Abstract
Bacillus methanolicus can efficiently utilize methanol as a sole carbon source and has an optimum growth temperature of 50°C. With the exception of mannitol, no sugars have been reported to support rapid growth of this organism, which is classified as a restrictive methylotroph. Here we describe the DNA sequence and characterization of a 19,167-bp circular plasmid, designated pBM19, isolated from B. methanolicus MGA3. Sequence analysis of pBM19 demonstrated the presence of the methanol dehydrogenase gene, mdh, which is crucial for methanol consumption in this bacterium. In addition, five genes (pfk, encoding phosphofructokinase; rpe, encoding ribulose-5-phosphate 3-epimerase; tkt, encoding transketolase; glpX, encoding fructose-1,6-bisphosphatase; and fba, encoding fructose-1,6-bisphosphate aldolase) with deduced roles in methanol assimilation via the ribulose monophosphate pathway are encoded by pBM19. A shuttle vector, pTB1.9, harboring the pBM19 minimal replicon (repB and ori) was constructed and used to transform MGA3. Analysis of the resulting recombinant strain demonstrated that it was cured of pBM19 and was not able to grow on methanol. A pTB1.9 derivative harboring the complete mdh gene could not restore growth on methanol when it was introduced into the pBM19-cured strain, suggesting that additional pBM19 genes are required for consumption of this carbon source. Screening of 13 thermotolerant B. methanolicus wild-type strains showed that they all harbor plasmids similar to pBM19, and this is the first report describing plasmid-linked methylotrophy in any microorganism. Our findings should have an effect on future genetic manipulations of this organism, and they contribute to a new understanding of the biology of methylotrophs.
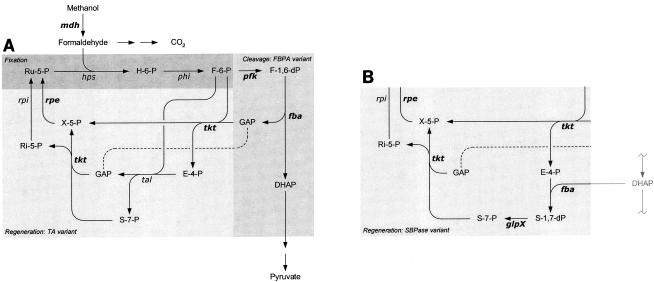
The methylotrophs constitute a diverse group of microorganisms that can utilize reduced one-carbon (C1) compounds, such as methanol, as sole carbon sources for growth (2, 3). The abundance, purity, and low price of methanol compared to sugars make methylotrophs interesting candidate organisms for production of amino acids, vitamins, cytochromes, coenzymes, single-cell proteins, and recombinant proteins (14, 21). The key intermediate for biological C1 fixation is formaldehyde, which may be assimilated via alternative biochemical routes. Bacteria that fix formaldehyde by the ribulose monophosphate (RuMP) pathway belong to three groups: gram-negative obligate methylotrophs, gram-positive facultative methylotrophs, and thermotolerant Bacillus species (14). The RuMP pathway (Fig. 1) has two unique enzymes, 3-hexulose-6-phosphate synthase (HPS) and 6-phospho-3-hexuloisomerase (PHI), which catalyze the two-step fixation phase of formaldehyde with ribulose-5-phosphate (Ru-5-P), which yields fructose 6-phosphate (F-6-P). RuMP pathway fixation operons, including the hps and phi genes, have been cloned from representatives of all three bacterial groups and characterized, and these operons have certain similarities in both gene organization and regulation (25, 31, 39). In addition to the fixation phase, a complete RuMP pathway includes the cleavage and rearrangement phases, which are thought to share enzymes with the pentose phosphate and glycolytic or Entner-Doudoroff pathways (Fig. 1). There are several RuMP pathways, and thermotolerant bacilli have been reported to use the fructose-1,6-bisphosphate aldolase (FBPA)-transaldolase (TA) variant (4, 16). In this pathway the F-6-P generated by the action of HPS and PHI is phosphorylated by 6-phosphofructokinase (PFK) before FBPA cleaves the resulting fructose 1,6-bisphosphate to form the triose phosphates glyceraldehyde 3-phosphate and dihydroxyacetone phosphate. Dihydroxyacetone phosphate enters the central pathway for synthesis of cell constituents, while glyceraldehyde 3-phosphate enters the final phase of the RuMP cycle, in which the formaldehyde acceptor Ru-5-P is regenerated. Important enzymes for the regeneration phase are transketolase (TKT), ribose-5-phosphate isomerase, TA, and Ru-5-P 3-epimerase (RPE). Another variant of the RuMP pathway, the FBPA-sedoheptulose-1,7-bisphosphatase (SBPase) variant, is found in certain facultative methylotrophic Bacillus species (16), in which the rearrangement phase includes SBPase instead of TA activity (Fig. 1).
FIG. 1.
Schematic representation of methanol assimilation via the RuMP pathway of thermotolerant methylotrophic B. methanolicus. (A) The RuMP pathway is divided into the fixation, cleavage, and regeneration phases, represented by the FBPA-TA variant. The dissimilatory pathway from formaldehyde to CO2 is indicated. (B) Regeneration phase of the FBPA-SBPase variant. Genes in boldface type are the genes identified on plasmid pBM19. mdh, MDH gene; hps, HPS gene; phi, PHI gene; pfk, PFK gene; fba, FBPA gene; tkt, TKT gene; tal, TA gene; glpX, fructose-1,6-bisphosphatase (and sedoheptulose-1,7-bisphosphatase) gene; rpi, ribose-5-phosphate isomerase gene; rpe, RPE gene; H-6-P, hexulose 6-phosphate; F-1,6-dP, fructose 1,6-bisphosphate; GAP, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; X-5-P, xylulose 5-phosphate; E-4-P, erythrose 4-phosphate; S-1,7-dP, sedoheptulose 1,7-bisphosphate; S-7-P, sedoheptulose 7-phosphate, Ri-5-P, ribose 5-phosphate.
Large plasmids, typically ranging in size from 50 to 200 kb, are commonly present in methylotrophic bacteria, including some methanotrophs (14, 22), yet our knowledge concerning the biological significance of any of these replicons in methylotrophs is limited. Methylotrophy does not correlate well with traditional methods of bacterial classification (2, 21), and it is tempting to speculate that methylotrophy might be plasmid linked. Hybridization experiments with several plasmids isolated from a range of methylotrophs revealed no homology, which argues against an episomal location of genes involved in methanol metabolism (14). To date, there is no evidence for a direct role of plasmid-borne genes in the C1 metabolism of any methylotrophic organism.
A number of thermotolerant and methylotrophic gram-positive Bacillus strains have been isolated from different locations, and based on physiological and 16S rRNA sequence analyses these organisms are collectively classified as Bacillus methanolicus (4, 33). B. methanolicus uses an NAD(P)-dependent methanol dehydrogenase (MDH) to oxidize methanol to formaldehyde, and in addition to entering the RuMP pathway, a linear branch for dissimilation of formaldehyde to CO2 was recently demonstrated (29). It was previously shown that this bacterium can secrete 55 g of glutamate per liter at 50°C by using methanol as a carbon source in fed-batch fermentation (11), and a homoserine dehydrogenase mutant (13A52-8A66) that secreted up to 35 g of l-lysine per liter at 50°C was described (18). In a previous study (13) it was found that the B. methanolicus NOA2 mutant 13A52 possesses a plasmid with an estimated size of 17 kb, and a plasmid of a similar size was later identified (N. Tsujimoto, H. Yasueda, and S. Sugimoto, 24 October 2000, Japanese patent application JP2000295988) in B. methanolicus PB1 (= NCIMB 13113). No sequence information is available for any of these DNA molecules, and the biological significance of them remains unknown. In this report we describe the DNA sequence and characterization of a 19,167-bp circular plasmid, designated pBM19, isolated from B. methanolicus MGA3. Remarkably, both mdh and five putative RuMP pathway genes were identified in this plasmid, and we found that pBM19 is essential for growth of this bacterium on methanol.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Recombinant Escherichia coli cells were grown at 37°C in Luria-Bertani medium (32) supplemented with ampicillin (100 μg/ml). For preparation of protoplasts B. methanolicus cells were grown at 50°C in SOB medium (which contains tryptone and yeast extract) (Difco) supplemented with 0.25 M sucrose, and the resulting recombinant cells were grown in the same medium supplemented with neomycin (25 μg/ml). For preparation of crude extracts B. methanolicus strains were grown in SOB containing 0.25 M sucrose, and methanol was added at a concentration of 150 mM 1 h prior to harvest to induce MDH expression. For all other purposes B. methanolicus cells were grown at 50°C in methanol minimal vitamin medium (MVcMY medium) containing 1 mM MgSO4, high-salt buffer, vitamins, 0.025% yeast extract (Difco), 200 mM methanol, and trace metals essentially as described previously (11). Mannitol medium was MVcMY medium without methanol supplemented with d-mannitol (10 g/liter; Sigma) as the sole carbon source. Bacterial growth was monitored by measuring the optical density at 600 nm.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Referenceb |
|---|---|---|
| B. methanolicus strains | ||
| MGA3 | Wild-type strain, MeOH+ | 33 |
| NOA2 | Wild-type strain, MeOH+ | 33 |
| PB1 NCIMB 13113 | Wild-type strain, MeOH+ | NCIMB |
| HEN9 | Wild-type strain, MeOH+ | R. Dillingham |
| TSL32 | Wild-type strain, MeOH+ | R. Dillingham |
| DFS2 | Wild-type strain, MeOH+ | R. Dillingham |
| CFS | Wild-type strain, MeOH+ | R. Dillingham |
| RCP | Wild-type strain, MeOH+ | R. Dillingham |
| SC6 | Wild-type strain, MeOH+ | R. Dillingham |
| NIWA | Wild-type strain, MeOH+ | R. Dillingham |
| BVD | Wild-type strain, MeOH+ | R. Dillingham |
| DGS | Wild-type strain, MeOH+ | R. Dillingham |
| JCP | Wild-type strain, MeOH+ | R. Dillingham |
| N2 | Wild-type strain, MeOH+ | R. Dillingham |
| MGA3C-A6 | MGA3 cured of pBM19, MeOH− | This study |
| E. coli strain | ||
| DH5α | General cloning host | BRL |
| Plasmids | ||
| pUC19 | General cloning vector, Apr | NEB |
| pGEM-3zf | General cloning vector, Apr | Promega |
| pGEM-11zf | General cloning vector, Apr | Promega |
| pLITMUS28 | General cloning vector, Apr | NEB |
| pDQ508 | E. coli-B. methanolicus shuttle plasmid, Neor Apr | 13 |
| pJB658 | Low-copy-number cloning vector, Apr | 9 |
| pBM19 | Endogenous B. methanolicus plasmid | This study |
| pTB1.9 | E. coli-B. methanolicus shuttle vector, Neor Apr | This study |
| pTB3.3H | 3,289-bp HindIII fragment from pBM19 cloned into pGEM-3zf | This study |
| pTB1.9mdh | pTB1.9 harboring mdh gene and 237 bp of upstream sequence | This study |
| pTB1.9mdhL | pTB1.9 harboring mdh gene and 1,125 bp of upstream sequencec | This study |
| pRMP1 | 910-bp PCR fragment encompassing the 3′ end of hps and the 5′ end of phi cloned into the XbaI/NcoI sites of pLITMUS28 | This study |
| pRMP2 | 1,056-bp PCR fragment encompassing the hps gene and upstream regions cloned into the HindIII/SalI sites of pGEM-11zf | This study |
| pRMP3 | 920-bp PCR fragment encompassing the 3′ end of hps and the phi gene cloned into the HindIII/SalI sites of pGEM-11zf | This study |
Apr, ampicillin resistance; Neor, neomycin resistance; MeOH+, grows on methanol; MeOH−, does not grow on methanol.
NCIMB, National Collection of Industrial and Marine Bacteria; BRL, Bethesda Research Laboratories; NEB, New England Biolabs.
See Materials and Methods.
DNA manipulations.
All recombinant E. coli procedures (plasmid preparation, restriction analysis, ligation, and transformation) were performed as described by Sambrook et al. (32). Transformation of B. methanolicus MGA3 strains was performed by using the protoplast method developed for B. methanolicus NOA2 mutant 13A52, essentially as described by Cue et al.(13). Plasmid and total DNA were prepared from B. methanolicus by using Qiagen Midi Prep and Dneasy tissue kits (Qiagen Gmbh, Hilden, Germany), respectively, according to the manufacturer's instructions. DNA fragments used for probes (Table 2) were isolated from agarose gels by using a Qiaex kit (Qiagen Gmbh), labeled by using a digoxigenin (DIG) kit from Boehringer Mannheim, and used for Southern hybridization analysis according to the manufacturer's instructions. DNA sequencing was performed by the Advanced Genetic Analysis Center (University of Minnesota, St. Paul), and the sequence data were analyzed by using the online programs Pfam (http://www.sanger.ac.uk/Software/Pfam/), BlastP (http://www.ncbi.nlm.nih.gov/BLAST/), Fasta (http://www.ebi.ac.uk/fasta33/), and Multialin (http://prodes.toulouse.inra.fr/multalin/multalin.html).
TABLE 2.
DNA probes used in this study
| Probe | Descriptiona |
|---|---|
| mdh-P | 817-bp SpeI fragment of pBM19 covering 774 bp of the 5′-terminal region of mdh and upstream sequences |
| pfk-P1 | 766-bp EcoRV-BglII fragment of pBM19 covering 625 bp of the 5′-terminal region of pfk and upstream sequences |
| tkt-P | 1,008-bp PstI fragment of pBM19 covering the central coding region of tkt |
| fba-P | 561-bp AvaI fragment of pBM19 covering the central coding region of fba |
| rpe-P | 526-bp ApoI fragment of pBM19 covering 494 bp of the 5′-terminal region of rpe and upstream sequences |
| glpX-P | 483-bp HindIII fragment of pBM19 covering the central region of glpX |
| pfk-P2 | 800-bp PCR fragment obtained by using primers pfk-F and pfk-R covering the central region of the pBM19 pfk gene |
| repB-P | 989-bp PCR fragment obtained by using primers repB-F and repB-R covering the central region of the pBM19 repB gene |
| tkt/fba-P | 780-bp PCR fragment obtained by using primers fba/tkt-F and fba/tkt-R covering the 3′ end of tkt and the 5′ end of fba |
| rmp-P | Corresponding to the 925-bp RMP insert of plasmid pRMP3 |
The DNA probes were labeled with DIG as described in Materials and Methods. The AT contents of the resulting DNA probes were between 58.9 and 63.9%.
Preparation of crude cell extracts and enzyme assays.
Crude extracts were prepared essentially as described by Arfman et al. (5). Late-exponential-phase cell cultures were harvested by centrifugation (Sorvall GSA rotor; 4.500 rpm, 10 min, 4°C) and washed twice in 50 mM potassium phosphate buffer (pH 7.5) containing 5 mM MgSO4. E. coli cells were disrupted by sonication (Branson Sonifer 250) as described previously (11). B. methanolicus cells were incubated at 37°C for 60 min in the presence of lysozyme (5 mg/ml) and 25 U of mutanolysin (10 U/ml; Sigma) before sonication. Cell debris was removed by centrifugation (13,000 × g, 30 min, 4°C), and the supernatants were collected as crude extracts. NAD(P)-dependent MDH activity was measured in the reverse direction by the formaldehyde reductase assay by monitoring the decrease in absorbance at 340 nm (Shimadzu UV-160A) due to formation of NAD+ at 50°C (5). All experiments were performed in duplicate. Polyacrylamide gels (15%, wt/vol) were stained with Coomassie brilliant blue.
Construction of pBM19-based shuttle vectors pTB1.9, pTB1.9mdh, and pTB1.9mdhL.
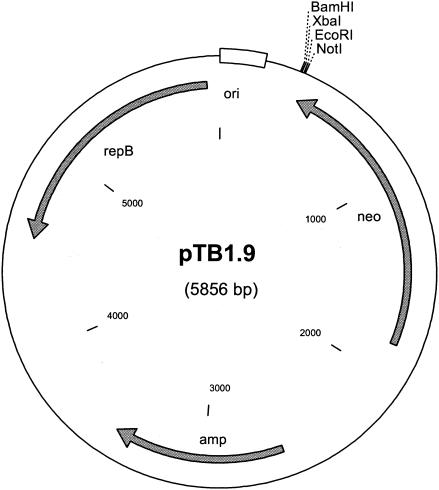
The 5,220-bp BamHI/PstI fragment of pBM19 was cloned into the corresponding sites of pUC19. The resulting plasmid was digested with AflIII, and the 4.2-kb fragment including the pUC19 vector backbone, as well as the repB-ori region of pBM19, was religated. The resulting construct was linearized with BamHI/SacI, and the cohesive SacI end was blunted with T4 DNA polymerase to obtain fragment 1. The 1.6-kb BamHI/PstI fragment of plasmid pDQ508 (encoding the Neor gene) was isolated, and the cohesive PstI end was blunted as described above to obtain fragment 2. Fragments 1 and 2 were ligated to obtain plasmid pTB1.9 (see Fig. 5). A DNA fragment including the mdh gene and its 237-bp upstream sequences was PCR amplified from pBM19 by using primers mdh-F and mdh-R (Table 3), and the resulting 1,580-bp PCR product was end digested with XbaI/SacI and cloned into the corresponding sites of plasmid pJB658. From the resulting construct the 1,575-bp XbaI/BamHI insert was cloned into the corresponding sites of pTB1.9, yielding vector pTB1.9mdh (Table 1). The 3,289-bp HindIII fragment of pBM19 was cloned into pGEM-3zf, yielding plasmid pTB3.3H. The 2,115-bp SacI/PstI fragment was isolated from this construct and used to replace the corresponding 1,192-bp SacI/PstI fragment of pTB1.9mdh, which yielded the MDH expression vector pTB1.9mdhL. All constructs were verified by DNA sequencing.
FIG. 5.
Physical map of shuttle vector pTB1.9. Shuttle plasmid pTB1.9 is a pUC19 derivative that carries the Neor gene and the ori- repB region from B. methanolicus MGA3 plasmid pBM19.
TABLE 3.
PCR primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| rmp-F | TTTTAAGCTTCCCCTGTCGCGCC (forward) |
| rmp-R4 | TTTTGTCGACAATGACAAGATCCGG (reverse) |
| rmp-F5 | TTTTGCATGCAACTTCAATTAGCTCTAG (forward) |
| rmp-R3 | TTTTGAATTCCAAGCCCTTTTTTCTCCATG (reverse) |
| rmp-F4 | TTTTAAGCTTAAGGGTGCTGTAGAAGAAGCG (forward) |
| rmp-R5 | TTTTGTCGACTACTCGAGATTGGCATGTC (reverse) |
| repB-F | CGTCAAGGCCTCATTTTTCTCC (forward) |
| repB-R | CTGGCTAGTAACATTCGGCCG (reverse) |
| pfk-F | TCCACGGCTTTTGCACCC (forward) |
| pfk-R | GGCGGGGATGCACCAGG (reverse) |
| rpe-F | GTTGAACGGGGCGGAGCCG (forward) |
| rpe-R | GGCTCCTGCTTCTACGCAA (reverse) |
| tkt/fba-F | TCAGCCATCGCCATCCC (forward) |
| tkt/fba-R | GACTGCTTTGCACCTGCG (reverse) |
| glpX-F | GAGGAGCAAGTCTTCCTTGG (forward) |
| glpX-R | GGGGAAATGGACGAAGCTCC (reverse) |
| mdh-F | TTTTCTGCAGCCCTTCCACCTTAACC (forward) |
| mdh-R | TTTTTCTAGACCTATGGCGGGATTCG (reverse) |
| tba42 | GTGGAAGATGGGAACC (forward) |
| tba15 | CTCACCAAGTAGGTGG (reverse) |
The underlined nucleotides are restriction sites used for simplified cloning of PCR products (see Materials and Methods and Table 1 for details).
PCR analysis of B. methanolicus strains for pBM19 DNA.
For analysis of MGA3 strains for chromosomal copies of pBM19 genes, total DNA were isolated and used as templates for PCR by using the following oligonucleotide primer pairs: repB-F plus repB-R, pfk-F plus pfk-R, rpe-F plus rpe-R, tkt/fba-F plus tkt/fba-R, glpX-F plus glpX-R, and mdh-F plus mdh-R (Table 3). These primer pairs correspond to the amplified regions of the repB, pfk, rpe, tkt and fba, glpX, and mdh genes of pBM19, respectively. To screen B. methanolicus wild-type strains for pBM19-like plasmids, both plasmid and total DNA were isolated and used as templates for PCR performed with primers tba15 and tba42 (Table 3). In all cases the 100-μl PCR mixture contained 0.1 to 0.5 μg of DNA, 50 pmol of the forward primer, 50 pmol of the reverse primer, 350 μmol of each deoxynucleoside triphosphate, 1× PCR buffer (GIBCO), and 2 U of the Taq DNA polymerase from the same system. The PCR was performed with a Perkin-Elmer GeneAmp PCR system 2400 by using the following program: one cycle of denaturation 94°C for 3 min, 30 cycles of denaturation at 94°C for 60 s, annealing at 55°C for 60 s, and synthesis at 68°C for 2 min, and one cycle of synthesis at 68°C for 7 min. The DNA fragments obtained were verified by DNA gel electrophoresis and partial DNA sequencing.
PCR cloning of the MGA3 RuMP pathway fixation operon.
Based on the previously published DNA sequence of the RuMP pathway fixation operon of Bacillus brevis S1 (39), the PCR primer pairs rmp-F plus rmp-R4, rmp-F5 plus rmp-R3, and rmp-F4 plus rmp-R5 were designed (Table 3). By using these primers, three DNA fragments were PCR amplified from MGA3 total DNA and individually cloned into pLITMUS28 or pGEM-11zf, which yielded plasmids pRMP1, pRMP2, and pRMP3, respectively (Table 1). Both strands of the cloned inserts of these plasmids were sequenced.
Estimation of plasmid copy number.
One DNA probe of chromosomal origin (rmp-P) and three DNA probes of pBM19 origin (repB-P, fba/tkt-P, and pfk-P2) were designed (Table 2). Coupled amplification and DIG labeling of these probes were performed by using a PCR-DIG probe synthesis kit (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's instructions. The DNA concentrations of the resulting probes were analyzed by gel electrophoresis and standardized. Total DNA was isolated from MGA3 cells grown in MVcMY medium at 50°C to the late exponential phase and was digested with SacI. Various dilutions of digested DNA were separated by gel electrophoresis and used for three independent two-probe Southern hybridizations with one chromosome-derived probe and one pBM19-derived probe. The hybridization bands obtained with both probes were scanned, and three-dimensional graphs of the intensity profiles of each band were generated by digital image analysis. Each graph was integrated, which gave the corresponding intensity volume. The pBM19 copy number was calculated by comparing the intensity volumes of chromosomal and plasmid bands at different dilutions.
Nucleotide sequence accession numbers.
The DNA sequences of plasmid pBM19 and the RuMP pathway fixation operon reported in this paper have been deposited in the GenBank nucleotide sequence database under accession numbers AY386314 and AY386313, respectively.
RESULTS
DNA sequencing of the B. methanolicus MGA3 plasmid pBM19.
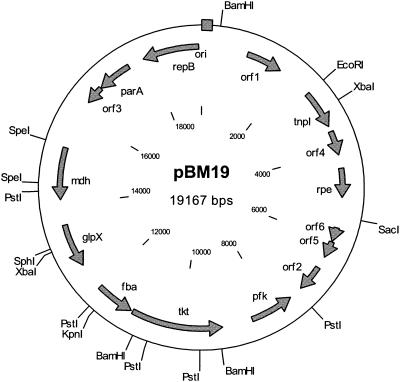
By analyzing B. methanolicus MGA3 DNA we isolated a plasmid with an estimated size of 19 kb, designated pBM19. Overlapping fragments representing the entire pBM19 DNA were cloned in E. coli, and both strands of the cloned inserts were sequenced by the primer extension method. PCR analysis was used when appropriate to verify the overlap between assumed neighboring inserts. Using this strategy, we determined the complete pBM19 DNA sequence consisting of 19,167 bp (Fig. 2). The overall G+C content was 36.7%, and computer-assisted analysis of the DNA sequences led to identification of the putative genes shown in Fig. 2 and Table 4. Interestingly, the pBM19 plasmid is composed of two regions encoding genes arranged in opposite orientations, suggesting that the present form of this plasmid might have been generated by fusion between separate DNA molecules.
FIG. 2.
Physical map of B. methanolicus plasmid pBM19. Genes and open reading frames are indicated by arrows, and the putative origin of replication (ori) is indicated by a box. All the genes and open reading frames are described in Table 4.
TABLE 4.
Putative genes and open reading frames identified in the pBM19 plasmid
| Gene or open reading frame | Region of pBM19a | No. of codons | Start codon | Ribosome binding site | Deduced productb | Putative function |
|---|---|---|---|---|---|---|
| orf1 | 1003-1801 | 266 | ATG | AAGGAGA | Hypothetical protein | Unknown |
| tnp1 | 2564-3416 | 284 | ATG | AAGGAG | TNP1 | Transposition |
| orf4 | 3527-4070 | 181 | ATG | GGAGGG | Hypothetical protein | Unknown |
| rpe | 4355-4997 | 214 | ATG | GAGG | RPE | RuMP pathway |
| orf6 | 5659-5974 | 105 | ATG | AGAAA | Transposase | Transposition |
| orf5 | 5983-6355 | 124 | ATG | GAAG | Transposase | Transposition |
| orf2 | 6612-7206 | 198 | ATG | AGGA | Hypothetical protein | Unknown |
| pfk | 7491-8457 (C) | 322 | ATG | GGAGGA | PFK | RuMP pathway |
| tkt | 9113-11114 (C) | 667 | GTG | GGGAGGG | TKT | RuMP pathway |
| fba | 11147-11999 (C) | 284 | ATG | AAGGAG | Class II FBPA | RuMP pathway |
| glpX | 12595-13555 (C) | 320 | ATG | AGGAGG | Class II FBPase | RuMP pathway |
| mdh | 14105-15251 (C) | 382 | ATG | AGGAGG | MDH | Methanol oxidation |
| orf3 | 16340-16754 (C) | 138 | ATG | AGGAGG | Hypothetical protein | Unknown |
| parA | 16756-17524 (C) | 256 | ATG | GGAGGA | ParA | Partition protein |
| repB | 17868-19104 (C) | 412 | GTG | AGGAGGG | RepB | Replication |
C in parentheses refers to the complementary strand.
All deduced products are based on similarity with sequences in the databases.
(i) Key gene for methanol oxidation, methanol dehydrogenase, is encoded by pBM19.
Remarkably, the mdh gene, including its ribosome binding site coding region and proposed promoter region, which exhibited 96.8% overall identity to the active mdh gene in B. methanolicus C1 (= NCIMB 13114) (15), was localized on pBM19. The deduced primary sequences of the mdh gene products (381 and 382 amino acids [aa]) of B. methanolicus MGA3 and C1 are 97.9% identical. MDH, which belongs to the family III group of NAD(P)-dependent alcohol dehydrogenases (6), catalyzes the oxidation of methanol to formaldehyde (Fig. 1) and thus plays a key role in methylotrophic growth of B. methanolicus.
(ii) Genes with deduced roles in methanol assimilation via the RuMP pathway.
Five putative genes with assigned roles in methanol assimilation via the RuMP pathway (glpX, fba, tkt, pfk, and rpe) were identified on pBM19 (Fig. 1 and Table 4). Except for rpe, these genes and mdh are arranged in the same orientation and occupy one continuous region of the pBM19 plasmid (Fig. 2). The fba and tkt coding sequences are separated by 12 nucleotides, suggesting that they may be translationally coupled. The remaining three genes are most probably transcribed from individual promoters.
The deduced glpX gene product is a 321-aa protein that exhibits the highest overall level of identity (55%) to the Bacillus halodurans class II fructose-1,6-bisphosphatase (FBPase) protein encoded by glpX (accession number BAB07502.1). Bacterial FBPase are bifunctional enzymes with both FBPase and SBPase activities. The class II variants typically have high SBPase-to-FBPase ratios (34) and play a role in one variant of the rearrangement phase of the RuMP pathway.
The deduced fba gene product is a 285-aa protein which is 79% identical to the B. halodurans class II FBPA protein (accession number NP_244653). Such aldolases are typically found in many bacterial autotrophs, including the chemoautotroph Xanthobacter flavus, in which its expression is induced during growth on methanol by the ribulose bisphosphate pathway (35). A number of catalytically important residues in the E. coli class II FBPA protein have been identified (28, 40), and sequence comparisons confirmed that these residues are conserved in the deduced B. methanolicus fba gene product.
The tkt gene encodes a 667-aa deduced protein whose primary sequence is 75% identical to the sequence of the TKT protein of B. halodurans (accession number Q9KAD7). TKT activity is needed in the RuMP pathway rearrangement phase (Fig. 1), and in X. flavus TKT activity is induced sixfold upon growth on methanol (35). A number of residues found to be critical for catalytic activity in the Saccharomyces cerevisiae TKT protein (24, 27, 37) are conserved in the pBM19-encoded TKT protein.
The deduced pfk gene product is a 322-aa protein that exhibits 62% overall identity with the PFK protein of B. halodurans (accession number Q9K843). In addition, it is 51% identical to the extensively characterized ATP-dependent PFK enzyme of E. coli (7). The active site motif TIDND, as well as the catalytically important residues R72, D103, R162, and R252, are 100% conserved in these two proteins, indicating that the putative pfk gene of pBM19 encodes an ATP-dependent PFK protein presumably involved in the RuMP pathway cleavage phase in B. methanolicus (Fig. 1). Interestingly, in the methylotrophic bacterium Amycolatopsis methanolica the ATP-dependent PFK protein is specifically induced upon growth on methanol (1).
The rpe gene on pBM19 is separated from the other putative metabolic genes (Fig. 2). The deduced 214-aa rpe gene product is 85% identical to the RPE enzyme of Bacillus anthracis (accession number NP_846240.1). RPE catalyzes the interconversion of R-5-P and xylulose-5-phosphate and thus plays a role in the rearrangement phase of the RuMP pathway (Fig. 1). Certain motifs and residues important for RPE activity have been reported (12), and these features, including the active site motif DGG, are conserved in the deduced B. methanolicus rpe gene product.
(iii) Genetic elements for plasmid replication and maintenance.
The repB gene encodes a 412-aa putative protein, and the 200-aa N-terminal sequence of this gene product is 36% identical to the replication initiator protein RepB (accession number CAA71788) encoded by the Pseudomonas alcaligenes plasmid pECB2. Immediately upstream of the repB gene is a distinct region with numerous direct repeats, which may represent the pBM19 origin of replication (ori). Another gene (parA) with a proposed function related to plasmid replication and maintenance is located 344 bp downstream of repB. The primary sequence of the deduced parA gene product (256 aa) exhibits 37% overall identity to the chromosome partition protein ParA (accession number NP_624291) from Thermoanaerobacter tengcongensis. ParA is an ATPase involved in active partitioning of bacterial chromosomes and plasmids during cell division (8). Together, repB, parA, and ori probably constitute genetic elements for pBM19 replication and segregational stability.
(iv) Mobile element-related genes.
The deduced tnpI gene product is a 284-aa polypeptide exhibiting 51% overall identity to a site-specific recombinase (TnpI) encoded by the Bacillus thuringiensis plasmid pGI2 (23). TnpI belongs to the phage integrase family of resolvases, and these proteins mediate transposition processes by catalyzing the site-specific recombination of the cointegrated replicon, yielding the final transposition product. Two more genes (orf5 and orf6) with proposed functions related to the tnpI function are located downstream of the rpe gene (Fig. 2). The deduced gene products of orf5 and orf6 are 37 and 79% identical to transposase proteins of Helicobacter pylori and thermophilic bacterium PS3 (26), respectively. Four more open reading frames (orf1 to orf4) were identified, and the deduced gene products exhibited no significant similarity with proteins in the databases.
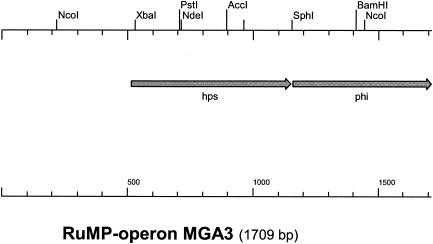
Cloning and sequencing of a chromosomal RuMP pathway fixation operon encoding HPS and PHI from MGA3.
Recently, the RuMP pathway fixation operons including HPS (hps) and PHI (phi) genes from B. subtilis (38) and thermotolerant B. brevis S1 (39) were described. These genes are critical for the fixation phase of the RuMP pathway (Fig. 1), and based on the previously published DNA sequences we designed PCR primers for amplification of the corresponding operon from B. methanolicus MGA3. Three PCR fragments of the expected lengths were obtained and cloned to obtain plasmids pRMP1, pRMP2, and pRMP3 (Table 1). The inserts of these plasmids were sequenced and were found to represent a putative RuMP pathway fixation operon, as shown in Fig. 3. Two genes, hps and phi, were identified, and the overall level of identity between the MGA3 and B. brevis S1 operons at the DNA level was 96%, suggesting that the cloned operon represented the active RuMP pathway fixation genes of B. methanolicus MGA3. The high level of DNA sequence identity between these two operons is in agreement with previous reports which suggested that B. brevis S1 should be classified as a B. methanolicus strain (4).
FIG. 3.
Putative RuMP pathway fixation operon of B. methanolicus MGA3. The hps and phi genes are presumably transcribed from a single promoter located upstream of hps.
Analysis of B. methanolicus MGA3 for chromosomal copies of pBM19 genes.
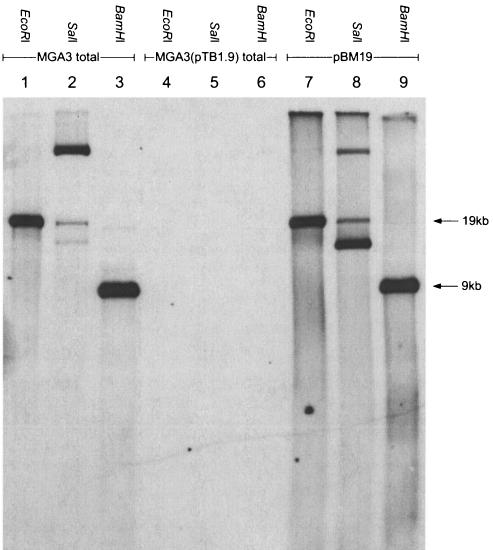
After mdh and five putative RuMP pathway genes were found to be located on plasmid pBM19, it was of interest to investigate whether chromosomal copies of these genes are present in B. methanolicus MGA3. In Methylomonas aminofaciens a copy of the three-member RuMP pathway gene cluster was discovered (31), and two copies of the cbb operon are present in some autotrophs; one of these copies is located on the chromosome, and one is plasmid borne (10, 17). Therefore, six different DNA probes representing individual pBM19 genes (mdh-P, rpe-P, tkt-P, glpX-P, pfk-P1, and fba-P) (Table 2) were prepared. Both plasmid DNA and total DNA isolated from MGA3 were digested with restriction enzymes having no (SalI), one (EcoRI), and three (BamHI) recognition sites within the pBM19 plasmid, separated by gel electrophoresis, and used for Southern hybridization experiments. The results of these experiments are shown in Fig. 4. The signal patterns obtained for plasmid DNA and total DNA were similar in all cases, indicating that none of these genes exists as a chromosomal copy in B. methanolicus MGA3. However, we cannot rule out the possibility that isogenes with different sequences that do not hybridize to the pBM19-derived DNA probes are present in this organism.
FIG. 4.
Southern analysis of B. methanolicus DNA by using pBM19-derived DNA as probes. DNA were digested with restriction enzymes EcoRI, SalI, and BamHI, which had one, no, and three recognition sites in pBM19, respectively (Fig. 2). The diagram shows the results obtained when probe pfk-p was used. With all the probes used (see text and Table 2) no additional bands were detected in the lanes loaded with total DNA compared to the lanes loaded with only plasmid DNA.
Curing of pBM19 by using the shuttle vector pTB1.9.
A shuttle vector, pTB1.9, harboring a 1.9-kb DNA fragment of pBM19 covering the putative ori-repB region, was constructed (Fig. 5). By using the protoplast method previously developed for B. methanolicus NOA2 mutant 13A52 (13), pTB1.9 was transformed into MGA3. Gel electrophoresis of plasmid DNA isolated from MGA3(pTB1.9) confirmed the presence of pTB1.9, whereas pBM19 was not detected. To analyze whether pBM19 was lost or chromosomally integrated, total DNA from MGA3(pTB1.9) was isolated and analyzed in a series of Southern hybridization experiments by using six different pBM19-derived DNA probes (mdh-P, rpe-P, fba-P, glpX-P, tkt-P, and pfk-P) (Table 2), as described above. No hybridization signals were obtained with any of the probes tested, suggesting that the recombinant strain was cured of pBM19 (Fig. 4). Next, a series of PCRs for amplification of regions representing six pBM19 genes (mdh, rpe, tkt, fba, pfk, and glpX) were performed by using MGA3(pTB1.9) total DNA as the template. No bands appeared when we analyzed the resulting PCR products by gel electrophoresis, which is in agreement with the Southern hybridization results (see above). As a control, total DNA from wild-type strain MGA3 was used as a template in similar experiments, and in this case all the desired DNA fragments were amplified (data not shown). Three additional MGA3(pTB1.9) transformants were analyzed in a similar manner, and in all cases the results obtained were the same. Thus, we concluded that MGA3(pTB1.9) was cured of pBM19, probably due to plasmid incompatibility.
MGA3 strains cured of pBM19 cannot grow on methanol but can grow on mannitol.
The pBM19-cured strain MGA3(pTB1.9) was used to investigate the effect of pBM19 on the ability to utilize methanol. Mid-log-phase cultures of recombinant and wild-type MGA3 cells grown at 50°C in SOB medium containing 0.25% sucrose were diluted 100-fold in 50°C prewarmed MVcMY medium for continued growth. Whereas the wild-type grew well under these conditions, the recombinant strain was unable to grow, supporting the hypothesis that B. methanolicus is dependent on pBM19 for methanol utilization. We next compared these two strains in similar experiments in which methanol was replaced with mannitol as the sole carbon source. This sugar was presumably taken up by the cells as F-6-P (Fig. 1), similar to what occurs in other Bacillus species (36). Both strains grew well on this sugar, suggesting that pBM19 genes are not critical for mannitol consumption in B. methanolicus. To rule out the possibility that there were any unwarranted effects caused by the presence of vector pTB1.9, we cured the recombinant strain of this plasmid. MGA3(pTB1.9) cells were cultivated at 50°C in SOB medium containing 0.25% sucrose for approximately 80 generations and plated on solid SOB medium containing 0.25% sucrose without antibiotic selection. Using replica plating, we identified neomycin-sensitive colonies, and one strain, designated MGA3C-A6, was isolated and characterized. Analysis by gel electrophoresis confirmed that pTB1.9 was not present in MGA3C-A6. As expected, this strain did not grow on methanol, whereas it grew well on mannitol. To completely exclude the possibility that the apparently cured strain was a contaminant, total DNA of MGA3C-A6 was isolated and used as template for PCR amplification of the RuMP pathway fixation operon (Fig. 3) by using PCR primers rmp-F and rmp-R3 (Table 3). One strong band of the expected size appeared upon analysis of the PCR product by gel electrophoresis, and partial DNA sequencing of the purified fragment confirmed that it represented the expected region (data not shown).
Introduction of the mdh gene is not sufficient to restore methanol growth of the pBM19-cured strain MGA3C-A6.
Besides mdh, it was unclear whether other pBM19 genes are involved in methanol assimilation in B. methanolicus. To investigate this, the mdh gene was introduced into MGA3C-A6 to test whether this gene is sufficient to restore the ability of this mutant strain to utilize methanol. The mdh gene and 237 bp of upstream sequence covering the deduced promoter region (15) was PCR amplified from pBM19 and cloned into the shuttle plasmid pTB1.9 to obtain plasmid pTB1.9mdh (Table 1). Surprisingly, neither enzyme assays nor sodium dodecyl sulfate-polyacrylamide gel electrophoresis of crude extracts prepared from E. coli DH5α harboring pTB1.9mdh revealed any MDH protein. We hypothesized that the endogenous mdh promoter region in pTB1.9mdh is not complete, and the analogous vector pTB1.9mdhL was constructed. This plasmid harbored the mdh gene and 1,125 bp of upstream sequence, including the entire intergenic region between mdh and orf3 (Fig. 2). When pTB1.9mdhL was used, MDH activity was expressed in E. coli, similar to findings reported previously (15). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of crude extracts prepared from E. coli cells harboring this plasmid produced a strong band at a position corresponding to the predicted MDH mass that was not present in crude extracts prepared from cells harboring pTB1.9mdh (data not shown). Plasmid pTB1.9mdhL was therefore used to transform strain MGA3C-A6, and the resulting recombinant strain was tested for growth on methanol. The strain could still not grow on this carbon source. As reported by other workers (15), MDH activity was virtually absent from B. methanolicus cells grown in complex media without methanol. This was also found to be the case for MGA3 strains (data not shown). Therefore, as a genetic control, plasmid pTB1.9mdhL was isolated from recombinant MGA3C-A6 and transformed into E. coli, and the resulting recombinant strain was shown to express a high level of MDH activity. We believe that these results indicate that pBM19 genes besides mdh are required for methanol consumption under the conditions tested.
Estimation of the pBM19 copy number in B. methanolicus MGA3.
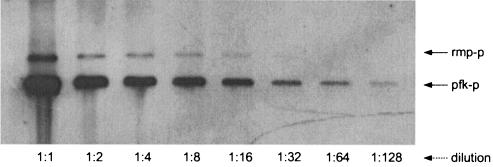
Genes encoded on plasmids are often present at elevated doses compared to the doses of chromosomal genes, and this could potentially have biological significance. After identification of putative RuMP pathway genes having both chromosomal and plasmid origins, it was therefore of interest to determine the pBM19 copy number in MGA3. Two-probe Southern hybridization experiments, in which one probe had a chromosomal origin and one probe had a pBM19 origin, were used to estimate the pBM19 copy number in B. methanolicus MGA3 grown on methanol. Care was taken to design all probes so that their lengths were similar (0.78 to 0.99 kb) and their G+C contents were similar (40.6 to 41.4%), and the concentrations of the probes were standardized. Moreover, by using SacI-digested total DNA we managed to ensure that the sizes of target DNA fragments for both the chromosome- and plasmid-directed probes were similar. A total of three independent Southern experiments were performed by using chromosomal probe rmp-P together with pBM19-derived probes repB-P, pfk-P2, and tkt/fba-P. The signal intensities in all experiments were similar (Fig. 6). The intensity volume of each band was calculated by digital image analysis. Plots of both chromosomal and plasmid intensity volumes versus DNA concentration showed that there was a good correlation (data not shown). The dilution ratio that gave the same intensity volumes for chromosomal and plasmid bands could then be calculated. By using this method, the pBM19 copy number in MGA3 was estimated to be 10 to 16 copies per chromosome.
FIG. 6.
Estimation of the pBM19 copy number. Total DNA isolated from B. methanolicus MGA3 was digested with SacI, and a series of dilutions of digested material were used for two-probe Southern hybridization analysis. The gel shows the results obtained with the DNA probes rmp-P (chromosomal target) and pfk-P (pBM19 target). The hybridization signals were scanned and used to estimate the pBM19 copy number.
Screening of B. methanolicus strains for pBM19-like plasmids.
We had access to 11 different thermotolerant and methylotrophic B. methanolicus wild-type strains (designated DFS2, HEN9, TSL32, CFS, RCP, SC6, NIWA, BVD, DGS, JCP, and N2) (Table 1) isolated at the University of Minnesota. Previous preliminary characterizations of these strains at the University of Minnesota indicated that they exhibit considerable physiological variation (Rick Dillingham, unpublished results). Thus, it was of interest to see whether pBM19-like plasmids are present in these wild-type strains. In addition, B. methanolicus PB1 (= NCIMB 13113), isolated elsewhere, as well as the previously characterized strain NOA2 (33), were included in this analysis. Restriction analysis of plasmid DNA isolated from all the strains indicated that they all harbor pBM19-like plasmids. However, the restriction patterns obtained were not identical. Therefore, each plasmid was used as a template for PCR analysis with primers tba42 and tba15 (see Materials and Methods). These two primers amplify a 2,796-bp DNA fragment including the 3′-terminal and 5′-terminal parts of the mdh and fba structural genes, respectively, as well as the entire glpX structural gene, of pBM19. Gel electrophoresis analysis of the PCR products demonstrated that there was a single strong band at the same predicted size in all cases (data not shown). Partial DNA sequencing showed that the PCR fragments were similar, demonstrating that pBM19-like plasmids or parts of such plasmids are present in all of the B. methanolicus strains tested. These results indicate that plasmid-dependent methylotrophy is not restricted to strain MGA3 and is widespread in nature.
DISCUSSION
Until recently it was assumed that HPS and PHI activities are restricted to methylotrophic organisms, and the presence of these activities was regarded as indicative of such organisms (16). However, the identification of such enzymes in nonmethylotrophic bacteria suggests that the RuMP pathway is common in prokaryotes (30, 38) and presumably is used for detoxification of formaldehyde. Three genes, ywjI, fbaA, and ywjH, encoding class II FBPase, class II FBPA, and TA, respectively, are clustered in the B. subtilis chromosome (accession numbers Z99104 to Z99124). In the obligate methylotroph M. aminofaciens 77a (31) and the facultative methylotroph Mycobacterium gastri MB19 (25) the gene for TA was found in the same cluster as the genes for hexulose phosphate synthase and hexulose phosphate isomerase.
The remarkable finding that mdh and five putative RuMP pathway genes (pfk, rpe, tke, glpX, and fba) are present on a B. methanolicus plasmid provides crucial information regarding the genetic basis for methanol metabolism in this organism. Chromosomal copies were not detected for any of these genes in B. methanolicus MGA3, and the complete inability to grow on methanol of pBM19-cured strain MGA3C-A6 confirms that this plasmid is essential for methanol consumption under the conditions tested. In B. methanolicus C1 (= NCIMB 13114) it has been unequivocally demonstrated that methanol oxidation is catalyzed by the NAD(P)-dependent MDH encoded by mdh (4, 15). Despite the extensive biochemical characterization of this protein (15, 19, 20), the mdh gene has never been reported to originate on a plasmid. The failure to complement growth of MGA3C-A6 on methanol by introducing the MDH expression plasmid pTB1.9mdhL implies that additional pBM19 genes are required for methanol consumption. The ability of MGA3C-A6 to grow rapidly on mannitol suggests that B. methanolicus has isoenzymes of both PFK and FBPA to metabolize F-6-P (Fig. 1), and it also implies that MGA3C-A6 and MGA3 are similar in other respects.
The gene encoding the MDH activator protein ACT was not found on pBM19, yet expression of this protein and expression of MDH in B. methanolicus have been reported to be regulated coordinately (19, 20). This suggests that methanol oxidation in B. methanolicus may be governed by the concerted action of both chromosomally and plasmid-borne genes. The latter notion is supported by our finding that the formaldehyde fixation genes hps and phi are present on the B. methanolicus chromosome. Also not present on pBM19 are genes for the RuMP pathway enzymes TA and ribose-5-phosphate isomerase (Fig. 1). Although low levels of activity of both proteins have been detected in crude extracts of this bacterium (4), it is not known whether the proteins are crucial for methanol consumption. This fact, together with the presence of the glpX gene on pBM19, suggests that the SBPase variant, and not the TA variant, is the relevant RuMP pathway in this organism (Fig. 1). The advantage of possessing certain RuMP pathway genes on a multicopy plasmid is unknown. However, based on the present results it is tempting to speculate that methylotrophy may be a transferable metabolic property in nature.
Previous reports have shown that B. methanolicus is sensitive to rapid changes in methanol concentrations, presumably due to toxic intracellular formaldehyde accumulation (29). Although HPS synthesis has been reported to be induced by formaldehyde, there is noncoordinate expression of this enzyme and MDH in B. methanolicus, and cells may have high MDH levels and low HPS levels (5) during growth on methanol. Based on the present findings it is tempting to speculate that this feature is partially caused by the multiple copies of the mdh gene compared to the chromosomal hps gene (and the phi gene). It is possible that engineering of a pBM19 derivative that includes these two genes may result in improved formaldehyde tolerance, as well as higher methanol assimilation rates if the derivative is introduced into B. methanolicus strains.
Acknowledgments
This work was supported by a grant from the Research Council of Norway.
We are grateful to Rick Dillingham for isolation of B. methanolicus strains, and we thank Trine Aakvik for helping with the cloning and sequencing of the RuMP pathway fixation operon. Also, we thank Sergey B. Zotchev for carefully reading the manuscript and Arne Strøm and Kjell Josefsen for encouraging discussions during the course of this work.
REFERENCES
- 1.Alves, A. M. C. R., G. J. W. Euverink, H. Santos, and L. Dijkhuizen. 2001. Different physiological roles of ATP- and PPi-dependent phosphofructokinase isoenzymes in the methylotrophic actinomycete Amycolatopsis methanolica. J. Bacteriol. 183:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, Inc. (London), Ltd., London, United Kingdom.
- 3.Anthony, C. 1991. Assimilation of carbon by methylotrophs, p. 70-109. In I. Goldberg and J. S. Rokem (ed.), Biology of methylotrophs. Butterworth-Heinemann, Boston, Mass.
- 4.Arfman, N., E. M. Watling, W., Clement, R. J. van Oosterwijk, G. E. De Vries, W. Harder, M. M. Attwood, and L. Dijkhuizen. 1989. Methanol metabolism in thermotolerant methylotrophic Bacillus strains involving a novel catabolic NAD-dependent methanol dehydrogenase as a key enzyme. Arch. Microbiol. 152:280-288. [DOI] [PubMed] [Google Scholar]
- 5.Arfman, N., L. Dijkhuizen, G. Kirchof, W. Ludwig, K.-H. Schleifer, E. S. Bulygina, K. M. Chumakov, N. I. Govorhukhina, Y. A. Trotsenko, D. White, and R. J. Sharp. 1992. Bacillus methanolicus sp. nov., a new species of thermotolerant, methanol-utilizing, endospore-forming bacteria. Int. J. Syst. Bacteriol. 42:439-445. [DOI] [PubMed] [Google Scholar]
- 6.Arfman, N., H. J. Hektor, L. V. Bystrykh, N. I. Govorukhina, L. Dijkhuizen, and J. Frank. 1997. Properties of an NAD(H)-containing methanol dehydrogenase and its activator protein from Bacillus methanolicus. Eur. J. Biochem. 244:426-433. [DOI] [PubMed] [Google Scholar]
- 7.Berger, S. A., and P. R. Evans. 1992. Site-directed mutagenesis identifies catalytic residues in the active site of Escherichia coli phosphofructokinase. Biochemistry 31:9237-9242. [DOI] [PubMed] [Google Scholar]
- 8.Bignell, C., and C. M. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1-34. [DOI] [PubMed] [Google Scholar]
- 9.Blatny, J. B., T. Brautaset, H. C. Winther-Larsen, P. Karunakarunakaran, and S. Valla. 1997. Improved broad-host-range vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 38:35-51. [DOI] [PubMed] [Google Scholar]
- 10.Bowien, B., and B. Kusian. 2002. Genetics and control of CO2 assimilation in the chemoautotroph Ralstonia eutropha. Arch. Microbiol. 178:85-93. [DOI] [PubMed] [Google Scholar]
- 11.Brautaset, T., M. D. Williams, R. D. Dillingham, C. Kaufmann, A. Bennaars, E. Crabbe, and M. C. Flickinger. 2003. The role of Bacillus methanolicus citrate synthase II gene, citY, in regulating the secretion of glutamate in lysine-secreting mutants. Appl. Environ. Microbiol. 69:3986-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, Y. R., F. W. Larimer, E. H. Serpersu, and F. C. Hartman. 1999. Identification of a catalytic aspartyl residue of d-ribulose 5-phosphate 3-epimerase by site-directed mutagenesis. J. Biol. Chem. 274:2132-2139. [DOI] [PubMed] [Google Scholar]
- 13.Cue, D., H. Lam, R. L. Dillingham, R. S. Hanson, and M. C. Flickinger. 1997. Genetic manipulation of Bacillus methanolicus, a gram-positive thermotolerant methylotroph. Appl. Environ. Microbiol. 63:1406-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Vries, G. E., U. Kües, and U. Stahl. 1990. Physiology and genetics of methylotrophic bacteria. FEMS Microbiol. Rev. 6:57-102. [DOI] [PubMed] [Google Scholar]
- 15.De Vries, G. E., N. Arfman, P. Terpstra, and L. Dijkhuizen. 1992. Cloning, expression, and sequence analysis of the Bacillus methanolicus C1 methanol dehydrogenase gene. J. Bacteriol. 174:5346-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dijkhuizen, L., P. R. Levering, and G. E. De Vries. 1992. The physiology and biochemistry of aerobic methanol-utilizing gram negative and gram positive bacteria, p. 149-181. In J. C. Murrell and H. Dalton (ed.), Methane and methanol utilizers. Plenum Press, New York, N.Y.
- 17.Gibson, J. L., J. M. Dubbs, and F. R. Tabita. 2002. Differential expression of the CO2 operons of Rhodobacter sphaeroides by the Prr/Reg two-component system during chemoautotrophic growth. J. Bacteriol. 184:6654-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson, R. S., R. L. Dillingham, P. Olson, G. H. Lee, D. Cue, F. J. Schendel, C. Bremmon, and M. C. Flickinger. 1996. Production of l-lysine and some other amino acids by mutants of B. methanolicus, p. 227-234. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 19.Hektor, H. J., H. Kloosterman, and L. Dijkhuizen. 2002. Identification of a magnesium-dependent NAD(P)(H) binding domain in the nicoprotein methanol dehydrogenase from Bacillus methanolicus. J. Biol. Chem. 277:46966-46973. [DOI] [PubMed] [Google Scholar]
- 20.Kloosterman, H., J. W. Vrijbloed, and L. Dijkhuizen. 2002. Molecular, biochemical, and functional characterization of a nudix hydrolase protein that stimulates the activity of a nicotinoprotein alcohol dehydrogenase. J. Biol. Chem. 277:34785-34792. [DOI] [PubMed] [Google Scholar]
- 21.Large, P. J., and C. W. Bamforth. 1988. Methylotrophy and bio/technology. Longman Scientific & Technical, Harlow, Essex, England.
- 22.Lidstrom, M. E., and A. E. Wopat. 1984. Plasmids in methylotrophic bacteria: isolation, characterization, and hybridization analysis. Arch. Microbiol. 140:27-33. [DOI] [PubMed] [Google Scholar]
- 23.Mahillon, J., and D. Lereclus. 1988. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 7:1515-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meshalkina, L., U. Nilsson, C. Wikner, T. Kostikowa, and G. Schneider. 1997. Examination of the thiamine diphosphate binding site in yeast transketolase by site-directed mutagenesis. Eur. J. Biochem. 244:646-652. [DOI] [PubMed] [Google Scholar]
- 25.Mitsui, E., Y. Sakai, H. Yasueda, and N. Kato. 2000. A novel operon encoding formaldehyde fixation: the ribulose monophosphate pathway in the gram-positive facultative methylotrophic bacterium Mycobacterium gastri MB19. J. Bacteriol. 182:944-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murai, N., H. Kamata, Y. Nagashima, H. Yagisawa, and H. Hirata. 1995. A novel insertion sequence (IS)-like element of the thermophilic bacterium PS3 promotes expression of the alanine carrier protein-encoding gene. Gene 163:103-107. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson, U, L. Meshalkina, Y. Lindquist, and G. Schneider. 1997. Examination of substrate binding in thiamine diphosphate-dependent transketolase by protein crystallography and site-directed mutagenesis. J. Biol. Chem. 272:1864-1869. [DOI] [PubMed] [Google Scholar]
- 28.Plater, A. R., S. M. Zgiby, G. J. Thomsen, S. Qamar, C. W. Wharton, and A. Berry. 1999. Conserved residues in the mechanism of the E. coli class II FBP-aldolase. J. Mol. Biol. 285:843-855. [DOI] [PubMed] [Google Scholar]
- 29.Pluschkell, S. B., and M. C. Flickinger. 2002. Dissimilation of [13C]methanol by continuous cultures of Bacillus methanolicus MGA3 at 50oC studied by 13C NMR and isotope-ratio mass spectrometry. Microbiology 148:3223-3233. [DOI] [PubMed] [Google Scholar]
- 30.Reizer, J., A. Reizer, and M. H. Saier, Jr. 1997. Is the ribulose monophosphate pathway widely distributed in bacteria? Microbiology 143:2519-2520. [DOI] [PubMed] [Google Scholar]
- 31.Sakai, Y., R. Mitsui, Y. Katayama, H. Yanase, and N. Kato. 1999. Organization of the genes involved in the ribulose monophosphate pathway in an obligate methylotrophic bacterium, Methylomonas aminofaciens 77a. FEMS Microbiol. Lett. 176:125-130. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Schendel, F. J., C. E. Bremmon, M. C. Flickinger, M. Guettler, and R. S. Hanson. 1990. l-Lysine production at 50°C by mutants of a newly isolated and characterized Bacillus sp. Appl. Environ. Microbiol. 56:963-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamoi, M., A. Murakami, T. Takeda, and S. Shigeoka. 1998. Acquisition of a new type of fructose-1,6-bisphosphatase with resistance to hydrogen peroxide in cyanobacteria: molecular characterization of the enzyme from Synechococcus PCC 6803. Biochim. Biophys. Acta 1383:232-244. [DOI] [PubMed] [Google Scholar]
- 35.Van den Bergh, E. R. E., S. C. Baker, R. J. Taggers, P. Terpstra, E. C. Woudstra, L. Dijkhuizen, and W. G. Meijer. 1996. Primary structure and phylogeny of the Calvin cycle enzymes transketolase and fructose bisphosphate aldolase of Xanthobacter flavus. J. Bacteriol. 178:888-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe, S., M. Hamano, H. Kakeshita, K. Bunai, S. Tojo, H. Yamaguchi, Y. Fujita, S.-L. Wong, and K. Yamane. 2003. Mannitol-1-phosphate dehydrogenase (MtlD) is required for mannitol and glucitol assimilation in Bacillus subtilis: possible cooperation of mtl and gut operons. J. Bacteriol. 185:4816-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wikner, C., U. C. Nilsson, L. Meshalkina, C. Udekwu, Y. Lindquist, and G. Schneider. 1997. Identification of catalytically important residues in yeast transketolase. Biochemistry 36:15643-15649. [DOI] [PubMed] [Google Scholar]
- 38.Yasueda, H., Y. Kawahara, and S. I. Sugimoto. 1999. Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. J. Bacteriol. 181:7154-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yurimoto, H., R. Hirai, H. Yasueda, R. Mitsui, Y. Sakai, and N. Kato. 2002. The ribulose monophosphate pathway operon encoding formaldehyde fixation in a thermotolerant methylotroph, Bacillus brevus S1. FEMS Microbiol. Lett. 214:189-193. [DOI] [PubMed] [Google Scholar]
- 40.Zgiby, S., A. R. Plater, M. A. Bates, G. J. Thomsen, and A. Berry. 2002. A functional role for a flexible loop containing Glu182 in the class II fructose-1,6-bisphosphate aldolase from Escherichia coli. J. Mol. Biol. 315:131-140. [DOI] [PubMed] [Google Scholar]