Abstract
The integral inner membrane resistance-nodulation-division (RND) components of three-component RND-membrane fusion protein-outer membrane factor multidrug efflux systems define the substrate selectivity of these efflux systems. To gain a better understanding of what regions of these proteins are important for substrate recognition, a plasmid-borne mexB gene encoding the RND component of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa was mutagenized in vitro by using hydroxylamine and mutations compromising the MexB contribution to antibiotic resistance identified in a ΔmexB strain. Of 100 mutants that expressed wild-type levels of MexB and showed increased susceptibility to one or more of carbenicillin, chloramphenicol, nalidixic acid, and novobiocin, the mexB genes of a representative 46 were sequenced, and 19 unique single mutations were identified. While the majority of mutations occurred within the large periplasmic loops between transmembrane segment 1 (TMS-1) and TMS-2 and between TMS-7 and TMS-8 of MexB, mutations were seen in the TMSs and in other periplasmic as well as cytoplasmic loops. By threading the MexB amino acid sequence through the crystal structure of the homologous RND transporter from Escherichia coli, AcrB, a three-dimensional model of a MexB trimer was obtained and the mutations were mapped to it. Unexpectedly, most mutations mapped to regions of MexB predicted to be involved in trimerization or interaction with MexA rather than to regions expected to contribute to substrate recognition. Intragenic second-site suppressor mutations that restored the activity of the G220S mutant version of MexB, which was compromised for resistance to all tested MexAB-OprM antimicrobial substrates, were recovered and mapped to the apparently distal portion of MexB that is implicated in OprM interaction. As the G220S mutation likely impacted trimerization, it appears that either proper assembly of the MexB trimer is necessary for OprM interaction or OprM association with an unstable MexB trimer might stabilize it, thereby restoring activity.
Bacterial antimicrobial resistance is attributable in part to the activity of antimicrobial efflux transporters, which are grouped into five families based primarily upon amino acid sequence homology: the ATP-binding cassette (ABC) family, the major facilitator (MF) family, the small multidrug resistance family, the multidrug and toxic compound extrusion family, and the resistance-nodulation-division (RND) family (43, 44, 51). Despite structural differences, multidrug efflux pumps have in common the ability to accommodate and expel from the bacterial cell a wide variety of structurally unrelated antimicrobial compounds. RND family efflux systems, in particular, play an important role in the export of and resistance to clinically significant antimicrobials (43).
In Pseudomonas aeruginosa, an opportunistic human pathogen whose innate resistance to antimicrobials has long complicated antipseudomonal chemotherapy, RND-type multidrug efflux systems contribute significantly to intrinsic, as well as acquired, resistance (42, 48). To date, seven RND multidrug efflux systems have been characterized in P. aeruginosa, including MexAB-OprM (14, 27, 46, 47), MexCD-OprJ (45), MexEF-OprN (24), MexXY-OprM (2, 36, 63), MexJK-OprM (4), and, most recently, MexHI-OpmD (1, 56) and MexVW-OprM (31). The major system contributing to intrinsic multidrug resistance is encoded by the mexAB-oprM operon, which is also hyperexpressed in so-called nalB-type multidrug-resistant mutants (18, 19, 23, 33, 34, 41, 49, 52, 68). These mutants carry mutations in the mexR gene (19, 49, 53, 60, 68), which encodes a repressor of mexAB-oprM (60). MexAB-OprM accommodates a broad range of structurally diverse antimicrobials, including dyes and detergents (26, 59, 61), organic solvents (28, 30), disinfectants (55), and a number of clinically relevant antibiotics (5, 23, 29, 35, 59). This efflux system is also implicated in the export of homoserine lactones involved in quorum sensing (11, 40) and may export virulence factors (17).
RND transporters such as MexB, an inner membrane-spanning drug-H+ antiporter, function in association with accessory proteins that include a channel-forming outer membrane factor (OMF) (e.g., OprM) and a periplasmic membrane fusion protein (MFP) (e.g., MexA); the latter is believed to facilitate association of the RND and OMF components during efflux (51, 66, 67). Together, these proteins form a tripartite efflux system that is responsible for the transport of substrates from the periplasm and/or cytoplasm of P. aeruginosa to the external environment in a single step. MexB, like other RND family multidrug efflux transporters, consists of 12 transmembrane segments (TMSs) with extensive periplasmic loops between TMS-1 and -2 and between TMS-7 and -8, with both termini residing on the cytoplasmic side of the inner membrane (15). Three charged amino acids (D407 and D408 of TMS-4 and K939 of TMS-10) have been identified by site-directed mutagenesis as essential for MexB pump function, possibly participating in proton translocation or energy coupling (16).
The RND components of tripartite RND-MFP-OMF multidrug efflux systems (e.g., MexB) are known to determine substrate specificity (39), but until recently, the regions involved in substrate recognition had not been identified. Construction and characterization of AcrB-MexB (62) and AcrB-AcrD (10) chimeric RND transporters from Escherichia coli and P. aeruginosa recently confirmed that the extensive periplasmic loops in RND transporters determine substrate selectivity. In agreement with this, mutant studies confirmed the importance of specific amino acid residues within the large periplasmic loops (LPLs) of the P. aeruginosa RND transporter MexD in substrate selectivity (32). Eda et al. (9) also demonstrated that both the N- and C-terminal halves of MexB are required for efflux of its antibiotic substrates, implying that both LPLs are essential for transporter activity.
Little is known, however, about the nature and number of substrate-binding sites in RND-type multidrug transporters or the identity of residues involved in substrate selectivity, although substrate recognition by other families of multidrug transporters has been examined in some detail. Mutations differentially impacting the substrate profile of Bmr, an MF family multidrug transporter from Bacillus subtilis, for example, were identified by a random mutagenesis approach, and the existence of multiple substrate-binding sites was postulated (22). Additional studies support the occurrence of multiple substrate-binding sites in MF family multidrug efflux transporters in E. coli (MdfA) (25), Lactococcus lactis (LmrP) (50), and Staphylococcus aureus (QacA) (37) and in human ABC multidrug transporter P-glycoprotein (6, 13). Crystal structures of multidrug-binding regulators of multidrug efflux systems (e.g., the QacR regulator of qacA gene expression [54a]) reveal a large generalized binding “site” with different residues involved in binding different substrates. Still, it is unclear whether multidrug efflux systems themselves use a similar strategy. In this study, we used chemical mutagenesis to identify random mutations in MexB that do not alter protein production but compromise resistance to one or more antimicrobial substrates of MexAB-OprM, in hopes of identifying residues important for individual substrate recognition.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. P. aeruginosa K1589 was constructed by introduction of in-frame mexR (60) and mexB (17) deletions into strain K870 as described previously. Elimination of the chromosomal mexB gene was necessary to permit screening of mutated plasmid-borne mexB genes for changes impacting substrate selectivity (see below), while elimination of mexR served to ensure high-level expression of the chromosomally encoded MexA and OprM components. Together with the high-level expression of the plasmid-encoded MexB (under plac control [see below]), this served to ensure maximal production of a MexAB-OprM efflux system and thus ready assessment of its activity (by using MIC assays) and any changes in activity resulting from mutations in mexB. E. coli and P. aeruginosa were routinely cultured in Luria-Bertani (LB) broth (15.5 g of Miller's Luria broth base [Difco] and 2 g of NaCl per liter of H2O) at 37°C with shaking (180 rpm), except during growth of the conjugation recipient strains (42°C without shaking) and during incubation of organic solvent susceptibility testing plates (30°C). Strains carrying pRSP35, pJKM14, and their derivatives were cultured in the presence of tetracycline (10 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | endA hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR [φ80 dlacΔ(lacZ)M15] | 3 |
| S17-1 | thi pro hsdR recA Tra+ | 57 |
| P. aeruginosa | ||
| K767 | PAO1 wild type | 33 |
| K870 | Strr derivative of K767 | 49 |
| K1589 | K870 ΔmexR ΔmexB | 17 |
| Plasmids | ||
| pRK415 | Broad-host-range cloning vector; plac MCS, Tetr | 21 |
| pAK1900 | E. coli-P. aeruginosa shuttle cloning vector; Apr Carr | A. Kropinski, Queen's University |
| pPV6 | pAK1900::mexAB; mexAB in opposite orientation to plac | 46 |
| pRSP19 | pRK415::mexAB; mexAB in same orientation as plac | 58 |
| pRSP35 | pRK415::mexB; mexB in same orientation as plac | 59 |
| pJKM1 | pRK415::mexB* (T60I) | This study |
| pJKM2 | pRK415::mexB* (S183L) | This study |
| pJKM3 | pRK415::mexB* (M395I) | This study |
| pJKM4 | pRK415::mexB* (A618T) | This study |
| pJKM5 | pRK415::mexB* (R716H) | This study |
| pJKM6 | pRK415::mexB* (S450L) | This study |
| pJKM7 | pRK415::mexB* (E946K) | This study |
| pJKM8 | pRK415::mexB* (C966Y) | This study |
| pJKM9 | pRK415::mexB* (G51D) | This study |
| pJKM10 | pRK415::mexB* (G754D) | This study |
| pJKM11 | pRK415::mexB* (S977F) | This study |
| pJKM12 | pRK415::mexB* (S462F) | This study |
| pJKM13 | pRK415::mexB* (V928M) | This study |
| pJKM14 | pRK415::mexB* (G220S) | This study |
| pJKM15 | pRK415::mexB* (T578I) | This study |
| pJKM16 | pRK415::mexB* (E864K) | This study |
| pJKM17 | pRK415::mexB* (G1002D) | This study |
| pJKM18 | pRK415::mexB* (D407N) | This study |
| pJKM19 | pRK415::mexB* (D408N) | This study |
| pJKM20 | pRK415::mexB* (G220S E796K)b | This study |
| pJKM21 | pRK415::mexB* (G220S V203M G581D)b | This study |
| pJKM22 | pRK415::mexB* (G220S A737V)b | This study |
| pJKM23 | pRK415::mexB* (G220S L738F)b | This study |
| pJKM24 | pRK415::mexB* (G220S D793N)b | This study |
Mutated plasmid-borne mexB genes are indicated by asterisks, and the nature of the mutation(s) is given in parentheses. Strr, streptomycin resistant; Tetr, tetracycline resistant; Apr, ampicillin resistant; Carr, carbenicillin resistant.
Derivative of pJKM14 obtained following a second round of mutagenesis.
Plasmid protocols.
Electrocompetent E. coli DH5α and S17-1 cells were prepared as described by Dower et al. (7). Electroporation of plasmids pRSP35, pJKM14, and their derivatives into 100 μl of electrocompetent E. coli was performed with the ECM399 electroporator (BTX, Holliston, Mass.) at 2,500 V for 5 ms. Plasmid DNA used for nucleotide sequencing was isolated from E. coli hosts by using plasmid midikits (Qiagen Inc., Mississauga, Ontario, Canada) according to a protocol provided by the company for isolation of low-copy-number plasmids.
Hydroxylamine hydrochloride mutagenesis.
Chemical mutagenesis with hydroxylamine hydrochloride was adapted from the method of Garinot-Schneider et al. (12). Briefly, plasmid pRSP35 (10 μg) carrying the mexB gene was incubated at 70°C for 1.5 h in 500 μl of potassium phosphate buffer (pH 6.0; 44 mM) containing EDTA (5 mM) and hydroxylamine hydrochloride (0.46 M). A 20-μl sample was removed at 1.5 h, and hydroxylamine hydrochloride was inactivated by addition of Tris-HCl (pH 8.0; 90 mM final concentration) and EDTA (9 mM final concentration). Plasmid DNA was precipitated by using a standard protocol (54), washed in 70% (vol/vol) ethanol, and resuspended in 20 μl of H2O. E. coli S17-1 was electroporated with 1 to 3 μl of hydroxylamine hydrochloride-treated pRSP35 and, following addition of 1 ml of LB broth, was permitted to recover at 37°C with shaking for 1 h in the absence of antibiotic selective agents. Electroporated cells (1 ml) were then added to 9 ml of LB broth containing tetracycline (10 μg/ml) and cultured overnight at 37°C with shaking.
Mutagenized pRSP35 was mobilized into P. aeruginosa from E. coli S17-1 via conjugation as described previously (47), with modifications. Briefly, a 1.0-ml overnight culture of plasmid-containing E. coli S17-1 was mixed with 0.3 ml of an overnight LB culture of P. aeruginosa K1589 and pelleted in a microcentrifuge tube. Following resuspension in 150 μl of LB broth and spotting onto the center of an LB agar plate, cells were incubated for 4 h at 37°C. Bacterial growth was then resuspended in 1 ml of LB broth, and appropriate dilutions were plated on LB plates containing tetracycline (10 μg/ml) and streptomycin (250 μg/ml), the latter to counterselect the donor E. coli S17-1. Colonies of P. aeruginosa K1589 with plasmid-borne mutated mexB were recovered from LB plates containing tetracycline and streptomycin and screened for loss of MexB function by picking isolated colonies onto master plates of the same formulation and replica plating onto each of four LB plates containing chloramphenicol (64 μg/ml), carbenicillin (64 μg/ml), nalidixic acid (128 μg/ml), or novobiocin (128 μg/ml). Antibiotics were chosen to be at the highest concentration at which P. aeruginosa K1589(pRSP35) containing wild-type mexB could grow. pRSP35-carrying P. aeruginosa K1589 organisms that were unable to grow on one or more of the antibiotic plates were recovered and stored at −70°C.
Hydroxylamine mutagenesis of plasmid pJKM14 expressing MexBG220S was performed as described above, with modifications. Plasmid pJKM14 DNA (7.5 μg) was incubated at 70°C for 1 h in 150 μl of potassium phosphate buffer (pH 6; 75 mM) containing EDTA (0.75 mM) and hydroxylamine hydrochloride (0.4 M). DNA was then precipitated without addition of Tris-EDTA and resuspended in 20 μl of H2O as described above. Following electroporation into E. coli S17-1, plasmid pJKM14 and its derivatives were then mobilized into P. aeruginosa K1589 as described above, although imipenem (0.5 μg/ml) served as counterselection in this instance. P. aeruginosa K1589 containing pJKM14 with compensatory mutations which partially or completely restored wild-type MexB function was selected on LB broth with tetracycline (10 μg/ml) and carbenicillin (8 or 16 μg/ml) and stored at −70°C.
Antibiotic susceptibility testing.
The susceptibility of P. aeruginosa K1589 carrying pRSP35, pJKM14, and their derivatives was assessed by using the twofold serial microtiter broth dilution method described previously (20), with an inoculum of 5 × 105 cells per ml. MICs were recorded as the lowest concentration of antibiotic inhibiting visible growth after an 18-h incubation at 37°C.
Organic solvent susceptibility testing.
The ability of P. aeruginosa K1589 carrying mutated mexB plasmids to grow in the presence of the organic solvent n-hexane was assessed by the efficiency-of-plating approach outlined previously (28). Briefly, 100 μl of a suspension of approximately 107 cells/ml was spread to cover the surface of a glass plate containing LB agar and overlaid with 1 ml of 100% n-hexane. The plates were sealed and incubated for 20 h at 30°C, at which point the presence or absence of growth was recorded.
SDS-PAGE and Western immunoblotting.
Whole-cell extracts of P. aeruginosa K1589 harboring pRSP35, pJKM14, and their derivatives were prepared by harvesting 1 ml of overnight LB cultures (18 h, 37°C) and resuspending the cells in 300 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (SDS [4% wt/vol], glycerol [20% vol/vol], Tris-HCl [pH 6.8; 0.25 M]) containing 2-mercaptoethanol (10%, vol/vol). Samples were heated at 95°C for 5 min, sonicated 20 s at 50% power with a Vibra Cell sonicator (Sonics and Materials, Inc., Danbury, Conn.), and centrifuged at 12,000 × g for 2 min to remove debris. Protein samples were analyzed by SDS-10% (vol/vol) PAGE and subsequently electroblotted and screened with MexB-specific antibodies as described previously (58), with the exception that proteins were transferred onto Immobilon-P transfer membranes (Millipore, Bedford, Mass.) by using a Trans-Blot SD semidry electrophoretic transfer cell (Bio-Rad Laboratories, Hercules, Calif.) at a 25-mA constant current for 1 h at room temperature. Blots were developed by using Western Lightning chemiluminescence reagents (Perkin-Elmer Life Sciences, Boston, Mass.), according to the manufacturer's instructions.
Nucleotide sequencing.
The mexB genes of pRSP35, pJKM14, and their derivatives were sequenced by Cortec DNA Service Laboratories, Inc., Kingston, Ontario, Canada, by using custom primers mexB forward2 (5′-GAAGAATGTCGCGTCCGC-3′), mexB forward3 (5′-CAACGCGCAGTTCAACGG-3′), mexB forward4 (5′-CAGGCGCAGAACGTGCAG-3′), mexB reverse (5′-GGGGATCAGCGAGCAGC-3′), mexB reverse2 (5′-GCGTCACCGGAACTCAGG-3′), and mexB reverse3 (5′-GTACGCCCTGGTCCTCGTC-3′), which were synthesized by the company. Mutations in mexB were identified following sequence alignment with the wild-type gene by using DNAMAN version 4.11 software.
Protein modeling.
The three-dimensional structure of MexB was modeled (by M. Kuiper of the Department of Biochemistry, Queen's University, Kingston, Ontario, Canada) with SYBYL version 6.5 molecular modeling software (Tripos Associates, St. Louis, Mo.) by threading the MexB protein sequence onto the crystal structure of the homologous RND transporter, AcrB, of E. coli (38) and refining the structure by minimization and dynamics. Locations of mutations within MexB were pinpointed within the larger three-dimensional structure of MexB and hydrogen bonds were predicted, using both DeepView/Swiss-PdbViewer version 3.7 and POV-RAY for Windows.
RESULTS
Isolation of MexB mutants exhibiting reduced antibiotic resistance.
To identify mutations in mexB that compromised MexB activity, ca. 4,000 colonies of a ΔmexB derivative of PAO1 (strain K1589) harboring hydroxylamine-mutated, plasmid-borne mexB were screened for increased susceptibility to one or more of four antibiotics chosen as representative MexB antimicrobial substrates. Initially, 249 colonies showed increased susceptibility to one or more of chloramphenicol, carbenicillin, nalidixic acid, or novobiocin on plates. Following susceptibility testing in broth cultures, these were grouped into six families based upon common patterns of susceptibility or resistance to these four agents (Table 2). Western immunoblotting subsequently confirmed that in ca. 100 instances (studied from this point forth), wild-type levels of MexB were being produced (representative examples are shown in Fig. 1) despite the increased susceptibility of the mutant MexB-producing P. aeruginosa K1589. Some defects in MexB impacted resistance to a single (family A, chloramphenicol; family C, carbenicillin; and family D, nalidixic acid); or only two antibiotics used in the original screen (family B, chloramphenicol and carbenicillin), while others impacted three of four (family E, all but novobiocin) or all four (family F) antibiotics, with a subgroup of F being totally compromised for resistance (family F2) (Table 2). As we were interested in the possibility of separate binding sites for periplasmically acting β-lactams from those for antimicrobials with cytoplasmic targets, the lone representative of family C (encoded by pJKM9 [Table 2]) exhibiting increased susceptibility to carbenicillin alone was tested in broth culture for susceptibility to three other β-lactams, i.e., piperacillin, meropenem, and aztreonam. Although it exhibited increased susceptibility to meropenem and aztreonam compared to that provided by wild-type MexB, its resistance to the β-lactam piperacillin was not impacted (Table 2). Thus, MexB produced by pJKM9 was not generally compromised for β-lactam resistance.
TABLE 2.
Antimicrobial and organic solvent susceptibilities of P. aeruginosa K1589 expressing MexB mutant proteinsa
| Familyb | Plasmid designationc | MexB mutationd | Location of mutatione | MIC (μg/ml)f
|
Growthg with HEX | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAM | CAR | NAL | NOV | PIP | MER | AZT | |||||
| — | pRSP35 | Wild type | 256 | 256 | 1,024 | 1,024 | 16 | 1 | 32 | + | |
| pRK415 | No MexB | 4 | 2 | 16 | 32 | 0.5 | 0.125 | 0.5 | − | ||
| A | pJKM1 | T60I | LPL-1 | 64 | 128 | 512 | 512 | NDh | ND | ND | + |
| pJKM2 | S183L | LPL-1 | 64 | 128 | 512 | 1,024 | ND | ND | ND | + | |
| pJKM3 | M395I | TMS-4 | 64 | 128 | 1,024 | 1,024 | ND | ND | ND | + | |
| pJKM4 | A618T | LPL-2 | 64 | 128 | 512 | 1,024 | ND | ND | ND | + | |
| pJKM5 | R716H | LPL-2 | 64 | 128 | 512 | 1,024 | ND | ND | ND | + | |
| B | pJKM6 | S450L | TMS-5 | 32-64 | 64 | 512 | 1,024 | ND | ND | ND | + |
| pJKM7 | E946K | CL between TMS-10 and -11 | 32-64 | 64 | 512 | 1,024 | ND | ND | ND | + | |
| pJKM8 | C966Y | CL between TMS-10 and -11 | 32-64 | 64 | 512 | 1,024 | ND | ND | ND | + | |
| C | pJKM9 | G51D | LPL-1 | 128 | 32-64 | 512 | 1,024 | 8 | 0.25 | 4 | + |
| D | pLKM10 | G754D | LPL-2 | 128 | 128 | 128-256 | 1,024 | ND | ND | ND | + |
| pLKM11 | S977F | TMS-11 | 128 | 128 | 128-256 | 512 | ND | ND | ND | + | |
| E | pJKM12 | S462F | PL between TMS-5 and -6 | 32-64 | 64 | 256 | 1,024 | ND | ND | ND | − |
| pJKM13 | V928M | TMS-10 | 32-64 | 32 | 256 | 512 | ND | ND | ND | + | |
| F1 | pJKM14 | G220S | LPL-1 | 16-32 | 32 | 64 | 256 | 2-4 | 0.125-0.25 | 1-2 | + |
| pJKM15 | T578I | LPL-2 | 32-64 | 32-64 | 32 | 256 | 1-2 | 0.125 | 1-2 | + | |
| pJKM16 | E864K | LPL-2 | 8 | 32 | 128 | 256 | 4 | 0.25 | 2-4 | − | |
| pJKM17 | G1002D | TMS-12 | 64 | 64 | 256 | 256 | 4 | 0.25 | 4 | − | |
| F2 | pJKM18 | D407N | TMS-4 | 4 | 2 | 16 | 32 | 1 | 0.125 | 0.5 | − |
| pJKM19 | D408N | TMS-4 | 8 | 2 | 16 | 32 | 0.5-1 | 0.125 | 0.5 | − | |
Following mutagenesis of pRSP35-carried mexB and subsequent introduction of mutated pRSP35 into the ΔmexB strain K1589, selection was made for increased susceptibility to one or more of chloramphenicol (CAM), carbenicillin (CAR), nalidixic acid (NAL), or novobiocin (NOV) in K1589 harboring the mutant mexB plasmids and expressing wild-type levels of MexB protein.
Mutations in MexB that yielded the same pattern of antimicrobial susceptibility in the initial screen of K1589 harboring mutant mexB plasmids were grouped into families, arbitrarily designated A to F; the following antibiotics were those for which resistance was compromised in each family: A, CAM; B, CAM and CAR; C, CAR; D, NAL; E, CAM, CAR, and NAL; and F, CAM, CAR, NAL, NOV. F1 and F2 represent MexB mutations which partially and completely compromised resistance, respectively, to all substrates tested. Agents not included in the initial screen were piperacillin (PIP), meropenem (MER), aztreonam (AZT), and n-hexane (HEX). MexB+ [i.e., K1589(pRSP35)] and MexB− [i.e., K1589(pRK415)] control strains (—) are included for comparison.
Plasmid designations refer to descriptions in Table 1.
Point mutations within each MexB mutant are as identified.
Locations of MexB mutations with respect to MexB topology are as indicated. LPL-1, LPL between TMS-1 and -2; LPL-2, LPL between TMS-7 and -8; CL, cytoplasmic loop; PL, periplasmic loop.
MICs are representative of those determined a minimum of two trials performed on different days for each mutant within a given family. MexB mutations were classified as causing susceptibility to a particular antimicrobial agent or solvent (and MICs are in boldface) if a ≥4-fold reduction in MIC from that for the MexB+ control strain K1589(pRSP35) was demonstrated in K1589 expressing the mutant MexB protein.
+, growth; −, no growth.
ND, not determined.
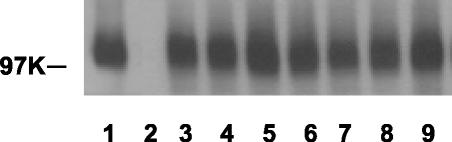
FIG. 1.
Production of mutant MexB proteins representative of families A to F (Table 2) in P. aeruginosa K1589. Whole-cell extracts of ΔmexB P. aeruginosa strain K1589 carrying the plasmids indicated below (the mutation present in the MexB protein produced by each plasmid and the family designation is indicated in parentheses) were prepared, electrophoretically separated by SDS-PAGE, electroblotted, and developed with antibodies directed against MexB. Lane 1, pRSP35 (wild-type MexB); lane 2, pRK415 (no MexB; vector control); lane 3, pJKM1 (T60I, A); lane 4, pJKM6 (S450L, B); lane 5, pJKM9 (G51D, C); lane 6, pJKM10 (G754D, D); lane 7, pJKM12 (S462F, E); lane 8, pJKM14 (G220S, F1); lane 9, pJKM18 (D407N, F2). The molecular mass marker is indicated (in kilodaltons).
To assess the impact of MexB defects on resistance to nonantibiotic antimicrobials, we tested susceptibility to an organic solvent by measuring the presence or absence of growth on plates overlaid with n-hexane. In general, only hydroxylamine hydrochloride-induced mutations in mexB that impacted resistance to all or almost all antibiotics (Table 2, families E and F) also compromised solvent tolerance.
Identification of MexB mutations impacting antimicrobial resistance.
To identify the mutations in the mexB gene that are responsible for the compromised resistance seen in families A to F (Table 2), plasmid-borne mexB was recovered from representatives of each family and sequenced. Of 46 sequenced mutants, 28 had a single MexB mutation, of which 19 were unique. While the majority of mutations occurred within the LPLs between TMS-1 and -2 (LPL-1) and between TMS-7 and -8 (LPL-2), mutations in the TMSs as well as other periplasmic and cytoplasmic loops were also seen (Table 2). With the exception of family B, mutations occurring in TMSs and smaller loops were found mostly in families E and F, which impacted larger numbers of substrates. Of interest is that our hydroxylamine mutagenesis approach identified two mutations within TMS-4, D407N and D408N (Table 2, family F2), that were previously identified as essential for MexB function and predicted to participate in proton translocation or energy coupling (16). Both mutants exhibited susceptibility to all tested antimicrobials at levels equivalent to that of the MexB-deficient strain.
Mapping of MexB mutations to a three-dimensional model of MexB.
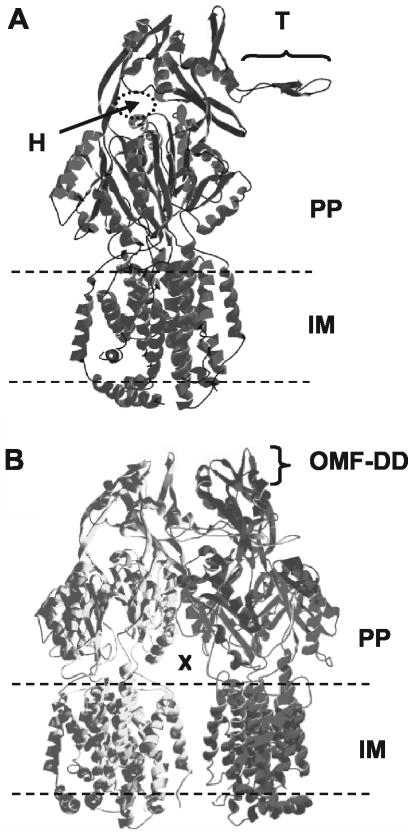
To gain a more complete understanding of the effects of these mutations on MexB function, we first modeled the MexB three-dimensional structure upon the X-ray crystal structure of AcrB, the homologous (70% amino acid identity) RND transporter from E. coli (38). Like AcrB, MexB appears to be a trimer formed by the interlocking of a protruding thumb region of one monomer with a hole in the upper periplasmic region of the neighboring monomer (Fig. 2A). The trimer is embedded in the inner membrane but forms a central cavity in the periplasmic domain which is fed by openings or vestibules that occur between adjacent monomers at the plane of the membrane and are contiguous with the periplasm (Fig. 2B). The vestibules are predicted to function as portals for substrates entering the AcrB-MexB central cavity from the periplasm and/or outer leaflet of the inner membrane. A pore at the distal end of the MexB trimer likely connects with the OMF component of the efflux system (i.e., OprM) (Fig. 2B) to permit passage of substrates captured by MexB across the outer membrane and into the extracellular milieu. With the exception of family B, mutations impacting one or a few antimicrobials (Table 2, families A, C, and D) tended to map towards the top of the MexB structure (periplasmic domain), while mutations impacting the majority of antimicrobials (Table 2, families E and F) tended to map to the TMSs or loop regions near the inner membrane plane (Fig. 3). Two exceptions were mutations G220S and T578I in MexB mutants of family F1, which mapped to the periplasmic domain (Fig. 3F1).
FIG. 2.
Three-dimensional model of MexB created by threading the MexB protein sequence onto the crystal structure of AcrB from E. coli (38). (A) MexB monomer. The presumed inner membrane (IM) and periplasmic (PP) domains of the protein are indicated, as are a thumb (T) and hole (H) region implicated in trimerization (see text). (B) MexB trimer with rear monomer removed for ease of viewing. A vestibule leading to a central cavity formed by the periplasmic domains of the MexB trimer is indicated (X), as is a presumed OprM docking domain (OMF-DD).
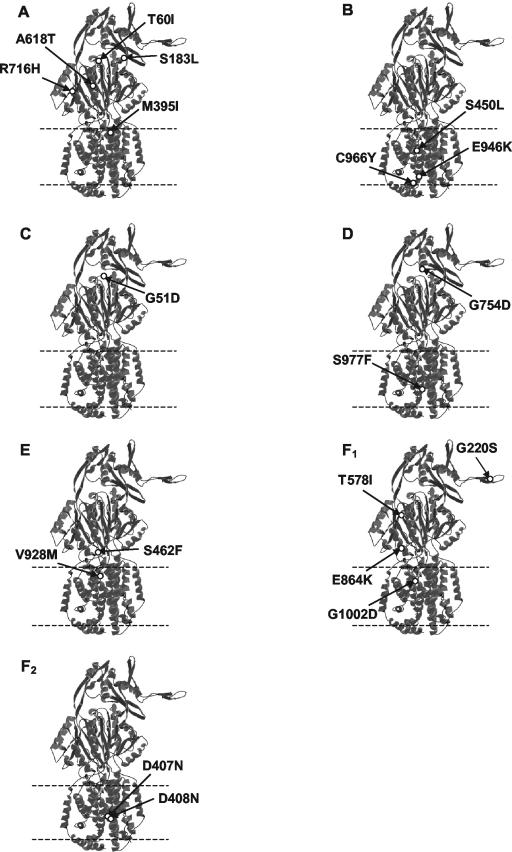
FIG. 3.
Location in the MexB monomer of mutations that compromise drug resistance. Mutations are grouped according to family (Table 2). The inner membrane region is defined by dashed lines.
(i) Family A.
Of the mutants with increased susceptibility to chloramphenicol alone, two (T60I and S183L) had mutations in the vicinity of the periplasmic hole (Fig. 3A) into which the thumb of a neighboring monomer is predicted to insert (see Fig. 2A). T60 bordered the hole itself and was oriented towards the center of the trimer, below the point of thumb insertion. While it is not entirely clear how mutation of this residue might have impacted MexB function, T60 may have formed hydrogen bonds with T56 (2.8 Å away) and V57 (3.2 Å away) in the wild-type protein, and a mutation to the hydrophobic isoleucine would have prevented this. As a result, a disruption of the MexB tertiary structure immediately below the thumb may have occurred, possibly interfering with thumb insertion. Indeed, T60 of one monomer is predicted to be within close proximity of residues on the thumb of a neighboring monomer (e.g., R239, 3.7Å away), and any changes in T60 may adversely affect thumb insertion. In contrast to T60, S183 occurred somewhat to the right of the hole region shown in Fig. 2A and towards the outer surface of MexB (Fig. 3A). Perhaps mutation to the hydrophobic leucine indirectly (i.e., at a distance) impacted MexB structure in the vicinity of the hole.
Two mutations in members of family A (A618T and R716H) occurred on an outward-facing portion of the MexB trimer, within a groove that extends from the hole where the thumb inserts to the inner membrane plane (Fig. 3A) and was previously predicted to be involved in MexA binding (38). Thus, these mutations may have adversely affected MexA binding to MexB. The final member of family A carried a M395I mutation within TMS-4, potentially altering the local conformation of MexB in the inner membrane (Fig. 3A). It is not clear how this might have impacted function, although given the apparent importance of TMS-4 and TMS-10 in proton translocation (16), it may have adversely affected this essential process in the operation of the MexB drug-H+ antiporter.
(ii) Family B.
In contrast to members of families A, C, and D (descriptions of families C and D are below), which showed susceptibility to one antimicrobial and carried MexB mutations that tended to map to the upper periplasmic region of the protein, MexB mutants belonging to family B (which also showed susceptibility to only a few agents) carried mutations that mapped to TMS-5 (S450L) and the cytoplasmic loops between TMS-10 and -11 (E946K and C966Y) (Fig. 3B). How these mutations might have impacted MexB function, particularly with respect to efflux of or resistance to these two substrates only, is unclear.
(iii) Family C.
Similar to the T60I mutation of family A, the G51D family C mutation that resulted in increased susceptibility to carbenicillin was located within the hole into which the thumb of the neighboring monomer was predicted to insert (Fig. 3C). G51 was, however, even closer (within 1.5 Å of S216) to the inserting thumb than T60. The change from a small and flexible residue (glycine) to one that is larger and negatively charged (aspartic acid) likely had a negative impact on thumb insertion and thus trimer formation.
(iv) Family D.
The G754D mutation of family D that caused increased susceptibility to nalidixic acid also occurred within the thumb insertion hole region (Fig. 3D). Again, too, the mutated residue is much closer to the inserting thumb of the neighboring monomer than is the wild type (e.g., G754 is within 3.3 Å of G217 of the thumb, while D754 is within less than 1 Å of G217). This family D mutation appears, therefore, also to negatively impact thumb insertion and thus trimerization. In contrast, a second family D mutation, S977F, occurs in TMS-11 and may have had an impact on local packing of TMSs, since following the mutation, potential hydrogen bonds with two residues in the carboxy-terminal tail (M1009 and T1003) would be lost. How this would impact function is at present unclear.
(v) Family E.
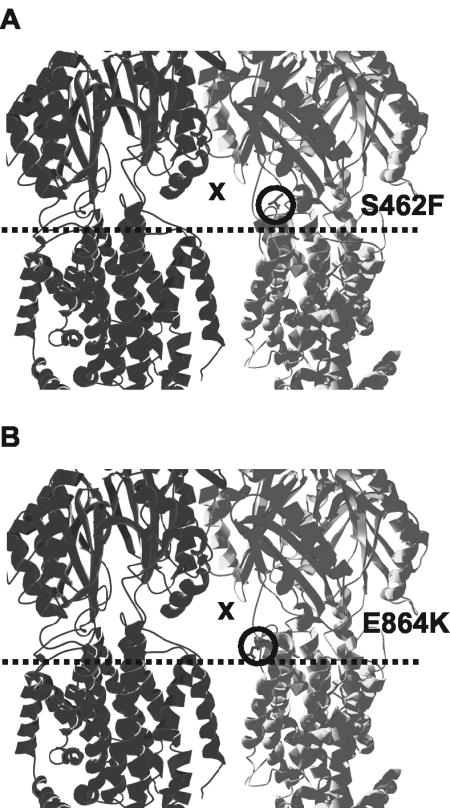
Mutants susceptible to all antimicrobials used in the initial screen except novobiocin (family E) possessed mutations within TMS-10 (V928) or the vestibule region (S462F) (Fig. 4A). While the nature of the defect caused by the V928M change is uncertain, the substitution of a bulky, hydrophobic residue (F) for a small, polar one (S) at position 462 in the vestibule region may have disrupted tertiary structure in such a way as to limit substrate recognition or entry, possibly through steric hindrance or loss of hydrogen bonds with the substrate itself. With all or most substrates predicted to enter the MexB exporter via the vestibules, it might not be surprising to find that some changes here impact a large number of substrates.
FIG. 4.
Mutations (circled) in the vestibule region (X) of the MexB trimer (the front monomer has been removed for ease of viewing) that compromise drug resistance. (A) S462F mutation of family E; (B) E864K mutation of family F1. Inner membrane leaflets are indicated by dashed lines.
(vi) Family F1.
The G220S mutation that compromised resistance to all tested antimicrobials occurred on the protruding thumb region of MexB (Fig. 3F1), which is predicted to insert into the periplasmic hole of the neighboring monomer during trimer formation. As G220 is within approximately 3.6 to 4.2 Å of Q622, R779, and M780 of the periplasmic hole region of the neighboring monomer, changes in thumb secondary structure as a result of the G220S change may, for steric reasons, have hindered thumb insertion and thus decreased efficiency of trimer formation. Similar to A618T and R716H of family A, the family F1 T578I mutation occurred within the vicinity of the outward-facing periplasmic groove of MexB, with which MexA could potentially interact (Fig. 3F1). This mutation might thus adversely impact MexA binding to the MexB trimer. The remaining two members of family F1 possessed mutations within TMS-12 (G1002D) or immediately above the membrane plane (E864K) (Fig. 3F1) in the vestibule region that occurs between neighboring monomers (Fig. 4B) and serves as a predicted entry point for substrates from the periplasm to a central collection cavity (38). The introduction of a large, negatively charged residue (D1002) into the largely hydrophobic TMS-12 may have interfered with the packing of the neighboring helices, thus having an indirect negative effect on proton translocation. Similarly, a mutation from negative (E864) to positive (K864) charge in the vestibule region between neighboring monomers may have disrupted the local tertiary structure. Again, if the vestibules are the portals of entry for most if not all substrates accommodated by MexB, changes here are expected to impact many or most substrates.
(vii) Family F2.
Each F2 mutant with completely compromised resistance to all tested antimicrobials contained a mutation in TMS-4 (D407N or D408N) (Fig. 3F2), which was previously identified as being essential for pump function (16). These residues are proposed to interact with K939 on TMS-10 and to participate in proton pumping by this drug-H+ antiporter, by a mechanism that has thus far not been characterized.
Identification of compensatory MexB mutations restoring antimicrobial resistance in a MexBG220S-expressing strain.
To identify second-site suppressor mutations in mexB that restore antimicrobial resistance to P. aeruginosa strain K1589 expressing the G220S family F1 mutant MexB protein (characterized by increased susceptibility to all tested antimicrobials), plasmid pJKM14, encoding MexBG220S, was subjected to a second round of hydroxylamine mutagenesis. Plasmid pJKM14-carrying K1589 that expressed MexB with restored function was then selected on plates with carbenicillin at levels where the G220S mutant could not survive unless a compensatory mexB mutation had occurred. In order to confirm a wild-type phenotype in the compensatory mutants, 24 compensatory mutants from the initial screen were subjected to susceptibility testing in broth culture with representative MexB antimicrobial substrates. Plasmid-borne mexB from 14 of these mutants that exhibited wild-type or near-wild-type levels of resistance to a number of the tested antimicrobials (Table 3) was recovered and subjected to nucleotide sequencing.
TABLE 3.
Antimicrobial susceptibilities of P. aeruginosa K1589 expressing MexBG220S with compensatory mutationsa
| MexB mutation(s)b | Plasmid designationc | Location of mutation(s)d | MIC (μg/ml)e
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| CAM | CAR | NAL | NOV | PIP | MER | AZT | |||
| Wild type | pRSP35 | 256 | 256 | 1,024 | 1,024 | 16 | 1 | 32 | |
| No MexB | pRK415 | 4 | 1 | 16 | 32 | 0.5 | 0.125 | 0.5 | |
| G220S | pJKM14 | LPL-1 | 16-32 | 32 | 32-64 | 256 | 2-4 | 0.25 | 2 |
| G220S, E796K | pJKM20 | LPL-1, LPL-2 | 256 | 256 | 1,024 | 1,024 | 16 | 1 | 32 |
| G220S, V203M, G581D | pJKM21 | LPL-1, LPL-1, LPL-2 | 128 | 128 | 512 | 1,024 | 8 | 1 | 16 |
| G220S, A737V | pJKM22 | LPL-1, LPL-2 | 256 | 128 | 256 | 1,024 | 8 | 0.25 | 8 |
| G220S, L738F | pJKM23 | LPL-1, LPL-2 | 128 | 128 | 512 | 512 | 8 | 0.5 | 8 |
| G220S, D793N | pJKM24 | LPL-1, LPL-2 | 128 | 128 | 256 | 1,024 | 8 | 0.25 | 8 |
Following mutagenesis of pJKM14, encoding the G220S MexB derivative, and screening for full or partial restoration of MexB-dependent antimicrobial resistance in K1589, plasmid DNA was recovered and mexB was sequenced to identify the compensatory mutations.
Point mutations within each MexB mutant protein are as identified. Compensatory mutations are indicated in boldface.
Plasmid designations refer to descriptions in Table 1.
Locations of MexB mutations with respect to MexB topology are as indicated. LPL-1, LPL between TMS-1 and -2; LPL-2, LPL between TMS-7 and -8. The location of the compensatory mutations is in boldface.
Mutant MexB proteins were considered to provide wild-type levels of antimicrobial resistance to a particular antimicrobial agent (indicated by boldface) if the MIC for K1589 expressing the particular MexB derivative was within twofold of that seen for the same strain expressing wild-type MexB from pRSP35. Abbreviations are as defined in footnotes a and b Table 2.
Nine of 10 mutants in which resistance to all tested antimicrobials was restored to wild-type levels contained the same compensatory mutation, E796K, which occurred within LPL-2. As Table 3 indicates, the remaining mutant that exhibited wild-type levels of resistance to all tested antimicrobials harbored two mutations, V203M and G581D, in LPL-1 and -2, respectively, although it was not clear if one or both were needed. Four additional compensatory mutants contained a single mutation in LPL-2, of which three were unique (A737V, L738F, and D793N) and showed only partially restored MexB function (i.e., resistance, although increased for most antimicrobials, was not restored to wild-type levels for all [Table 3].
Mapping of compensatory MexB mutations to a three-dimensional model of MexB.
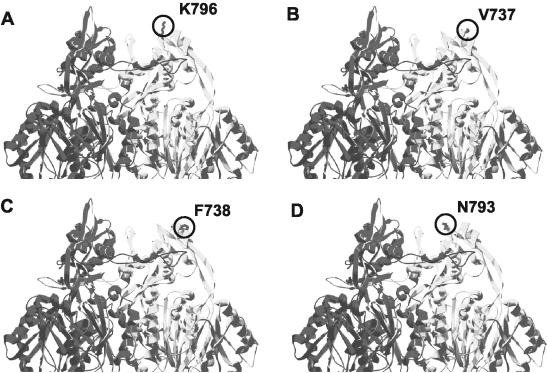
All single MexB mutations (Table 3) restoring antibiotic resistance of a G220S MexB mutant to wild-type levels for some or all of the tested antimicrobial substrates mapped to the upper periplasmic portion of MexB in the region predicted to interact with the OMF OprM (Fig. 5). The original G220S mutant may form altered trimers with which OprM docks with reduced efficiency, and compensatory mutations may enhance OprM docking with mutant trimers, restoring antimicrobial efflux. Alternatively, compensatory mutations may enhance OprM docking, facilitating more efficient MexB trimer formation in the face of the destabilizing G220S mutation, also restoring efflux.
FIG. 5.
Locations of single compensatory mutations within the MexB trimer (the front monomer has been removed for simplicity) that restore function of MexBG220S. Compensatory mutations are circled on one monomer only.
Conservation in other RND transporters of residues whose mutation in MexB compromises function.
In order to determine whether MexB residues whose mutation in this study compromised antimicrobial resistance were conserved in other RND transporters from P. aeruginosa as well representatives from other bacteria (and thus were of broad functional significance), we performed alignments of the protein sequences of these transporters. As we predict that many of the mutations identified in this study affect trimer formation and proton pumping, which should be conserved functions within RND transporters, it was expected that amino acid residues involved in these functions would be highly conserved. With the exception of T60 and S183 (which were conserved in only 4 and 1 of the 11 RND transporters examined, respectively), residues predicted to be involved in MexB trimer formation (G51, G754, and G220) were highly conserved (in 8 to 11 of the RND transporters to which they were compared) (Table 4). Indeed, when the analysis was extended to seven additional RND pumps found in Pseudomonas putida, Burkholderia pseudomallei, Campylobacter jejuni, Porphyromonas gingivalis, and Stenotrophomonas maltophilia, residues corresponding to G51, G220, and G754 in MexB were also very highly conserved (not shown). Similarly, MexB residues predicted to impact efficiency of proton pumping were generally well conserved, occurring in 6 to 11 of the RND transporters (M395, V928, E946, S977, G1002, D407, and D408) (Table 4). This was also true when the analysis was extended to the additional RND transporters described above, with residues corresponding to D407, D408, E946, G220, S977, and G1002 in particular being very well conserved. S450 and C966 were clearly exceptions, however, with serine and cysteine being very poorly represented at the corresponding positions of the RND transporters listed in Table 4. Interestingly, all of the exceptions mentioned above occurred at residues that, when mutated, decreased resistance only to the antimicrobials chloramphenicol and/or carbenicillin, which are predicted to be poorer substrates of MexB. Thus, although these residues are predicted to participate in conserved functions (e.g., trimer formation and proton pumping), they may play comparatively minor roles.
TABLE 4.
Conservation in RND multidrug transporters of amino acid residues whose mutation in MexB compromises drug resistancea
| Familyb | Mutated residue in MexBc | Corresponding residue ind:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MexD | MexF | MexY | MexK | AcrB | AcrF | YhiV | MtrD | AdeB | AmeB | CeoB | ||
| A | T60 | S | T | A | Q | T | S | T | S | S | S | T |
| S183 | A | S | A | K | A | A | A | A | A | A | A | |
| M395 | M | L | M | L | F | L | M | M | M | M | L | |
| A618 | S | N | Y | D | A | F | A | S | S | S | N | |
| R716 | M | F | M | N | R | D | R | R | W | R | F | |
| B | S450 | S | C | V | A | S | V | S | I | A | T | V |
| E946 | E | E | E | D | E | D | E | E | E | E | E | |
| C966 | A | C | A | T | V | C | V | A | A | A | S | |
| C | G51 | G | G | G | G | G | G | G | G | G | G | G |
| D | G754 | G | G | G | T | G | G | G | S | G | A | G |
| S977 | S | S | S | A | S | T | S | S | S | S | S | |
| E | S462 | S | L | A | G | S | L | S | A | S | S | L |
| V928 | V | I | V | L | V | I | V | V | I | V | I | |
| F1 | G220 | G | G | G | E | G | E | G | G | G | G | G |
| T578 | L | L | Q | L | L | A | L | L | L | T | L | |
| E864 | Q | Q | E | D | Q | V | Q | E | Q | Q | Q | |
| F2 | G1002 | G | G | G | A | G | G | G | G | G | G | G |
| D407 | D | D | D | D | D | D | D | D | D | D | D | |
| D408 | D | D | D | D | D | D | D | D | D | D | D | |
Amino acid sequences of the listed RND-type multidrug transporters were aligned with MexB by using DNAMAN version 4.11 software, and residues of these transporters aligning MexB residues whose mutation in this study was shown to compromise drug resistance were identified.
Families are as defined in footnote b of Table 2.
The original MexB amino acid residue whose mutation in this study compromised drug resistance. See Table 2 for a description of the mutations and the corresponding drug resistance phenotypes.
Corresponding amino acid residues in other RND multidrug transporters from P. aeruginosa (MexD [accession number AAB41957], MexF [CAA67864], MexY [BAA34300], and MexK [also called PA3676] [A83186]), E. coli, (AcrB [P31224], AcrF [P24181], and YhiV [P37637]), Neisseria gonorrhoeae (MtrD [AAC45560]), Acinetobacter baumannii (AdeB [AAL14440]), Agrobacterium tumefaciens (AmeB [AAG09746]), and Burkholderia cepacia (CeoB [AAB5816]). Residues that are conserved in MexB are indicated in boldface type.
Mutations predicted to impact the interaction of MexB with the MFP MexA (T578, A618, and R716) occurred at residues that were poorly conserved in the 11 RND transporters in Table 4, and this held true for T5781 and A618 when the analysis was extended to the above-mentioned 7 additional transporters (arginines corresponding to R716 were present in 5 of these transporters, making this residue somewhat better conserved in the RND transporters). This is not entirely unexpected, since subunit swapping studies have indicated that MFP association with its cognate RND transporter is very specific and that MFPs cannot generally be exchanged and efflux function still retained (64). Thus, it is probable that each RND transporter has unique amino acid residues in those regions with which the MFP interacts. Similar to the situation with mutations potentially impacting association of MexB with MexA, the two MexB mutations (S462F and E864K) within predicted substrate-binding sites in the vestibule regions occurred at amino acid residues that were less well conserved (E864; conserved in 2 of 11) and somewhat less well conserved (S462; conserved in 5 of 11) in the other RND transporters to which they were compared (Table 4). This observation was again supported by comparisons with the seven additional RND transporters and may reflect an expected variability in the vestibule regions of different RND transporters that allows different pumps to accommodate different antimicrobial substrates. In any case, the two MexB mutations (listed above) affected susceptibilities to almost all tested antimicrobials (Table 2, families E and F1), which is evidence that although these residues are not highly conserved, they are important for substrate accommodation by MexB.
DISCUSSION
Although RND transporters have the ability to accommodate a surprisingly vast array of substrates, little is known about the basis of their broad substrate specificity. The two substantial periplasmic loops in RND transporters MexB, MexD, AcrB, and AcrD have been implicated in substrate selectivity by others (8, 10, 32, 62). Our hydroxylamine hydrochloride mutagenesis approach also identified a range of mutations within the two LPLs of MexB that negatively impacted the MexB contribution to resistance, consistent with their having a role in substrate selectivity. In contrast to the conclusions of the aforementioned studies, however, we contend that most of our mutations in the major periplasmic loop regions did not directly impact substrate binding. Instead, these mutations appeared to affect pump assembly and/or activity, although how is not entirely clear. That such mutations were, nonetheless, somewhat substrate specific might be explained by any reduction in pump assembly or activity having a greater impact on those substrates that are not as efficiently accommodated by the MexB transporter. Thus, chloramphenicol is the only substrate affected by the family A mutations (T60I and S183L) that are predicted to impact trimerization, possibly because it is a poorer substrate for MexAB-OprM and a reduction in efficiency of trimer formation affects its export to a greater extent. Similarly, the family B mutations that apparently impact the transmembrane region of MexB, the lone family C mutation that possibly impacts trimer formation, and the family D mutations that impact trimerization or proton pumping are all fairly substrate selective, despite the apparent impact on MexB assembly and/or function and not substrate recognition per se. Again, the fact that mutations impacting assembly and/or activity of MexB do not affect all substrates suggests that that they must have only a modest impact on MexB assembly or function, with the compromised MexB able to accommodate some substrates better than others. Still, other mutations predicted to impact trimerization (e.g., G220S) do compromise resistance for all substrates, presumably because trimerization and thus pump assembly are impacted to a greater extent.
Beyond their contribution to substrate selectivity, the two large loops comprising the periplasmic domain of an RND transporter are presumably also involved in interaction with the cognate MFP. Using AcrB-MexB chimeric RND transporters, Tikhonova et al. demonstrated that the N-terminal half of AcrB is likely involved in determining the specificity of interaction with the MFP AcrA, with residues 590 to 612 apparently of particular importance (62). When these residues are mapped to our model of MexB, they occur on the outward-facing surface of each monomer, to the right of a large groove that extends from the mid-periplasmic domain to the inner membrane and was previously postulated to be involved in binding (in the case of AcrB) to its cognate MFP, AcrA (38). Interestingly, two of our mutations that are predicted to impact interaction of MexA with MexB (i.e., T578I and A618T) occur in a region of MexB that, in general, corresponds to the MFP-binding region of AcrB (62). A third mutation in MexB that is also predicted to impact MexA association (R716H) occurs outside this. It is, however, in close proximity to T578 and A618 in the three-dimensional structure of MexB and, as such, may also play a role in determining MexA binding. Still, unlike T578 and A618, which are poorly conserved in other RND transporters (consistent with a role in determining the specificity of MexA binding by MexB), R716 is modestly conserved and may not, in fact, determine the specificity of MexA interaction with MexB. Nonetheless, it may be important for MexA binding.
Yu et al. (65) have very recently crystallized AcrB in the presence of several antimicrobial substrates and demonstrated that different substrates bind to different amino acid residues in AcrB, all of which, however, occur within the vestibule-central cavity region, at or immediately above the plane of the inner membrane. Only two of the mutations characterized here for MexB occurred in the vestibule region (and none occurred in the central cavity), although these did result in increased susceptibility to all (E864K) or most (S462F) tested substrates. In all cases, however, antimicrobial resistance was decreased from wild-type levels but not completely abolished. As mutation of a residue within a specific substrate-binding site of MexB would generally be expected to completely abolish resistance to the substrate(s) it accommodates, it seems likely that MexB, like AcrB, has a large and flexible binding pocket with which any given substrate has multiple points of interaction. A single mutation in the binding region, then, is unlikely to eliminate binding or export and thus will have only a modest effect on resistance.
As in our study, mutations in a second RND transporter of P. aeruginosa, MexD of MexCD-OprJ, have also been reported to impact transport of numerous substrates. Indeed, MexD mutations affecting susceptibility to a broad range of β-lactams as well as several cytoplasmically acting antimicrobials were identified and shown to occur within the two LPL regions of this RND transporter (32). Upon mapping of these MexD mutations to our model of MexB (corresponding to Q34K, E89K, A292V, and P328L in MexB), it is evident that they occur at the beginning or end of LPL-1 and line the vestibule or central cavity region. These residues in MexD are thus likely involved in substrate recognition. Interestingly, these MexD mutations affected periplasmically and cytoplasmically acting antimicrobials. Similarly, we noted here that MexB mutations near the vestibules (S462F and E864K) also negatively impact resistance to both periplasmically (carbenicillin, meropenem, and aztreonam) and cytoplasmically (chloramphenicol and nalidixic acid) acting antimicrobials. Thus, it does not appear that there are separate substrate-binding sites for antimicrobials with periplasmic versus cytoplasmic targets.
Unexpectedly, we found that a possible decreased efficiency of MexB trimer formation in the G220S mutant may also have had an indirect negative effect on the ability of MexB to form a functional association with the trimeric OprM component of the MexAB-OprM multidrug efflux system. Since the majority of mutations compensating for the original G220S mutation on the thumb region occurred in the upper docking domain of MexB, it seems plausible that a reduction in efficiency of trimer formation (caused by the G220S mutation) reduces the ratio of functional to nonfunctional couplings of MexB with OprM, its outer membrane counterpart. Potentially, trimer formation in wild-type MexB induces a conformational change in MexB that permits OprM recruitment and subsequent export of antimicrobials across the outer membrane.
Acknowledgments
This work was supported by an operating grant from the Canadian Cystic Fibrosis Foundation (CCFF). J.K.M. was supported by a studentship from the Natural Sciences and Engineering Research Council. K.P. is a CCFF Scholar.
REFERENCES
- 1.Aendekerk, S., B. Ghysels, P. Cornelis, and C. Baysse. 2002. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 148:2371-2381. [DOI] [PubMed] [Google Scholar]
- 2.Aires, J. R., T. Köhler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. John Wiley & Sons, Inc., New York, N.Y.
- 4.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 184:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean, C. R., M. A. Visalli, S. J. Projan, P.-E. Sum, and P. A. Bradford. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47:972-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dey, S., M. Ramachandra, I. Pastan, M. M. Gottesman, and S. V. Ambudkar. 1997. Evidence for two nonidentical drug-interaction sites in the human P-glycoprotein. Proc. Natl. Acad. Sci. USA 94:10594-10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eda, S., H. Maseda, and T. Nakae. 2003. An elegant means of self-protection in Gram-negative bacteria by recognizing and extruding xenobiotics from the periplasmic space. J. Biol. Chem. 278:2085-2088. [DOI] [PubMed] [Google Scholar]
- 9.Eda, S., H. Yoneyama, and T. Nakae. 2003. Function of the MexB efflux-transporter divided into two halves. Biochemistry 42:7238-7244. [DOI] [PubMed] [Google Scholar]
- 10.Elkins, C. A., and H. Nikaido. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 184:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans, K., L. Passador, R. Srikumar, E. Tsang, J. Nezezon, and K. Poole. 1998. Influence of the MexAB-OprM multidrug efflux system on quorum-sensing in Pseudomonas aeruginosa. J. Bacteriol. 180:5443-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garinot-Schneider, C., A. J. Pommer, G. R. Moore, C. Kleanthous, and R. James. 1996. Identification of putative active-site residues in the DNase domain of colicin E9 by random mutagenesis. J. Mol. Biol. 260:731-742. [DOI] [PubMed] [Google Scholar]
- 13.Garrigos, M., L. M. Mir, and S. Orlowski. 1997. Competitive and non-competitive inhibition of the multidrug-resistance-associated P-glycoprotein ATPase—further experimental evidence for a multisite model. Eur. J. Biochem. 244:664-673. [DOI] [PubMed] [Google Scholar]
- 14.Gotoh, N., H. Tsujimoto, K. Poole, J.-I. Yamagishi, and T. Nishino. 1995. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob. Agents Chemother. 39:2567-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan, L., M. Ehrmann, H. Yoneyama, and T. Nakae. 1999. Membrane topology of the xenobiotic-exporting subunit, MexB, of the MexA, B-OprM extrusion pump in Pseudomonas aeruginosa. J. Biol. Chem. 274:10517-10522. [DOI] [PubMed] [Google Scholar]
- 16.Guan, L., and T. Nakae. 2001. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J. Bacteriol. 183:1734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirakata, Y., R. Srikumar, K. Poole, N. Gotoh, T. Suematsu, S. Kohno, S. Kamihira, R. E. Hancock, and D. P. Speert. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J Exp. Med. 196:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jalal, S., O. Ciofu, N. Hoiby, N. Gotoh, and B. Wretlind. 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis. Antimicrob. Agents Chemother. 44:710-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jalal, S., and B. Wretlind. 1998. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microbiol. Drug Resist. 4:257-261. [DOI] [PubMed] [Google Scholar]
- 20.Jo, J. T., F. S. Brinkman, and R. E. Hancock. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 47:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 22.Klyachko, K. A., S. Schuldiner, and A. A. Neyfakh. 1997. Mutations affecting substrate specificity of the Bacillus subtilis multidrug transporter Bmr. J. Bacteriol. 179:2189-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohler, T., M. Kok, M. Michea-Hamzehpour, P. Plesiat, N. Gotoh, T. Nishino, L. Kocjanici Curty, and J.-C. Pechere. 1996. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:2288-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohler, T., M. Michea-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty, and J.-C. Pechere. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 25.Lewinson, O., and E. Bibi. 2001. Evidence for simultaneous binding of dissimilar substrates by the Escherichia coli multidrug transporter MdfA. Biochemistry 40:12612-12618. [DOI] [PubMed] [Google Scholar]
- 26.Li, X.-Z., K. Poole, and H. Nikaido. 2003. Contributions of MexAB-OprM and an EmrE homologue to intrinsic resistance of Pseudomonas aeruginosa to aminoglycosides and dyes. Antimicrob. Agents Chemother. 47:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, X.-Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, X.-Z., L. Zhang, and K. Poole. 1998. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J. Bacteriol. 180:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, X.-Z., L. Zhang, R. Srikumar, and K. Poole. 1998. β-Lactamase inhibitors are substrates of the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, X. Z., and K. Poole. 1999. Organic solvent-tolerant mutants of Pseudomonas aeruginosa display multiple antibiotic resistance. Can. J. Microbiol. 45:18-22. [DOI] [PubMed] [Google Scholar]
- 31.Li, Y., T. Mima, Y. Komori, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. A new member of the tripartite multidrug efflux pumps, MexVW-OprM, in Pseudomonas aeruginosa. J Antimicrob. Chemother. 52:572-575. [DOI] [PubMed] [Google Scholar]
- 32.Mao, W., M. S. Warren, D. S. Black, T. Satou, T. Murata, T. Nishino, N. Gotoh, and O. Lomovskaya. 2002. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: the large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol. Microbiol. 46:889-901. [DOI] [PubMed] [Google Scholar]
- 33.Masuda, N., and S. Ohya. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda, N., E. Sakagawa, and S. Ohya. 1995. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mine, T., Y. Morita, A. Kataoka, T. Mitzushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell, B. A., I. T. Paulsen, M. H. Brown, and R. A. Skurray. 1999. Bioenergetics of the staphylococcal multidrug export protein QacA. Identification of distinct binding sites for monovalent and divalent cations. J. Biol. Chem. 274:3541-3548. [DOI] [PubMed] [Google Scholar]
- 38.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 39.Murata, T., M. Kuwagaki, T. Shin, N. Gotoh, and T. Nishino. 2002. The substrate specificity of tripartite efflux systems of Pseudomonas aeruginosa is determined by the RND component. Biochem. Biophys. Res. Commun. 299:247-251. [DOI] [PubMed] [Google Scholar]
- 40.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piddock, L. J. V., M. C. Hall, F. Bellido, M. Bains, and R. E. W. Hancock. 1992. A pleiotropic, posttherapy, enoxacin-resistant mutant of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1057-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poole, K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3:255-264. [PubMed] [Google Scholar]
- 43.Poole, K. 2001. Multidrug resistance in Gram-negative bacteria. Curr. Opin. Microbiol. 4:500-508. [DOI] [PubMed] [Google Scholar]
- 44.Poole, K. 2002. Outer membranes and efflux: the path to multidrug resistance in gram-negative bacteria. Curr. Pharm. Biotechnol. 3:77-98. [DOI] [PubMed] [Google Scholar]
- 45.Poole, K., N. Gotoh, H. Tsujimoto, Q. Zhao, A. Wada, T. Yamasaki, S. Neshat, J.-I. Yamagishi, X.-Z. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB multidrug resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 46.Poole, K., D. E. Heinrichs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10:529-544. [DOI] [PubMed] [Google Scholar]
- 47.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poole, K., and R. Srikumar. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 1:59-71. [DOI] [PubMed] [Google Scholar]
- 49.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Putman, M., L. A. Koole, H. W. van Veen, and W. N. Konings. 1999. The secondary multidrug transporter LmrP contains multiple drug interaction sites. Biochemistry 38:13900-13905. [DOI] [PubMed] [Google Scholar]
- 51.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rella, M., and D. Haas. 1982. Resistance of Pseudomonas aeruginosa PAO to nalidixic acid and low levels of β-lactam antibiotics: mapping of chromosomal genes. Antimicrob. Agents Chemother. 22:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saito, K., H. Yoneyama, and T. Nakae. 1999. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 179:67-72. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54a.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 294:2158-2163. [DOI] [PubMed] [Google Scholar]
- 55.Schweizer, H. P. 1998. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob. Agents Chemother. 42:394-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sekiya, H., T. Mima, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Functional cloning and characterization of a multidrug efflux pump, MexHI-OpmD, from a Pseudomonas aeruginosa mutant. Antimicrob. Agents Chemother. 47:2990-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon, R., U. Priefer, and A. Puehler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 58.Srikumar, R., T. Kon, N. Gotoh, and K. Poole. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 42:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srikumar, R., X.-Z. Li, and K. Poole. 1997. Inner membrane efflux components are responsible for the β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srikumar, R., and K. Poole. 1999. Demonstration of ethidium bromide efflux by multiresistant pumps of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 5:5S58-5S59. [Google Scholar]
- 62.Tikhonova, E. B., Q. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from Gram-negative bacteria. J. Bacteriol. 184:6499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoneyama, H., A. Ocaktan, N. Gotoh, T. Nishino, and T. Nakae. 1998. Subunit swapping in the Mex-extrusion pumps in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 244:898-902. [DOI] [PubMed] [Google Scholar]
- 65.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976-980. [DOI] [PubMed] [Google Scholar]
- 66.Zgurskaya, H. I. 2002. Molecular analysis of efflux pump-based antibiotic resistance. Int. J. Med. Microbiol. 292:95-105. [DOI] [PubMed] [Google Scholar]
- 67.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]
- 68.Ziha-Zarifi, I., C. Llanes, T. Koehler, J.-C. Pechere, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]