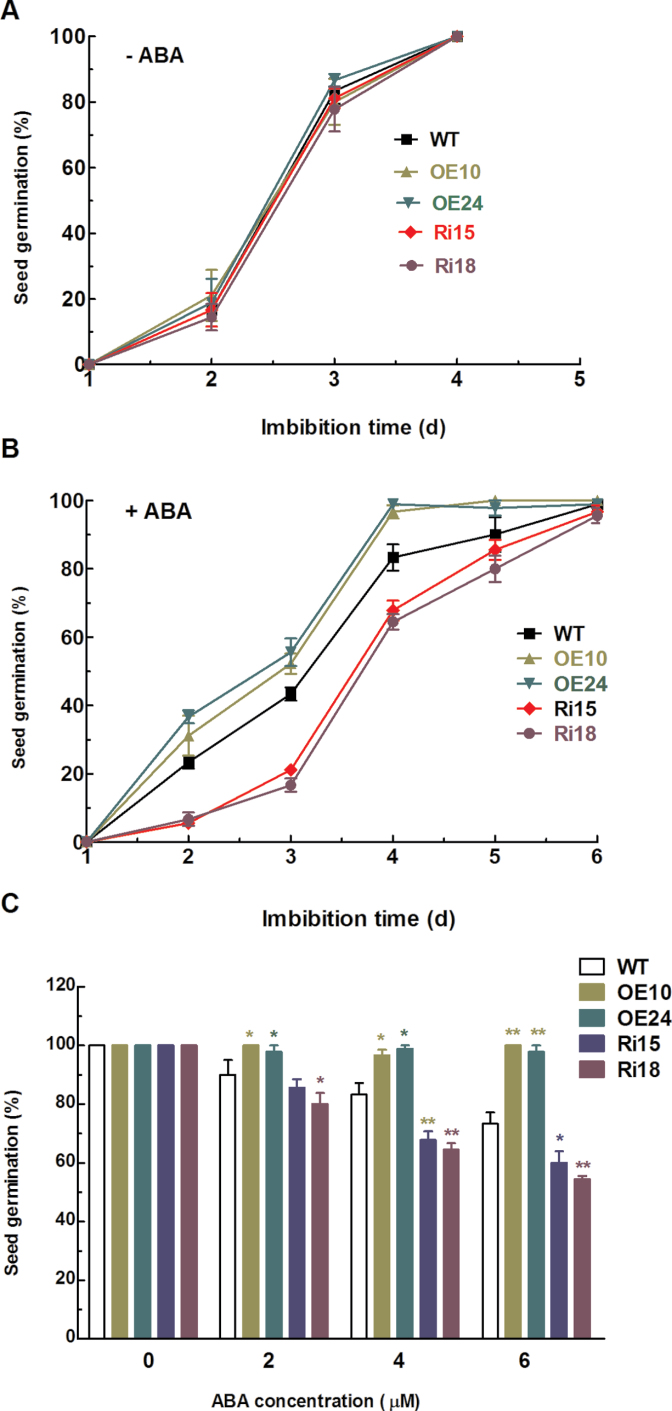
Given that seed germination is sensitive to ABA and expression of OsFbx352 was strongly induced by ABA, the role of OsFbx352 played in seed germination was evaluated. There were no differences in seed germination among WT and transgenic lines in the control medium, and both WT and transgenic seeds were fully germinated after 4 days of incubation in the control medium (Fig. 4A). Germination of WT and RNAi seeds was delayed when ABA was present in the incubation medium, while germination of seeds with overexpressing OsFbx352 was relatively insensitive to ABA (Fig. 4B). In addition, the inhibitory effect of ABA on germination of RNAi seeds was more evident than that of WT seeds (Fig. 4B). For instance, both WT and transgenic seeds were germinated after 4 days in the control medium, while the germination rate for WT and RNAi seeds was reduced to approximately 80 and 60%, respectively, after the same period incubation in the presence of ABA (Fig. 4A, 4B). Moreover, the inhibitory effect of ABA on seed germination of WT and RNAi seeds was increased with an increase in ABA concentrations (Fig. 4C). In addition to seed germination, overexpression of OsFbx352 also rendered the seedlings less sensitive to ABA, as evidenced by longer shoot length of overexpression lines than the WT in the presence of ABA while suppression of OsFbx352 led to a shorter shoot length than the WT (Supplementary Fig. S1, available at JXB online).
Fig. 4.
OsFbx352-overexpressing seeds exhibited decreased sensitivity to exogenous abscisic acid (ABA). (A, B) Seeds of wild-type (WT), OE10, OE24, Ri15, and Ri18 lines were germinated in control medium (A) or 4 µM ABA (B) for 4 days. (C) Seed germination rate for WT and transgenic rice seeds after incubation in without and with different concentrations of ABA for 4 days. Data are mean ± SE for three replicates with each replicate containing 30 seeds. Statistically significant difference from the wild type within a treatment: *, P < 0.05; **, P < 0.01(this figure is available in colour at JXB online).
Seed germination and seedling growth of transgenic seeds displayed different sensitivity to glucose
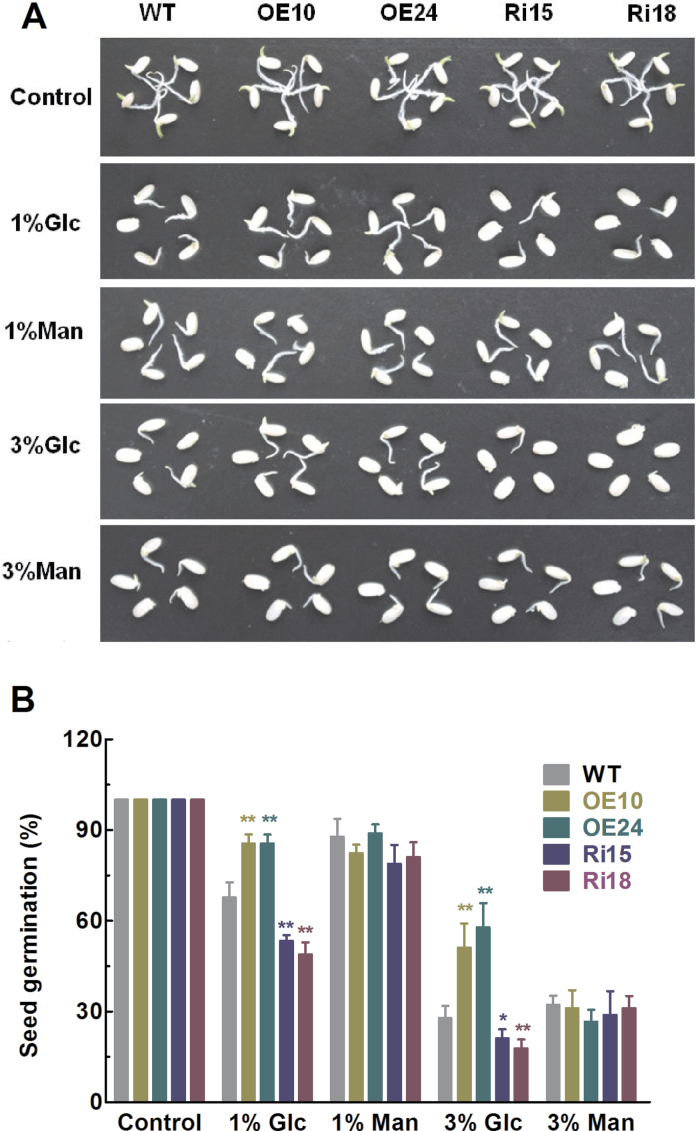
In addition to ABA, the sensitivity of seed germination to glucose was also investigated using WT and transgenic seeds with overexpression (OE10, OE24) and knockdown of OsFbx352 (Ri15, Ri18). As shown in Fig. 5A, there were no significant differences in seed germination among WT, overexpressing, and RNAi lines in the control medium. Addition of glucose to the incubation medium significantly suppressed seed germination for WT, overexpressing, and knockdown seeds (Fig. 5A). The inhibitory effect of glucose on seed germination in the overexpressing lines was significantly less than that of the WT, while the inhibitory effect on seed germination of the RNAi lines was greater than that of WT and overexpressing lines (Fig. 5). The differential effect of glucose on germination of WT, overexpressing, and RNAi seeds became more evident with an increase in glucose concentration from 1 to 3%. In contrast to glucose, addition of identical concentration of mannitol to the incubation medium had similar inhibitory effect on seed germination of both WT and transgenic seeds (Fig. 5), indicting the specific effect of glucose on seed germination.
Fig. 5.
Effects of glucose on wild-type (WT), OsFbx352-overexpressing, and RNAi seeds during germination.(A) Representative images of WT, OE10, OE24, Ri15, and Ri18 seeds on half-strength MS medium with or without glucose or mannitiol for 4 days. (B) Germination rate of WT and transgenic seeds in the indicated conditions at 4 days after initiation. Data are mean ± SE for three replicates with each replicate containing 30 seeds. Data were statistically analysed with t-test. Statistically significant difference from the wild type within a treatment:*, P < 0.05; **, P < 0.01.
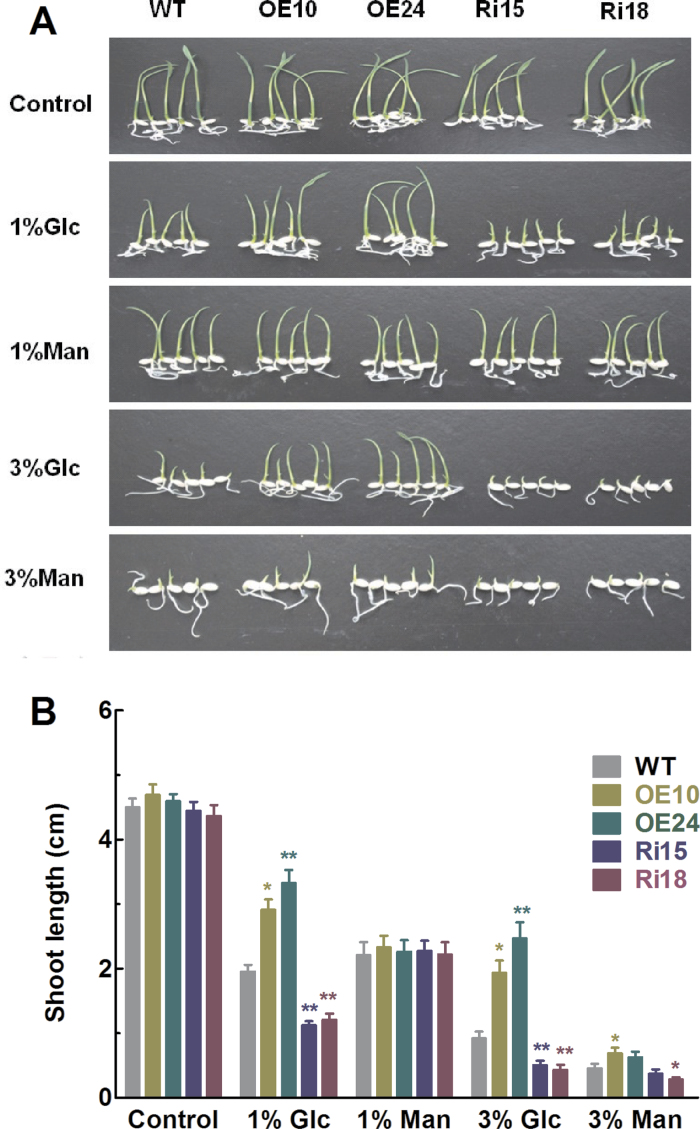
In addition to inhibition of seed germination, glucose can also arrest the post-germination growth (Jang and Sheen, 1997; Gibson, 2005). In the present study, no obvious differences in phenotypes were observed between WT and transgenic plants when they were grown on half-strength MS medium (Fig. 6A). There was a marked reduction in shoot growth for WT, overexpressing, and RNAi lines when glucose was added to the incubation medium, and the inhibitory effect was increased with an increase in glucose concentration (Fig. 6B). Similar to seed germination, glucose had less inhibitory effect on shoot growth in the OsFbx352-overexpressing lines than in the WT, while it had greater inhibitory effect on shoot growth in the OsFbx352-knockdown lines than the WT (Fig. 6B). The differential effect of glucose on shoot growth of WT, overexpressing, and RNAi lines was specific to glucose as shoot growth of WT, overexpressing, and RNAi lines was equally inhibited by 1% mannitol (Fig. 6). At a higher concentration of mannitol, shoot length of OE10 was significantly longer than that of WT, and shoot length of Ri18 was significantly shorter than that of WT in the presence of 3% mannitol (Fig. 6).
Fig. 6.
Effects of OsFbx352 on the post-germination development of the seedlings. (A) Representative images of seeds from wild-type (WT) and transgenic plants grown on half-strength MS medium with or without glucose or mannitiol at the post-germination stage. (B) Effect of glucose on shoot length of WT and transgenic seedlings. Data are mean ± SE for three replicates with each replicate containing 15 seeds. Data were statistically analysed with t-test. Statistically significant difference from the wild type within a treatment: *, P < 0.05;**, P < 0.01.
Overexpression of OsFbx352 suppressed glucose-induced increase in seed ABA content
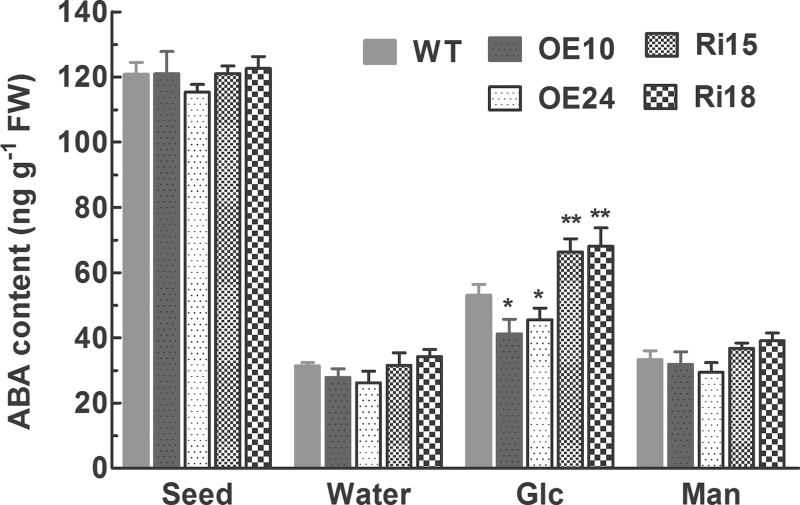
To investigate whether the altered sensitivity of seed germination in the OsFbx352-overexpressing and RNAi lines to glucose is related to ABA, changes in ABA contents in seeds of WT, overexpressing and RNAi lines were monitored during imbibition in the absence and presence of glucose. There were no differences in ABA contents in dry seeds among WT, overexpressing, and RNAi lines (Fig. 7). ABA contents in WT, overexpressing, and RNAi seeds were sharply reduced upon imbibition (Fig. 7). Moreover, ABA contents in the imbibed seeds were increased when glucose was present in the incubation medium, and the glucose-induced increase in ABA contents in the overexpressing seeds was significantly lower than in WT seeds, while ABA contents in the RNAi seeds were significantly higher than in WT seeds in the presence of glucose (Fig. 7). In contrast to glucose, addition of the same concentration of mannitol to the incubation medium had no effect on ABA levels in seeds of both WT and transgenic lines (Fig. 7), discounting the possibility that the glucose-induced increases in ABA levels in seeds are due to osmotic effect. These results suggest that overexpression of OsFbx352 disrupts glucose-dependent ABA metabolism, thus rendering the germination of overexpressing seeds less sensitive to glucose.
Fig. 7.
Effects of water, mannitol, and glucose treatments on endogenous ABA contents in germinating seeds. Seeds were treated with water, mannitol, or glucose for 6h at 28 °C and were subsequently collected for ABA quantification. Data are mean ±SE with three replicates. Data were statistically analysed with t-test. Statistically significant difference from the wild type within a treatment: *, P < 0.05; **, P < 0.01.
ABA biosynthesis and catabolism were affected by alterations in OsFbx352 expression
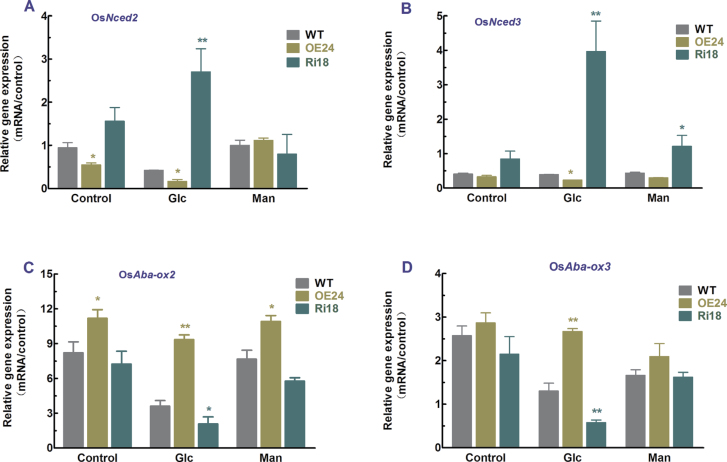
NCED and ABA8ox are key genes involved in regulating ABA biosynthesis and catabolism (Saito et al., 2004; Zhu et al., 2011, 2009). To further evaluate the role of OsFbx352 played in the regulation of ABA-dependent seed germination, the effect of glucose on ABA synthesis and metabolism at the transcriptional level was also studied. The abundance of OsNced2 in seeds of the overexpressing line OE24 was significantly lower than that in WT seeds imbibed in water (control) (Fig. 8A), while the abundance of OsNced2 in the Ri18 seeds was significantly higher than that in WT seeds under the control conditions (Fig. 8A), suggesting that overexpression of OsFbx352 may downregulate ABA levels in seeds by enhancing ABA catabolism and suppressing ABA synthesis at the transcriptional level. The expression of OsNced2 was suppressed in both WT and OE24 seeds by glucose (Fig. 8A). By contrast, glucose markedly upregulated expression of OsNced2 in seeds of the RNAi line (Fig. 8A). A similar effect of glucose on the expression of OsNced3 was also observed (Fig. 8B). In contrast to glucose, mannitol at the same concentration equally suppressed expression of OsNced2 and OsNced3 (Fig. 8A, 8B). Unlike OsNced genes, overexpression of OsFbx352 led to an increase in OsAba-ox2 transcripts, while OsAba-ox3 transcripts in WT, overexpressing, and RNAi seeds were not significantly different under control conditions (Fig. 8C, 8D). Expression of OsAba-ox2 was suppressed in seeds of WT, OE24, and Ri18 lines when challenged by glucose, but the glucose-induced suppression of OsAba-ox2 expression differed among WT, OE24, and Ri18 lines such that the expression of OsAba-ox2 was significantly higher and lower in seeds of the OE24 and Ri18 lines than the WT, respectively (Fig. 8C). A similar differential response of OsAba-ox3 expression among seeds of WT, OE24, and Ri18 lines to glucose was observed (Fig. 8D). In general, expression of OsAba-ox2 and OsAba-ox3 in seeds of WT, OE24, and Ri18 lines was relatively insensitive to the addition of the same concentrations of mannitol to the incubation medium (Fig. 8C, 8D).
Fig. 8.
Expression of ABA biosynthesis and catabolism genes of WT, OE24, and Ri18 rice seeds in response to glucose. Seeds were grown on plates containing water, mannitiol, or glucose for 6h and then harvested for RNA extraction. Data are mean ± SE with three replicates. Data were statistically analysed with t-test. Statistically significant difference from the wild type within a treatment: *, P < 0.05; **, P < 0.01 (this figure is available in colour at JXB online).
Discussion
F-box proteins comprise one of the large protein families in plants. There are more than 692 and 779 F-box genes in the A. thaliana and rice genomes, respectively (Xu et al., 2009). Previous studies demonstrated that F-box proteins are involved in the regulation of several biological processes, including circadian clock, self-incompatibility, photomorphogenesis, floral meristem, phytohormonal signalling, and responses to abiotic stresses (Sullivan et al., 2003; Moon et al., 2004; Ni et al., 2004; Smalle and Vierstra, 2004; Dreher and Callis, 2007). For instance, the F-box protein TIR1/AFB can bind to the transcriptional repressor protein Aux/IAA, thus promoting their proteasomal degradation and activating auxin-induced transcription (Dharmasiri et al., 2005; Tan et al., 2007). The involvement of F-box proteins in response to both biotic and abiotic stresses has also been reported. For example, four F-box proteins from different species (DOR, OsDRF1, PvFBS1, and MAIF1) have been shown to act as key regulators in responses to biotic (Cao et al., 2008) and abiotic stress (Zhang et al., 2008; Yan et al., 2011; Maldonado-Calderón et al., 2012). Kiba et al. (2007) reported that an F-box protein, ZTL, is a major factor to control the period of circadian and early photomorphogenesis by regulating PRR5 for proteasome-dependent degradation. As a primary sugar, glucose modulates many important physiological processes ranging from seed germination to seedling development (Smeekens, 2000; Dekkers et al., 2004; Gibson, 2005; Rolland et al., 2006). However, there has been no report on the involvement of F-box proteins in glucose-regulated physiological processes in general and seed germination in particular. The present study identified a novel F-box gene in rice, OsFbx352, and demonstrated that disruption of OsFbx352 expression altered the sensitivity of its seed germination and post-germination growth to glucose. The study further explored the physiological mechanism by which transgenic rice seeds with altered expression of OsFbx352 exhibited different sensitivity to glucose. One important finding is that disruption of OsFbx352 expression altered glucose-induced ABA content in seeds such that ABA content in seeds of the overexpressing and underexpressing OsFbx352 was lower and higher, respectively, than in WT seeds in the presence of glucose. To the best knowledge of the authors, this is the first report highlighting the involvement of F-box protein in suppression of seed germination and post-germination growth by glucose.
It has been well documented that the inhibitory effect of sugar on seed germination is due to the increase in the active endogenous ABA level (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Zhu et al., 2009, 2011; Carvalho et al., 2010). For example, Zhu et al. (2009) reported that a higher ABA content in imbibed seeds may account for the glucose-induced delay of rice seed germination. The present results showing that ABA contents were markedly increased in imbibed seeds of both WT and transgenic lines with altered expression of OsFbx352 when challenged with glucose are in line with those findings reported in the literature. In addition, this study found that the glucose-induced increase in ABA content in the OsFbx352-overexpressing seeds was lower than in WT seeds, while it was higher in the two RNAi seeds than in WT seeds (Fig. 7). These results suggest that overexpression of OsFbx352 disrupts glucose-induced ABA metabolism, thus rendering the germination of overexpressing seeds less sensitive to glucose.
The differential response of ABA contents in WT and transgenic seeds to glucose was further investigated by studying expression of Nced and Aba-ox genes that are responsible for ABA biosynthesis and catabolism (Cheng et al., 2002; Saito et al., 2004; Nambara and Marion-Poll, 2005; Zhu et al., 2009, 2011). The results revealed that the expression of OsNced2 and OsNced3 was suppressed in seeds of both the WT and the overexpressing line, while the expression of OsAba-ox2 and OsAba-ox3 were significantly higher and lower, in seeds of the OE24 line and the Ri18 line, respectively, than the WT. These results indicate that OsFbx352 may participate in glucose-induced suppression of seed germination by stimulating ABA biosynthesis and inhibiting ABA catabolism. A similar inhibition of ABA catabolism by glucose leading to delay of seed germination has been reported by Zhu et al. (2009). However, in contrast to those findings by Zhu et al. (2009), the present results showed that expression of OsNced2 and OsNced3 was suppressed by glucose (Fig. 8). The present data do not suggest an explanation for the difference, but it can be speculated that differences in the rice genotypes and the experimental protocols used in the two studies may partly account for the difference.
There are a number of reports showing that proteins such as G-protein (Chen et al., 2006), SR45 (Carvalho et al., 2010), and bZIP protein (Matiolli et al., 2011) are involved in regulation of glucose and ABA-dependent seed germination and early seedling development. There are some reports suggesting that F-box proteins may be involved in ABA biosynthesis and signalling. For example, Zhang et al. (2008) reported that an F-box protein, DOR, is involved in regulation of ABA-induced stomatal closure under drought stress by affecting ABA signalling. Yan et al. (2011) also showed that an F-box protein in rice, MAIF, has a negative role in ABA signalling during rice seed germination and root growth.
In summary, this study identified a gene encoding an F-box protein from rice plants, OsFbx352, and characterized its function in seed germination by generating transgenic rice with overexpression and knockdown of OsFbx352. One important finding is that expression of OsFbx352 in seeds was upregulated during imbibition and that the increase in OsFbx352 expression was markedly suppressed by glucose. This study further demonstrated that the glucose-induced inhibition of seed germination was negatively correlated with OsFbx352 expression level. The differential response of the transgenic and WT seeds to glucose may be accounted for by differences in ABA contents among overexpressing, RNAi, and WT seeds due to alterations of ABA synthesis and catabolism at the transcriptional level. These findings highlight the regulatory role of OsFbx352 in glucose-induced suppression of seed germination by targeting ABA metabolism. Identification of the targets of OsFbx352 and elucidating the signalling network may help to unravel the molecular mechanism underlying the OsFbx352-dependent insensitivity to glucose.
Supplementary Material
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Shoot length of 2-day-old rice seedlings in wild-type and transgenic lines treated with 0, 4, and 8 µM abscisic acid for 8 days.
Acknowledgements
This work was supported by the National Science Foundation of China (31071844 and 30788003). The authors acknowledge Ms Chen Jie and Ms Chen Ying for their help in isolation of the gene and thank the two anonymous reviewers for their constructive suggestions.
References
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P. 2000. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar Genes and Development 14 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yang Y, Zhang H, Li D, Zheng Z, Song F. 2008. Overexpression of a rice defense-related F-box protein gene OsDRF1 in tobacco improves disease resistance through potentiation of defense gene expression Physiological Plantarum 134 440–452 [DOI] [PubMed] [Google Scholar]
- Carvalho RF, Carvalho SD, Duque P. 2010. The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis Plant Physiology 154 772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ji FF, Xie H, Liang JS, Zhang JH. 2006. The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination Plant Physiology 140 302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, et al. 2002. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions The Plant Cell 14 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers B, Schuurmans J, Smeekens S. 2004. Glucose delays seed germination in Arabidopsis thaliana Planta 218 579–588 [DOI] [PubMed] [Google Scholar]
- Dekkers B, Schuurmans J, Smeekens S. 2008. Interaction between sugar and abscisic acid signaling during early seedling development in Arabidopsis Plant Molecular Biology 67 151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor Nature 435 441–445 [DOI] [PubMed] [Google Scholar]
- Dreher K, Callis J. 2007. Ubiquitin, hormones and biotic stress in plants Annals of Botany 99 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S. 2004. Sugar and phytohormone response pathways: navigating a signaling network Journal of Experimental Botany 55 253–264 [DOI] [PubMed] [Google Scholar]
- Gibson S. 2005. Control of plant development and gene expression by sugar signaling Current Opinion in Plant Biology 8 93–102 [DOI] [PubMed] [Google Scholar]
- Ho MS, Ou C, Chan YR, Chien CT, Pi H. 2008. The utility F-box for protein destruction Cell and Molecular Life Sciences 65 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S. 2000. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses The Plant Journal 23 577–585 [DOI] [PubMed] [Google Scholar]
- Jang JC, Sheen J. 1997. Sugar sensing in higher plants Trends in Plant Science 2 208–214 [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua NH. 2007. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana The Plant Cell 19 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H. 2002. Seed dormancy and germination Current Opinion in Plant Biology 5 33–36 [DOI] [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI. 2000. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response The Plant Journal 23 587–596 [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim WT. 2011. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis Molecular Cell 31 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo’n P, Sheen J. 2003. Sugar and hormone connections Trends in Plant Science 8 110–116 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Maldonado-Calderón MT, Sepúlveda-García E, Rocha-Sosa M. 2012. Characterization of novel F-box proteins in plants induced by biotic and abiotic stress Plant Science 185 208–217 [DOI] [PubMed] [Google Scholar]
- Matiolli CC, Tomaz JP, Duarte GT, et al. 2011. The Arabidopsis bZIP gene AtbZIP63 is a sensitive integrator of transient abscisic acid and glucose signals Plant Physiology 157 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M. 2004. The ubiquitin-proteasome pathway and plant development The Plant Cell 16 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism Annual Review of Plant Biology 56 165–185 [DOI] [PubMed] [Google Scholar]
- Ni W, Xie D, Hobbie L, Feng B, Zhao D, Akkara J, Ma H. 2004. Regulation of flower development in Arabidopsis by SCF complexes Plant Physiology 134 1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. 2006. Sugar sensing and signaling in plants: conserved and novel mechanisms Annual Review of Plant Biology 57 675–709 [DOI] [PubMed] [Google Scholar]
- Rook F, Hadingham SA, Li Y, Bevan MW. 2006. Sugar and ABA response pathways and the control of gene expression Plant Cell and Environment 29 426–434 [DOI] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M. 2004. Arabidopsis CYP707 as encode (+)-abscisic acid 8'-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid Plant Physiology 134 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. 2010. The ubiquitin-proteasome system regulates plant hormone signaling The Plant Journal 61 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. 2004. The ubiquitin 26S proteasome proteolytic pathway Annual Review of Plant Biology 55 555–590 [DOI] [PubMed] [Google Scholar]
- Smeekens S. 2000. Sugar-induced signal transduction in plant Annual Review of Plant Physiology and Plant Molecular Biology 51 49–81 [DOI] [PubMed] [Google Scholar]
- Smeekens S. 2003. Sugar-induced signal transduction in plants Annual Review of Plant Physiology and Plant Molecular Biology 51 49–81 [DOI] [PubMed] [Google Scholar]
- Song SY, Chen Y, Zhao MG, Zhang WH. 2012. A novel Medicago truncatula HD-Zip gene, MtHB2, is involved in abiotic stress responses Environmental and Experimental Botany 80 1–9 [Google Scholar]
- Sullivan JA, Shirasu K, Deng XW. 2003. The diverse roles of ubiquitin and the 26S proteasome in the life of plants Nature Review of Genetics 4 948–958 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase Nature 446 640–645 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. 2009. The ubiquitin-26S proteasome system at the nexus of plant biology Nature Review of Molecular and Cellular Biology 110 385–397 [DOI] [PubMed] [Google Scholar]
- Xu GX, Ma H, Nei M, Kong HZ. 2009. Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification Proceedings of National Academy of Sciences, USA 106 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan YS, Chen XY, Yang K, Sun ZX, Fu YP, Zhang YM, Fang RX. 2011. Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice Molecular Plant 4 190–197 [DOI] [PubMed] [Google Scholar]
- Yang YZ, Peng H, Huang HM, Wu JX, Ha SR, Huang DF, Lu TG. 2004. Large-scale production of enhancer trapping lines for rice functional genomics Plant Science 167 281–288 [Google Scholar]
- Yang JC, Zhang JH, Wang ZQ, Zhu QS, Wang W. 2001. Hormonal changes in the grains of rice subjected to water stress during grain filling Plant Physiology 127 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, Wysocka-Diller J. 2006. Phytohormone signalling pathways interact with sugars during seed germination and seedling development Journal of Experimental Botany 57 3359–3367 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu W, Li Z, Deng XW, Wu W, Xue Y. 2008. F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in Arabidopsis Plant Physiology 148 2121–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SF, Gao F, Cao XS, Chen M, Ye GY, Wei CH, Li Y. 2005. The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms Plant Physiology 139 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Liu Y, Ye N, Liu R, Zhang J. 2011. Involvement of the abscisic acid catabolic gene CYP707A2 in the glucose-induced delay in seed germination and post-germination growth of Arabidopsis Physiologia Plantarum 143 375–384. [DOI] [PubMed] [Google Scholar]
- Zhu G, Ye N, Zhang J. 2009. Glucose induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis Plant Cell Physiology 50 644–651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.