Abstract
CooA is a heme-containing transcriptional activator that enables Rhodospirillum rubrum to sense and grow on CO as a sole energy source. We have identified a number of CooA homologs through database searches, expressed these heterologously in Escherichia coli, and monitored their ability to respond to CO in vivo. Further in vitro analysis of two CooA homologs from Azotobacter vinelandii and Carboxydothermus hydrogenoformans corroborated the in vivo data by revealing the ability of CO to bind to these hemoproteins and stimulate their binding at specific DNA sequences. These data, as well as the patterns of conserved residues in the homologs, are compared to what is already known about functionally important residues in the CooA protein of R. rubrum. The results identify critical regions of CooA and indicate features that distinguish CooAs from the general family of cyclic AMP receptor proteins.
The CO-dependent anaerobic growth of Rhodospirillum rubrum relies on a CO oxidation system encoded by two CO-regulated transcriptional units, cooMKLXUH and cooFSCTJ (8, 9, 15-17, 32). The key products of the coo regulon are an O2-sensitive CO dehydrogenase (CooS), a CooS-associated Fe-S protein (CooF), and a CO-tolerant hydrogenase (CooH), and the expression of the genes depends upon the activity of the CooA protein, which senses CO under anaerobic conditions. CooA, a member of the CRP/FNR (cyclic AMP receptor protein/fumarate nitrate reductase) transcriptional regulator family, is a homodimer in which each monomer contains a b-type heme (34). Remarkably, CooA senses both the redox state of the cell and CO, since the heme undergoes reduction at approximately −300 mV (24) and only the reduced form [heme Fe(II)] of CooA binds the effector (30, 32). An unusual switch between the Cys75 and His77 heme ligands, which is presumably important for setting the proper heme iron redox poise, accompanies the oxidation-reduction of the CooA heme (2, 33).
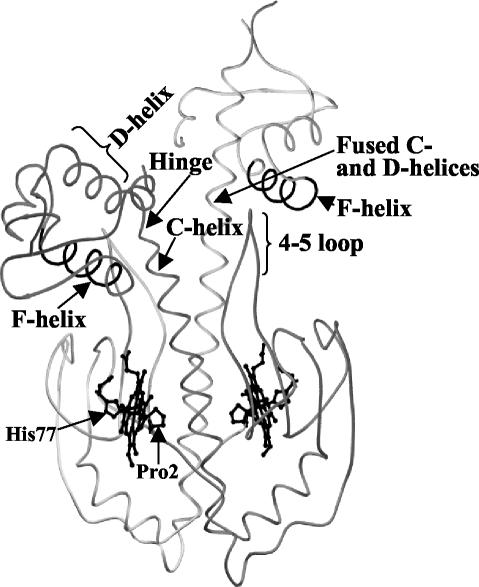
The crystal structure of Fe(II) CooA has been solved (Fig. 1) and provides a basis for comparing the various CooA homologs. That analysis revealed a novel subunit-swapped N-terminal Pro2 as the heme ligand trans to His77 in the homodimer (20). Nuclear magnetic resonance and resonance Raman studies indicate that CO displaces the Pro2 ligand of Fe(II) CooA (40, 43) and exposes the CO-bound heme to the long α-helices (termed the C-helices) that extend along the homodimer interface (4, 44, 45). Extensive mutagenic analysis suggests that CooA activation involves the monomer repositioning about the C-helices at the dimer interface (14, 44, 45). Therefore, our present hypothesis for CooA activation posits that CO binding displaces the endogenous Pro2 ligand, which allows an interaction of the CO-bound heme with the C-helices. This interaction stabilizes an alternative domain conformation about these helices which alters the hinge region (lying between C- and D-helices) that separates the DNA- and CO-binding domains (20) (Fig. 1). The alteration of the geometry of the hinge region destabilizes the inactive form of CooA and stabilizes the active form, which is presumably similar to the active form of CRP (26).
FIG. 1.
Inactive Fe(II) CooA structure adapted from that of the strain with PDB identification no. 1FT9. The protein consists of two monomers, shaded differently in this figure, which dimerize along the central C-helices of adjacent effector-binding domains. The solved structure is asymmetric, in which one monomer contains fused C- and D-helices (20). Nonetheless, both F-helices that interact with DNA in a sequence-specific manner are buried from the surface in the structure. The 4/5 loop is noted and so are the Pro2 and His77 heme Fe(II) ligands. In these representations, the DNA-binding domains form the upper part of each structure and the positions of the helices that specifically interact with DNA are designated as the F-helix.
CO has also been suggested to serve as a neural signal in vertebrates, perhaps by interacting with soluble guanylyl cyclase, although the matter remains controversial (3). Until this is understood, the CooA protein from R. rubrum remains the only naturally occurring CO sensor that has been proven to be physiologically relevant.
Similar CO oxidation systems have been reported for Carboxydothermus hydrogenoformans (37) and Desulfovibrio vulgaris, and a report of a CooA homolog in the latter organism (41) motivated us to examine the databases of completed and unfinished genomes for other CooA homologs. Eight homologs were identified in six organisms; all of these genomes also possess homologs to the CO oxidation system of R. rubrum, which suggests that the CooA homologs might well serve as CO sensors. The present work reports the functional characterization of the CooA homologs, which, together with previous work on CooA from R. rubrum, provides functional commonalities among the CO sensors.
MATERIALS AND METHODS
Sequence searches and alignment.
The R. rubrum CooA protein sequence was used as the query in a TBLASTN (version 2.2.4) search of the entire genome database of completed and unfinished microbial sequences (207 sequences) at the National Center for Biotechnology Information (1). Extracted sequences were aligned by using the T-COFFEE protocol (version 1.37) as implemented at http://www.cmbi.kun.nl/bioinf/tools/T_COFFEE/ (25).
Cloning of cooA homologs.
The cooA homologs were cloned into EcoRI-HindIII-digested pEXT20 (6) after genomic PCR amplification with 5′ (containing EcoRI) and 3′ (containing HindIII) primers designed according to each cooA sequence with previously described extensions (14). The cloned homologs included Azotobacter vinelandii CooA, C. hydrogenoformans CooA (C. hydrogenoformans 2350 CooA), and D. vulgaris Hildenborough CooA. For Desulfovibrio desulfuricans G20 CooA and Desulfitobacterium hafniense CooA, an EcoRI site inside the cooA gene was eliminated (see below) without changing the protein sequence before cloning the PCR product into the EcoRI/HindIII-digested vector. C. hydrogenoformans also contained a second cooA (C. hydrogenoformans 2340 CooA) as well as a second cluster of genes homologous to the coo genes of R. rubrum (37). This cooA also contained an internal EcoRI site and was therefore cloned as a blunt-ended (5′) and HindIII-digested (3′) PCR fragment into SmaI/HindIII-digested pEXT20. Genomic DNAs were kindly supplied by Luis M. Rubio, University of California, Berkeley (A. vinelandii); Frank Robb, University of Maryland, Baltimore (C. hydrogenoformans); Gerrit Voordouw, University of Calgary, Calgary, Alberta, Canada (D. vulgaris Hildenborough); Judy Wall, University of Missouri—Columbia, Columbia (D. desulfuricans G20); and Richard Villemur, INRS-Institut Armand-Frappier, Laval, Quebec, Canada (D. hafniense strain DCB-2). When necessary, mutations were introduced into the cooA genes by the QuikChange procedure (Stratagene, La Jolla, Calif.).
Reporter system for CooA transcription activity in vivo.
The in vivo activities of the CooA homologs were measured by their abilities to stimulate β-galactosidase synthesis with the E. coli reporter strain developed for the analysis of CooA of R. rubrum, which has been described previously (33). Anaerobic expression utilized 120-ml stoppered serum vials containing 20 ml of morpholinepropanesulfonic acid (MOPS)-buffered medium (38) supplemented with 100 μg of ampicillin (Na+ salt)/ml and 25 μM isopropyl-β-d-thiogalactopyranoside (IPTG). For CO-induced cultures, the headspace of the vials contained 2% (vol/vol) CO gas. Cultures were grown with shaking at 30°C to an optical density at 600 nm of 1.5 to 1.8, cell pellets were prepared and frozen, and β-galactosidase activities were measured by a standard protocol (28).
CooA homolog expression and purification.
For the partial purification of certain CooA proteins, the expression vectors were transferred into host strain VJS6737, which was a gift of Valley Stewart (35). For unknown reasons, the level of CooAs expressed in this strain often exceeds those of our standard reporter-containing host by 2- to 10-fold. Strains were cultivated at 30°C in 2× LC medium (14) supplemented with phosphate buffer (pH 7.0) to 10 mM, glucose to 10 mM, ampicillin (Na+ salt) to 100 μg/ml, and IPTG to 500 μM. Aerobic cultures involved the use of 40 ml of medium in 250-ml flasks agitated at 250 rpm; anaerobic cultures employed 200 ml of medium in 250-ml screw-cap bottles that were gently mixed at 80 rpm. R. rubrum CooA, A. vinelandii CooA, and C. hydrogenoformans 2340 CooA homologs were partially purified with a batch hydroxylapatite method as described previously (45), as spectral analysis showed that R. rubrum CooA and C. hydrogenoformans 2340 CooA were stable during aerobic purification (data not shown). The heme content of the CooA preparations was estimated by a modified reduced pyridine-hemochromogen method (42), and protein concentration was measured by the bicinchoninic acid assay (Pierce, Rockford, Ill.). UV-visible absorption spectroscopy of CooA samples in 25 mM MOPS buffer, pH 7.4, with 0.1 M NaCl, was performed at room temperature in quartz cuvettes with a Shimadzu UV-2401PC spectrophotometer. Samples were made anaerobic by flushing with argon and were reduced by the addition of an anaerobically prepared dithionite solution (final concentration, 2 mM). Anaerobic CO gas (final concentration, 20% [vol/vol]) was further added and mixed by gentle inversion for the CO-bound spectra. Potassium ferricyanide (20 μM final concentration) was used for the oxidation of isolated C. hydrogenoformans 2340 CooA.
In vitro DNA-binding assay.
In vitro DNA-binding assays of CooA homologs were performed by using the fluorescence polarization technique with a Beacon 2000 fluorescence polarization detector (PanVera Corp., Madison, Wis.) as described previously (22, 39). As a fluorescence probe, a 26-bp target DNA containing R. rubrum PcooF was labeled with Texas Red on one end of the duplex and used at a concentration of 6.4 nM. Binding assays were performed in 40 mM Tris-HCl (pH 8.0), 6 mM CaCl2, 50 mM KCl, 5% (vol/vol) glycerol, and 1 mM dithiothreitol with salmon sperm DNA added at 6.4 μM as a nonspecific competitor. The conditions for reduction and CO treatment of the samples were similar to those used for obtaining UV-visible spectra.
RESULTS AND DISCUSSION
Discovery and identification of CooA homologs.
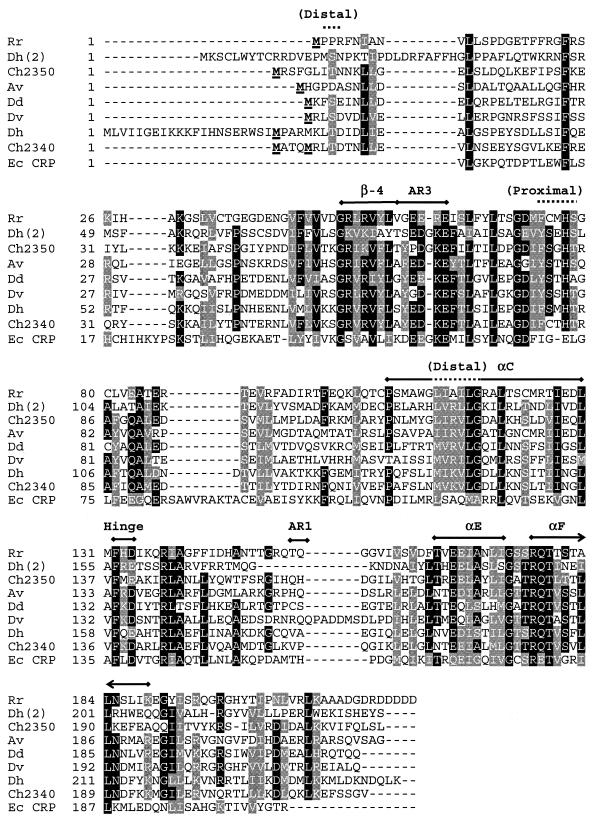
The report by Voordouw (41) of a gene encoding a CooA homolog in D. vulgaris prompted us to search the database for other cooA homologs. Eight genes from six organisms were identified (Table 1 and Fig. 2) that appeared to encode CO sensors based on the following criteria. (i) They showed substantial protein sequence similarity, ranging from 49 to 55%, compared to R. rubrum CooA. (ii) The homologs were found in genomes that also contained genes for CO oxidation systems homologous to that of R. rubrum. At least C. hydrogenoformans and D. vulgaris have also been shown to have CO dehydrogenase activity (5, 41). (iii) Near the invariant histidine proximal ligand, all homologs contained an internal deletion of eight or nine codons (with respect to the CRP) that appears to provide space for the heme in CooA of R. rubrum. (iv) The alignment showed significant conservation of R. rubrum CooA residues known to be critical for protein function (discussed below). Notably absent from the list are organisms wherein CO oxidation is an aerobic process catalyzed by a molybdenum-containing hydroxylase (23) as well as anaerobic organisms (e.g., methanogenic archaea) in which the capability of CO oxidation reflects an intrinsic function of a constitutively expressed metabolic process (7).
TABLE 1.
In vivo β-galactosidase-stimulating activities of CooA homologs
| CooA homolog | % β-Galactosidase activitya
|
Pellet colorb | |
|---|---|---|---|
| −CO | +CO | ||
| R. rubrum | 1.1 | 100 | Red |
| A. vinelandii | 11 | 500 | Light brown |
| A. vinelandii (S77C) | 56 | 493 | Light brown |
| C. hydrogenoformans 2340 | 2.0 | 1,268 | Red |
| C. hydrogenoformans 2340 (C80S) | 1.4 | 903 | Red |
| C. hydrogenoformans 2340 (S)c | 1.0 | 1,065 | Red |
| D. vulgaris | 1.5 | 3.8 | Pale brown |
| D. desulfuricans | 1.3 | 30 | Negatived |
| D. hafniense | 1.0 | 338 | Pale brown |
| C. hydrogenoformans 2350 | 1.0 | 1.5 | Negative |
Activity indicates the mean value of the results from two independent assays expressed relative to that of R. rubrum CooA in the presence of CO. The assays showed a variability of <10%.
Cell pellet color obtained from anaerobically grown cultures.
Shorter version of cloned C. hydrogenoformans 2340 CooA.
Negative indicates a similar pellet color (gray) to that of the control strain which contains only vector.
FIG. 2.
Sequence alignment of CooA homologs. Eight CooA homologs and CRP from E. coli were aligned by using the T-Coffee multiple sequence alignment tool (version 1.37) as implemented at http://www.cmbi.kun.nl/bioinf/tools/T_COFFEE/ (25). Putative CooA homologs were identified by a TBLASTN search of 207 completed and unfinished microbial genomes available at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) queried with the R. rubrum CooA protein. Designations of the aligned sequencesand their accession parameters are as follows: Rr, R. rubrum (gi no. 1498752); Dh(2), D. hafniense (gi no. 23114031); Ch2350, C. hydrogenoformans (gnl no. TIGR_129958, contig 2350:c_hydrogenoformans); Av, A. vinelandii (gi no. 23105899); Dd, D. desulfuricans G20 (gi no. 23473780); Dv, D. vulgaris (gnl no. TIGR_881, 1531); Dh, D. hafniense (gi no. 23111778); Ch2340, C. hydrogenoformans (gnl no. TIGR_129958, contig 2340:c_hydrogenoformans); Ec CRP, E. coli (gi no. 117484). Shaded portions indicate 70% conservation, with key structural elements indicated above sections of the alignment. These include residues that form the distal and proximal heme ligand environments (overlined with a dotted line) as well as conserved structural elements including the His heme ligand (His77 in R. rubrum CooA), the β-4 sheet, helices αC, αE, and αF, the hinge sequence between the effector and DNA-binding domains, and the AR1 and AR3 regions that interact with RNA polymerase (overlined with a thick line). Boldface underlined residues at or near the N terminus represent the cloned initiation of the proteins; for the CooA from R. rubrum, the terminal Met is removed posttranscriptionally.
C. hydrogenoformans and D. hafniense each contained two cooA homologs, consistent with the observation that each contains two clusters of other coo gene homologs as well. We cloned both of the CooA homologs from C. hydrogenoformans (designated C. hydrogenoformans 2340 and 2350 CooA) because they differ at a unique cysteine residue that serves as an Fe(III) heme ligand in R. rubrum CooA (Cys75 in R. rubrum CooA) (Fig. 2) but arbitrarily cloned only a single CooA homolog from D. hafniense (Fig. 2). The genes encoding D. hafniense CooA and C. hydrogenoformans 2340 CooA have the additional complication of more than one potential start codon, an issue of particular relevance for R. rubrum CooA, since the N-terminal Pro2 of one monomer is a heme ligand in the other. The cloned region of D. hafniense CooA was chosen in accordance with the terminus predicted by the ORF Finder utility (http://www.ncbi.nlm.nih.gov/gorf/gorf.html and its position homologous to the R. rubrum CooA terminus (Fig. 2). We created two clones for C. hydrogenoformans 2340 CooA, with start sites at residues −1 and −5 relative to the R. rubrum CooA sequence (Fig. 2). As shown below, both of these clones displayed substantial CooA activity, so the issue of the proper terminus remains unresolved.
CO-responsive in vivo activities of CooA homologs.
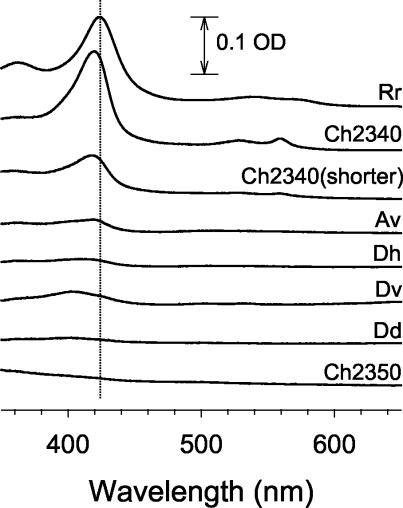
To determine if the CooA homologs function as CO sensors, we introduced each cloned cooA into the Escherichia coli reporter strain we employed for the analysis of R. rubrum CooA. This system has a CooA-binding site and a promoter cloned from R. rubrum placed in front of lacZ. We reasoned that since the DNA-binding F-helices are highly conserved among the CooA homologs (Fig. 2), a CO-dependent response would affect binding of the appropriate sequence in the reporter and, given appropriate levels of CooA accumulation and proper interaction with E. coli RNA polymerase, result in β-galactosidase accumulation. Indeed, most of the homologs displayed a significantly higher level of lacZ expression under anaerobic conditions in the presence of CO than in its absence, strongly suggestive that these cloned CooA homologs are capable of serving as CO sensors in vivo (Table 1). As mentioned before, both versions of C. hydrogenoformans 2340 CooA, which presumably differ in the length and identity of their N termini, displayed similar high activity. The only homologs that failed to show a CO response were C. hydrogenoformans 2350 CooA and D. vulgaris CooA. The former of these accumulated very poorly, but the basis of failure of the latter to show activity is unclear (Table 1 and Fig. 3). In the case of R. rubrum CooA, accumulation of heme-containing protein is revealed by a distinct reddish color of the cell pellet (Table 1). While D. desulfuricans CooA and D. hafniense CooA appeared to accumulate poorly (Table 1 and Fig. 3), they were active in response to CO in vivo (Table 1).
FIG. 3.
Cells containing most CooA homologs accumulate heme-containing proteins. CooA proteins were partially purified from anaerobically grown cells by the hydroxylapatite preparation described in Materials and Methods. The spectrum of each partially purified sample was corrected by subtracting the spectrum of a similarly treated control sample of the same protein concentration prepared from a strain containing only the pEXT20 vector. OD, optical density.
Although a positive response in this assay clearly implies the presence of a CO-sensing protein, comparisons of the magnitude of the response are more difficult to interpret because the assay reflects the combination of factors: the expression of CO-responsive heme-containing protein, the fraction of that population that exists in the proper form to bind DNA, interactions with a heterologous RNA polymerase, and the affinity of the CooA homolog for the R. rubrum PcooF promoter. Despite the complexity, we currently believe that many of the other homologs interact substantially better with E. coli RNA polymerase than does R. rubrum CooA, based on both the in vivo results and on in vitro analyses described below. The regions of CooA that interact with RNA polymerase are termed activating regions (AR), based on the large body of work done with CRP and FNR, another distantly related homolog (18, 19). This apparently better AR surface in the homologs was initially surprising, since R. rubrum CooA has been shown to function with the E. coli RNA polymerase in vivo and in vitro (12, 21). Nevertheless, there is no reason to suppose that R. rubrum CooA would interact any better with the heterologous RNA polymerase from E. coli than would any other homolog. Indeed, it is interesting that R. rubrum CooA variants with improved AR interactions were easily obtained in a screen for improved function (21), consistent with the notion that wild-type R. rubrum CooA might actually be rather poor in this regard. Based on this logic, we believe that A. vinelandii, C. hydrogenoformans 2340, and D. hafniense CooA possess enhanced AR surfaces (for interaction with RNA polymerase from E. coli) because the activity with CO is higher than that of R. rubrum CooA. This might also explain some of the activity without CO.
These results demonstrate that the majority of these homologs have a clear responsiveness to the presence of CO, consistent with the hypothesis that they serve as CO sensors in the organisms in which they are normally found. It is therefore appropriate to compare their properties with those of R. rubrum CooA to elucidate the necessary features of the generalized CooA family.
The CooA homologs accumulate poorly under aerobic growth conditions.
A preliminary experiment showed that the expressed CooA homologs accumulated heme-containing protein much less effectively than did R. rubrum CooA in aerobically grown cells (data not shown). We felt that this poor accumulation might reflect poor stability of the heme during aerobic growth because all but one of these homologs lacked a residue homologous to Cys75 (Fig. 2), one of the Fe(III) heme ligands in R. rubrum CooA (29), and it has already been shown that a C75S variant of R. rubrum CooA is unstable under aerobic conditions (33).
To determine if growth conditions perturb the level of heme-containing CooA homologs in E. coli, we prepared both aerobic and anaerobic cultures producing R. rubrum, A. vinelandii, and C. hydrogenoformans 2340 CooAs and compared their heme contents normalized to total extract protein. Consistent with the hypothesis of poorer accumulation of A. vinelandii and C. hydrogenoformans 2340 CooAs under aerobic conditions, anaerobic growth provided much higher heme accumulation for these homologs than for R. rubrum CooA (Table 2). However, these CooA homologs were stable during and after purification under aerobic conditions, suggesting that the higher yield of heme-containing protein from anaerobic expression might be related to external factors such as protease activity or better heme incorporation during anaerobic protein synthesis. A plausible explanation for this phenomenon is that under physiological conditions, the A. vinelandii and C. hydrogenoformans 2340 CooAs are never exposed to oxidizing conditions in their native hosts. While A. vinelandii is an aerobe, its internal milieu is sufficiently reducing to permit operation of strictly anaerobic nitrogenase systems (27), and C. hydrogenoformans is an obligate anaerobe isolated from a hydrothermal vent (36).
TABLE 2.
Heme accumulation and Soret maxima of selected CooA homologs
| CooA homolog | Concn (nM) of heme b per 1 mg of protein/mla
|
Soret maxima (nm) for form
|
|||
|---|---|---|---|---|---|
| Aerobic growth | Anaerobic growth | Fe(III) | Fe(II) | Fe(II)-CO | |
| pEXT20b | <50 | <50 | |||
| R. rubrum CooA | 4,292 | 1,362 | 424 | 425 | 422 |
| A. vinelandii CooA | 145 | 357 | 419 | 421.5 | 419.5 |
| C. hydrogenoformans 2340 CooA | 773 | 1,656 | 416 | 424.5 | 420.5 |
The heme b content was quantified by a modified pyridine-hemochromogen method with partially purified samples as described in Materials and Methods.
pEXT20 represents the same preparation of a control strain containing only the cloning vector.
Spectral comparison of R. rubrum, A. vinelandii, and C. hydrogenoformans 2340 CooAs reveals differences in Fe(III) and Fe(II) forms.
UV-visible spectra and in vitro DNA-binding activities were analyzed for R. rubrum, A. vinelandii, and C. hydrogenoformans 2340 CooAs that had been partially purified from anaerobically grown cells. The C. hydrogenoformans 2340 CooA was chosen in part because it is the only homolog that has a Cys residue at a position homologous to Cys75 of R. rubrum CooA, which serves as one of the heme ligands in the six-coordinate low-spin Fe(III) form (Fig. 2 and 4A). We were therefore interested to know whether C. hydrogenoformans 2340 CooA showed similar spectral properties to those of R. rubrum CooA.
FIG. 4.
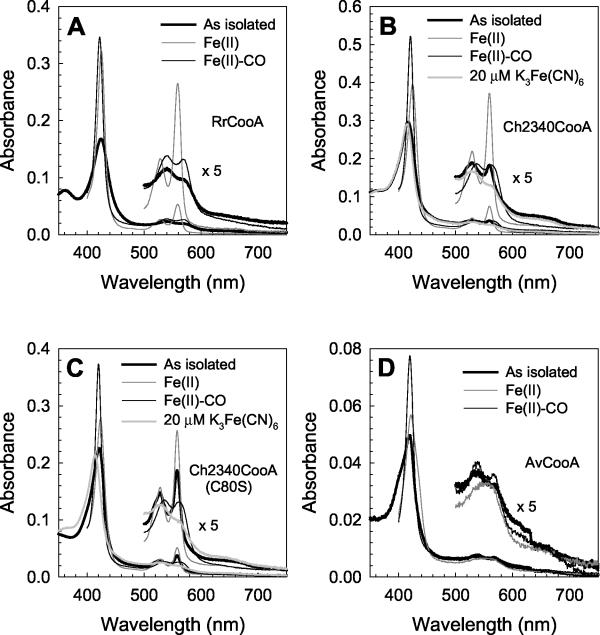
UV-visible spectra of the selected CooA homologs. (A) R. rubrum CooA; (B) C. hydrogenoformans 2340 CooA; (C) C80S C. hydrogenoformans 2340 CooA; (D) A. vinelandii CooA. CooA proteins were partially purified from anaerobically grown cells by the hydroxylapatite preparation described in Materials and Methods. Analyzed samples were diluted to 1 mg of protein/ml. For the spectra of C. hydrogenoformans 2340 CooA proteins, 20 μM potassium ferricyanide [K3Fe(CN)6] was added to the as isolated forms of the proteins.
Figure 4B presents the UV-visible spectra of the C. hydrogenoformans 2340 CooA in the “as isolated” form [expected to be Fe(III)] as well as the Fe(II), Fe(II)-CO, and chemically oxidized Fe(III) forms. Not surprisingly, CO addition to the Fe(II) form resulted in a spectral change indicative of CO binding. Overall spectral features of the CO-bound form are very similar to those of CO-bound R. rubrum CooA, presumably reflecting the invariant His77 that serves as the proximal ligand in that form of R. rubrum CooA (Fig. 2). In contrast, the Soret peak of C. hydrogenoformans 2340 CooA shows a reduced intensity ratio of the Fe(II) Soret band to the Fe(II)-CO Soret band, implying that the Fe(II) form in this homolog is altered relative to that of R. rubrum CooA. This spectral property is also a characteristic of Fe(II) ΔP3R4 R. rubrum CooA (46), in which the Pro2 ligand is perturbed by the deletion of the two penultimate residues, and suggests a weakened endogenous ligand trans to the invariant His in the C. hydrogenoformans 2340 CooA. Surprisingly, even aerobic purification yielded partially reduced C. hydrogenoformans 2340 CooA, indicated by sharply resolved α and β peaks in the spectrum of the as isolated form (Fig. 4B). These α and β peaks were eliminated by treatment with the oxidant potassium ferricyanide (Fig. 4B), confirming the partially reduced state of the as isolated protein. The as isolated R. rubrum CooA did not show any spectral difference with or without 20 μM potassium ferricyanide (data not shown). The redox potential of R. rubrum CooA has been determined to be approximately −300 mV, the threshold below which the CO oxidation catalytic enzyme, CO dehydrogenase, is active. This result suggests that C. hydrogenoformans 2340 CooA is shifted in its redox poise, presumably because of a less favorable Fe(III) state in this protein. The potassium ferricyanide-treated Fe(III) form of C. hydrogenoformans 2340 CooA appears to predominantly be the six-coordinate form (Fig. 4B) and not significantly different from that of R. rubrum CooA, but the natures of the endogenous ligands are yet to be determined.
To test whether or not Cys80 (Cys75 in R. rubrum CooA) serves as one ligand in the Fe(III) form of C. hydrogenoformans 2340 CooA, we introduced Ser at that position by site-directed mutagenesis. Both the heme accumulation and spectra of C80S C. hydrogenoformans 2340 CooA are remarkably similar to those of wild-type C. hydrogenoformans 2340 CooA (Fig. 4B and C), in contrast to the low heme accumulation and Fe(III) spectral perturbation of the R. rubrum CooA C75S variant (33). This indicates that Cys80 is not the ligand in the Fe(III) C. hydrogenoformans 2340 CooA. The most probable candidate for the proximal Fe(III) heme ligand in C. hydrogenoformans 2340 CooA is His82, which is homologous to the critical His77 of R. rubrum CooA.
A. vinelandii CooA was also analyzed spectrally because it represented a CooA homolog containing Ser at the position homologous to Cys75 in R. rubrum CooA (Fig. 2). Like C. hydrogenoformans 2340 CooA, A. vinelandii CooA displayed a typical Fe(II)-CO spectrum, but the spectra of its Fe(II) and as isolated forms were notably different from those of R. rubrum CooA (Fig. 4D) and are consistent with a significant fraction of five-coordinate heme ligation. As for C. hydrogenoformans 2340 CooA, we presume this reflects a weak endogenous ligand trans to the invariant His.
R. rubrum and C. hydrogenoformans 2340 CooA are the only two wild-type homologs that have Cys at the position homologous to Cys75 in R. rubrum CooA (Fig. 2), and this correlates with their excellent accumulation of heme-containing protein (Table 1; Fig. 3). However, the ability of C80S C. hydrogenoformans 2340 CooA to also accumulate heme well suggests that this is not a causal relationship. This notion was further tested by the creation and analysis of S77C A. vinelandii CooA. Neither protein accumulation nor spectra were significantly different from that of wild-type A. vinelandii CooA (data not shown). The above results suggest that the presence of Cys at this position is not correlated with the accumulation of heme-containing protein under these conditions.
As noted in the introduction, R. rubrum CooA undergoes a highly unusual ligand switch during oxidation and reduction and maintains a six-coordinate heme ligation under all conditions. One implication of the rather different redox behavior of the other homologs and the general absence in them of a strong ligand residue homologous to Cys75 [C. hydrogenoformans 2340 CooA contains a Cys residue, but it does not appear to serve as an Fe(III) ligand] is that these proteins probably do not undergo a similar redox change. Though this notion has not been experimentally tested, it is nevertheless consistent with the fact that R. rubrum is the only one of these organisms likely to face such a physiological decision.
The conclusion from these results is that the examined CooA homologs bind CO to create a species that is spectrally very similar to that of R. rubrum CooA. However, the homologs display some clear differences in the spectra of the Fe(II) and Fe(III) forms from that of R. rubrum CooA, though the molecular basis for these differences is unknown. The results suggest that it is not merely the presence or absence of a Cys75 analog that underlies these differences. Rather, we assume that these other homologs lack other residues in the heme vicinity that affect the properties of the Fe(II) and Fe(III) forms.
R. rubrum, A. vinelandii, and C. hydrogenoformans 2340 CooAs bind target DNA in a CO-dependent manner.
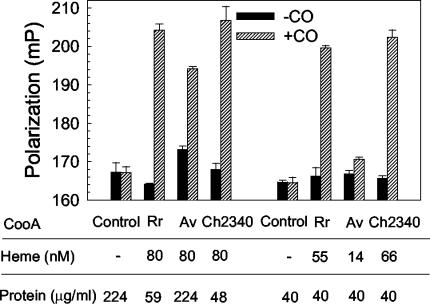
The DNA-binding properties of the partially purified CooA homologs were tested by a fluorescence polarization assay (39). This technique measures the ability of CooA to bind a known CooA binding site that is identical to that in the in vivo assay but without the complication of RNA polymerase interactions. As shown in Fig. 5, these CooA proteins showed substantial CO responsiveness regardless of whether the assay was normalized to sample heme or protein content, although the lower fraction of heme-containing CooA in the A. vinelandii CooA preparation reduces its activity in the latter analysis. The observation that these homologs display DNA-binding activity similar to that of R. rubrum CooA indicates that their very high in vivo activities (Table 1) reflect heterologous RNA polymerase interactions superior to those of R. rubrum CooA, insofar as the in vivo and in vitro assays depend on the same R. rubrum CooA DNA-binding sequences. Of course, conclusions cannot be extrapolated for relative affinities of these homologs for their native binding sites in their natural hosts. These results confirm the CO responsiveness of the tested homologs in terms of DNA binding, consistent with a CO-induced conformational change to reposition the F-helices, as envisioned for R. rubrum CooA (14).
FIG. 5.
In vitro DNA-binding activities of selected CooA homologs purified from anaerobically grown cells. The analyzed samples duplicated those prepared for spectral analyses (Fig. 4). The activities of the CooA homologs were measured at the same concentrations of either heme or protein. −, absent.
Sequence analysis of the regions 5′ to the coo structural genes in all the organisms from which cooA genes were isolated revealed reasonably positioned binding sites corresponding to the TGTC(A/G)N6(C/T)GACA consensus found in R. rubrum: 5′ of cooS in D. desulfuricans, 5′ of cooF in A. vinelandii, 5′ of cooF in D. hafniense, 5′ of cooS in D. vulgaris. Similar sequences are found in the 5′ regions of both cooF genes in C. hydrogenoformans. However, these sequences have not been confirmed experimentally as binding sites, nor is the R. rubrum CooA consensus a robust one, as it is based on the only two known binding sites in the genome (11). Nevertheless, examination of the F-helices of all the homologs (Fig. 2) reveals significant similarity with one another, consistent with the notion that they bind similar sequences. For example, they all have an Arg residue homologous to Arg180 of CRP that contacts the first G of the binding half-site (TGTGA) and they all have Gln at the position homologous to Glu181 of CRP. Because this Glu181 interacts with the second G of the CRP half-site (TGTGA), this replacement in the CooA homologs suggests that the Gln might interact with the C of the CooA half-site [TGTC(A/G)].
Critical residues in heme region of CooA.
The heme-binding region of CooA does not resemble those of other common heme regions, such as the PAS domain (10) or the heme domain found in globins (13). However, a comparison of the various CooA homologs, together with a substantial amount of mutational analysis of R. rubrum CooA, has revealed a number of critical residues in the vicinity of the heme.
All the homologs conserve a proximal heme environment (Fig. 2) that consists of an invariant His at position 77 (R. rubrum CooA numbering), small residues at position 75 and 78, and a Phe or Tyr at position 74. As already mentioned, Cys75 is a ligand for the Fe(III) heme in R. rubrum CooA (29) and its absence in most of the homologs is consistent with the strict anaerobic physiology of these organisms, whether a result of their environment or metabolic function. A C75S variant of R. rubrum CooA accumulates stable, active protein when expressed and analyzed under anaerobic conditions (33), consistent with the data for the homologs which all have Cys or Ser residues (32), while larger residues at this position preclude activity.
All the homologs possess a His residue homologous to His77 of R. rubrum CooA. The His77 residue of R. rubrum CooA serves as the proximal ligand in the Fe(II) and Fe(II)-CO forms and is critical for proper CO activation (33). We assume that its role is twofold. First, it must provide sufficient ligand strength to preclude its displacement by various small-molecule ligands, including CO, which itself must displace the trans ligand (43). Secondly, because it remains tethered to the CO-bound heme, it helps define the position that the heme can assume. As CooA activation depends on an interaction of the CO-bound heme with the C-helices, heme positioning is almost certainly critical for proper activation.
One of the striking observations from the Fe(II) structure of R. rubrum CooA was the evidence of Pro2 as the heme ligand trans to His77 (20), and indirect evidence also strongly suggests that it is the ligand in the Fe(III) form as well (24, 39, 45). An important role of this residue appears to be to maintain R. rubrum CooA in an inactive (non-DNA-binding) form in the absence of CO, and it also appears to provide at least one level of effector specificity, since other ligands such as CN− and imidazole are unable to displace it. However, it has little importance in the conversion to the active form when CO is present (39). Its dispensability is supported by the lack of conservation in the N-terminal region in the homologs (Fig. 2), where only one protein (D. hafniense CooA) potentially provides an N-terminal Pro ligand. The ligand trans to the His77 homologous position in the other CooAs is unknown, but it may be the terminal Met. In contrast to R. rubrum CooA, the large residues at position 2 should prevent the posttranslational processing that yields Pro2 as the R. rubrum CooA terminus. The various CooA homologs also differ in the apparent length of the N terminus, which is again consistent with mutational data for R. rubrum CooA, where increasing or shortening the N terminus yielded variants with substantial CO-responsive activity (39). Because of its reasonable level of accumulation, C. hydrogenoformans 2340 CooA was tested for imidazole and CN− binding but appeared to bind neither at concentrations up to 50 mM. This implies that the amino-terminal ligand of this homolog, at least, provides a roughly comparable type of exclusion to the binding of improper effectors, as does Pro2 of R. rubrum CooA.
In contrast to the case of Pro2, the C-helix residues Leu116 and Gly117 (Fig. 2) that form the distal heme pocket in CO-bound state of R. rubrum CooA are particularly critical for its response to CO (44, 45). Their conservation in all the homologs suggests that these residues are crucial for the CO response of the entire CooA family. Recent evidence also suggests that Leu120 is also important for this CO response (R. L. Kerby and G. P. Roberts, unpublished data), and this residue is again absolutely conserved in the CooA family.
Conservation of a signal transduction pathway within CooA.
The C-helices of the CRP/FNR family of regulators provide a stable hydrophobic dimer interface, but at the same time, they must allow sufficient mobility to permit an altered conformation, in at least CRP and CooA (14, 20, 26). This has been directly shown to be a critical signal pathway for CO binding (14) in R. rubrum CooA. It was therefore of great interest to see that the CooA homologs share with CRP the leucine zipper residues Ile113, Leu116, Leu120, Ile127, and Leu130 but invariably lack a typical d position residue at Cys123 (R. rubrum CooA numbering) (Fig. 2). Several residues around the interdomain hinge region are also conserved in both the CooA family and in CRP; these include Phe132, Asp134, Arg138, and Ala140 of R. rubrum CooA (Fig. 2). This extensive conservation suggests that the C-helix repositioning upon effector binding, as well as the mechanism for reorienting the DNA-binding domains, may be fundamentally similar in all these proteins, although this hypothesis must be experimentally tested.
The unusual C terminus of R. rubrum CooA is not conserved in the CooA homologs.
R. rubrum CooA not only possessed a unique N-terminal heme ligand but also contained a novel run of C-terminal Asp residues (Fig. 2) whose conformation, unfortunately, is unresolved in the structure (20). The deletion or substitution of these residues generally has little effect on CooA activity (H. Youn and G. P. Roberts, unpublished data), and their relative unimportance is supported by the absence of such residues in any of the homologs.
Surprising conservation of the 4/5 loop.
The 4/5 loop refers to the structure formed by the β-4 and β-5 sheets (Fig. 1) that extends from the effector-binding domain toward the DNA-binding domain, making contacts with this domain in the active form of CRP. In CRP, the tip of this loop has been shown to interact with the sigma subunit of RNA polymerase and is termed AR3 (31), but the 4/5 loop residues immediately preceding and subsequent to the AR3 region are not known to provide structural or functional importance. Surprisingly, inspection of the sequence alignments shows the β-4/5 loop to be highly conserved in the CooA homologs and notably different from CRP through the presence of two basic residues (Fig. 2). Indeed, the purified R51C R53C CooA double variant had low heme content and extremely poor DNA binding in response to CO (M. V. Thorsteinsson and G. P. Roberts, unpublished data). Given the structural connection of this region to the His77 region, the proximal heme ligand in the CO-bound form, the β-4/5 loop region containing these residues might therefore constitute a separate CO signal conduit within CooA in addition to the C-helix mechanism.
The results of the sequence comparison and functional and spectral analysis of the CooA homologs not only verifies their capability as CO sensors but have also corroborated the identification of critical features of CooA that had previously only been based on mutational and biochemical analysis of R. rubrum CooA. These features include the His ligand of the heme, the hydrophobic distal heme pocket, the negligible role of C-terminal poly-Asp tail in the CO-sensing function of R. rubrum CooA, the modified leucine zipper formed by the C-helices, residues in the hinge region between the effector- and DNA-binding domains, and the surprising conservation of the β-4 region. It is probably of significance that the residues in the C-helix and the hinge region are well conserved in CRP, suggesting a common pathway of activation that remains to be completely elucidated. An important difference between R. rubrum CooA and the homologs was the lack of redox-based ligand switch in the latter class, and it is clear that a Cys75 analog is necessary but not sufficient for this ability. Another interesting difference is in the N terminus, since it provides a heme ligand in R. rubrum CooA that serves as a factor in small-molecule specificity. It remains to be seen whether the CooA homologs have similar ligand specificities, and if so, what ligand supports this. The results provide a fairly clear idea of the functionally critical elements of the CooA family and, in most cases, the biochemical property that underlies the role of those elements.
Acknowledgments
This work was supported by the College of Agricultural and Life Sciences at the University of Wisconsin—Madison, Madison, National Institutes of Health Grant GM53228 (to G.P.R.).
We thank Luis Rubio, Frank Robb, Gerrit Voordouw, Judy Wall, and Richard Villemur for the generous provision of genomic DNA for the organisms that we examined.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aono, S., K. Ohkubo, T. Matsuo, and H. Nakajima. 1998. Redox-controlled ligand exchange of the heme in the CO-sensing transcriptional activator CooA. J. Biol. Chem. 273:25757-25764. [DOI] [PubMed] [Google Scholar]
- 3.Boehning, D., and S. H. Snyder. 2003. Novel neural modulators. Annu. Rev. Neurosci. 26:105-131. [DOI] [PubMed] [Google Scholar]
- 4.Coyle, C. M., M. Puranik, H. Youn, S. B. Nielsen, R. D. Williams, R. L. Kerby, G. P. Roberts, and T. G. Spiro. 2003. Activation mechanism of the CO sensor CooA: mutational and resonance Raman spectroscopic studies. J. Biol. Chem. 278:35384-35393. [DOI] [PubMed] [Google Scholar]
- 5.Dobbek, H., V. Svetlitchnyi, L. Gremer, R. Huber, and O. Meyer. 2001. Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster. Science 293:1281-1285. [DOI] [PubMed] [Google Scholar]
- 6.Dykxhoorn, D. M., R. St. Pierre, and T. Linn. 1996. A set of compatible tac promoter expression vectors. Gene 177:133-136. [DOI] [PubMed] [Google Scholar]
- 7.Ferry, J. G. 1999. Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol. Rev. 23:13-38. [DOI] [PubMed] [Google Scholar]
- 8.Fox, J. D., R. L. Kerby, G. P. Roberts, and P. W. Ludden. 1996. Characterization of the CO-induced, CO-tolerant hydrogenase from Rhodospirillum rubrum and the gene encoding the large subunit of the enzyme. J. Bacteriol. 178:1515-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox, J. D., Y. He, D. Shelver, G. P. Roberts, and P. W. Ludden. 1996. Characterization of the region encoding the CO-induced hydrogenase of Rhodospirillum rubrum. J. Bacteriol. 178:6200-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong, W., B. Hao, S. S. Mansy, G. Gonzalez, M. A. Gilles-Gonzalez, and M. K. Chan. 1998. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. USA 95:15177-15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, Y., D. Shelver, R. L. Kerby, and G. P. Roberts. 1996. Characterization of a CO-responsive transcriptional activator from Rhodospirillum rubrum. J. Biol. Chem. 271:120-123. [DOI] [PubMed] [Google Scholar]
- 12.He, Y., T. Gaal, R. Karls, T. J. Donohue, R. L. Gourse, and G. P. Roberts. 1999. Transcription activation by CooA, the CO-sensing factor from Rhodospirillum rubrum. The interaction between CooA and the C-terminal domain of the α subunit of RNA polymerase. J. Biol. Chem. 274:10840-10845. [DOI] [PubMed] [Google Scholar]
- 13.Hou, S., T. Freitas, R. W. Larsen, M. Piatibratov, V. Sivozhelezov, A. Yamamoto, E. A. Meleshkevitch, M. Zimmer, G. W. Ordal, and M. Alam. 2001. Globin-coupled sensors: a class of heme-containing sensors in Archaea and Bacteria. Proc. Natl. Acad. Sci. USA 98:9353-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerby, R. L., H. Youn, M. V. Thorsteinsson, and G. P. Roberts. 2003. Repositioning about the dimer interface of the transcription regulator CooA: a major signal transduction pathway between the effector and DNA-binding domains. J. Mol. Biol. 325:809-823. [DOI] [PubMed] [Google Scholar]
- 15.Kerby, R. L., P. W. Ludden, and G. P. Roberts. 1995. Carbon monoxide-dependent growth of Rhodospirillum rubrum. J. Bacteriol. 177:2241-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerby, R. L., P. W. Ludden, and G. P. Roberts. 1997. In vivo nickel insertion into the carbon monoxide dehydrogenase of Rhodospirillum rubrum: molecular and physiological characterization of cooCTJ. J. Bacteriol. 179:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerby, R. L., S. S. Hong, S. A. Ensign, L. J. Coppoc, P. W. Ludden, and G. P. Roberts. 1992. Genetic and physiological characterization of the Rhodospirillum rubrum carbon monoxide dehydrogenase system. J. Bacteriol. 174:5284-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolb, A., S. Busby, H. Buc, G. Garges, and S. Adhya. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62:749-795. [DOI] [PubMed] [Google Scholar]
- 19.Lamberg, K. E., and P. J. Kiley. 2000. FNR-dependent activation of the class II dmsA and narG promoters of Escherichia coli requires FNR-activating regions 1 and 3. Mol. Microbiol. 38:1-12. [DOI] [PubMed] [Google Scholar]
- 20.Lanzilotta, W. N., D. J. Schuller, M. V. Thorsteinsson, R. L. Kerby, G. P. Roberts, and T. L. Poulos. 2000. Structure of the CO sensing transcription activator CooA. Nat. Struct. Biol. 7:876-880. [DOI] [PubMed] [Google Scholar]
- 21.Leduc, J., M. V. Thorsteinsson, T. Gaal, and G. P. Roberts. 2001. Mapping CooAXRNA polymerase interactions. Identification of activating regions 2 and 3 in CooA, the CO-sensing transcriptional activator. J. Biol. Chem. 276:39968-39973. [DOI] [PubMed] [Google Scholar]
- 22.Lundblad, J. R., M. Laurance, and R. H. Goodman. 1996. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 10:607-612. [DOI] [PubMed] [Google Scholar]
- 23.Meyer, O., L. Gremer, R. Ferner, M. Ferner, H. Dobbek, M. Gnida, W. Meyer-Klaucke, and R. Huber. 2000. The role of Se, Mo and Fe in the structure and function of carbon monoxide dehydrogenase. Biol. Chem. 381:865-876. [DOI] [PubMed] [Google Scholar]
- 24.Nakajima, H., Y. Honma, T. Tawara, T. Kato, S.-Y. Park, H. Miyatake, Y. Shiro, and S. Aono. 2001. Redox properties and coordination structure of the heme in the CO-sensing transcriptional activator CooA. J. Biol. Chem. 276:7055-7061. [DOI] [PubMed] [Google Scholar]
- 25.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 26.Passner, J. M., S. C. Schultz, and T. A. Steitz. 2000. Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 Å resolution. J. Mol. Biol. 304:847-859. [DOI] [PubMed] [Google Scholar]
- 27.Poole, R. K., and S. Hill. 1997. Respiratory protection of nitrogenase activity in Azotobacter vinelandii-roles of the terminal oxidases. Biosci. Rep. 17:303-317. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds, A., and V. Lundblad. 1995. Assay for β-galactosidase in liquid culture, p. 13-30. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Short protocols in molecular biology: a compendium of methods from current protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 29.Reynolds, M. F., D. Shelver, R. L. Kerby, R. B. Parks, G. P. Roberts, and J. N. Burstyn. 1998. EPR and electronic absorption spectroscopies of the CO-sensing CooA protein reveal a cysteine-ligated low-spin ferric heme. J. Am. Chem. Soc. 120:9080-9081. [Google Scholar]
- 30.Reynolds, M. F., R. B. Parks, J. N. Burstyn, D. Shelver, M. V. Thorsteinsson, R. L. Kerby, G. P. Roberts, K. M. Vogel, and T. G. Spiro. 2000. Electronic absorption, EPR, and resonance Raman spectroscopy of CooA, a CO-sensing transcription activator from R. rubrum, reveals a five-coordinate NO-heme. Biochemistry 39:388-396. [DOI] [PubMed] [Google Scholar]
- 31.Rhodius, V. A., and S. J. Busby. 2000. Transcription activation by the Escherichia coli cyclic AMP receptor protein: determinants within activating region 3. J. Mol. Biol. 299:295-310. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, G. P., M. V. Thorsteinsson, R. L. Kerby, W. N. Lanzilotta, and T. L. Poulos. 2001. CooA: a heme-containing regulatory protein that serves as a specific sensor of both carbon monoxide and redox state. Prog. Nucleic Acid Res. Mol. Biol. 67:35-63. [DOI] [PubMed] [Google Scholar]
- 33.Shelver, D., M. V. Thorsteinsson, R. L. Kerby, S.-Y. Chung, G. P. Roberts, M. F. Reynolds, R. B. Parks, and J. N. Burstyn. 1999. Identification of two important heme site residues (cysteine 75 and histidine 77) in CooA, the CO-sensing transcription factor of Rhodospirillum rubrum. Biochemistry 38:2669-2678. [DOI] [PubMed] [Google Scholar]
- 34.Shelver, D., R. L. Kerby, Y. He, and G. P. Roberts. 1997. CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc. Natl. Acad. Sci. USA 94:11216-11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart, V., Y. Lu, and A. J. Darwin. 2002. Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 184:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svetlichny, V. A., T. G. Sokolova, M. Gerhardt, M. Ringpfeil, N. A. Kostrikina, and G. A. Zavarzin. 1991. Carboxydothermus hydrogenoformans gen. nov., sp. nov., a CO-utilizing thermophilic anaerobic bacterium from hydrothermal environments of Kunashir island. Syst. Appl. Microbiol. 14:254-260. [Google Scholar]
- 37.Svetlitchnyi, V., C. Peschel, G. Acker, and O. Meyer. 2001. Two membrane-associated NiFeS-Carbon monoxide dehydrogenases from the anaerobic carbon-monoxide-utilizing eubacterium Carboxydothermus hydrogenoformans. J. Bacteriol. 183:5134-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorsteinsson, M. V., R. L. Kerby, and G. P. Roberts. 2000. Altering the specificity of CooA, the carbon monoxide-sensing transcriptional activator: characterization of CooA variants that bind cyanide in the FeII form with high affinity. Biochemistry 39:8284-8290. [DOI] [PubMed] [Google Scholar]
- 39.Thorsteinsson, M. V., R. L. Kerby, M. Conrad, H. Youn, C. R. Staples, W. N. Lanzilotta, T. J. Poulos, J. Serate, and G. P. Roberts. 2000. Characterization of variants altered at the N-terminal proline, a novel heme-axial ligand in CooA, the CO-sensing transcriptional activator. J. Biol. Chem. 275:39332-39338. [DOI] [PubMed] [Google Scholar]
- 40.Uchida, T., H. Ishikawa, K. Ishimori, I. Morishima, H. Nakajima, S. Aono, Y. Mizutani, and T. Kitagawa. 2000. Identification of histidine 77 as the axial heme ligand of carbonmonoxy CooA by picosecond time-resolved resonance Raman spectroscopy. Biochemistry 39:12747-12752. [DOI] [PubMed] [Google Scholar]
- 41.Voordouw, G. 2002. Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 184:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waterman, M. R. 1978. Spectral characterization of human hemoglobin and its derivatives. Methods Enzymol. 52:456-463. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto, K., H. Ishikawa, S. Takahashi, K. Ishimori, I. Morishima, H. Nakajima, and S. Aono. 2001. Binding of CO at the Pro2 side is crucial for the activation of CO-sensing transcriptional activator CooA. 1H NMR spectroscopic studies. J. Biol. Chem. 276:11473-11476. [DOI] [PubMed] [Google Scholar]
- 44.Youn, H., R. L. Kerby, M. V. Thorsteinsson, R. W. Clark, J. N. Burstyn, and G. P. Roberts. 2002. Analysis of the L116K variant of CooA, the heme-containing CO sensor, suggests the presence of an unusual heme ligand resulting in novel activity. J. Biol. Chem. 277:33616-33623. [DOI] [PubMed] [Google Scholar]
- 45.Youn, H., R. L. Kerby, M. V. Thorsteinsson, M. Conrad, C. R. Staples, J. Serate, J. Beack, and G. P. Roberts. 2001. The heme pocket afforded by Gly117 is crucial for proper heme ligation and activity of CooA. J. Biol. Chem. 276:41603-41610. [DOI] [PubMed] [Google Scholar]
- 46.Youn, H., R. L. Kerby, and G. P. Roberts. 2003. The role of the hydrophobic distal heme pocket of CooA in ligand sensing and response. J. Biol. Chem. 278:2333-2340. [DOI] [PubMed] [Google Scholar]