Abstract
An extracellular proteinase was purified from culture filtrates of Cryptococcus neoformans NHPY24 by DEAE ion-exchange chromatography and gelatin affinity column chromatography with azoalbumin as the substrate. The molecular mass of the purified enzyme was 43 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, its pH optimum was 7.0 to 8.0, and maximal activity was obtained at pH 7.5 and 37°C. By isoelectric focusing, the purified enzyme had a pI of 4.77. Enzyme activity was inhibited by serine proteinase inhibitors such as phenylmethylsulfonyl fluoride and diisopropylfluorophosphate. The purified enzyme was thus a serine proteinase. It hydrolyzed natural substrates including hemoglobin, β-casein, and gamma globulin.
Cryptococcus neoformans is an opportunistic yeast pathogen that causes life-threatening meningoencephalitis in 6 to 8% of patients with AIDS (6). Little is known about the proteins it secretes or releases, despite evidence that they elicit substantial immune responses (4). Several proteins of C. neoformans have been investigated (1, 3, 8, 9, 18, 19, 23, 24; J. M. Goodley and A. J. Hamilton, Abstr. 2nd Int. Conf. Cryptococcus Cryptococcosis, abstr. P1-3, 1993). Extracellular proteinases are especially important because they may play a role in penetration and virulence (4, 7). Staib (21) first described the extracellular proteolytic activity of Candida albicans, a member of the other medically important group of Candida species, in 1965. Since then much experimental evidence has accumulated pointing to extracellular proteinases as the most important virulence factors for this fungus (15, 17). In contrast to C. albicans, the proteolytic activity of C. neoformans has been only superficially investigated (1, 3, 4; Goodley and Hamilton, Abstr. 2nd Int. Conf. Cryptococcus Cryptococcosis), and characterization has been hampered by a lack of purified enzyme. In this study, we purified the extracellular serine proteinase from culture filtrates of C. neoformans by column chromatography and characterized the purified enzyme.
MATERIALS AND METHODS
Strain selection and culture.
Twelve isolates of C. neoformans were obtained from Korean patients and identified by using a Vitek instrument (Biomerieux, Co., Marcy l'Etoile, France) with a yeast biochemical card. To select a strain producing a high level of proteinase, we used yeast carbon base (YCB; Difco Laboratories, Detroit, Mich.) medium containing 1% bovine serum albumin (BSA; Sigma Co., St. Louis, Mo.), 0.1% polypeptone, and 1.8% agar (Difco). Inocula of the 12 isolates were adjusted to 105 CFU/10 μl, spread on plates, and incubated at 37°C for 14 days. The amount of proteinase produced by the strains was compared on the basis of the size of the zone of clearing around the colonies. The selected isolate was cultured in YCB broth medium containing 1% BSA and 0.1% polypeptone to harvest the extracellular proteinase.
Proteinase assay.
To select the optimum substrate, we compared 1% azoalbumin, 1% hemoglobin, and 1% azocasein (Sigma) as substrates. Ten microliters of crude enzyme solution was incubated with 100 μl of each substrate and 290 μl of buffer solution at 37°C for 16 h. Trichloroacetic acid (20%; Sigma) was added to stop the reaction, and the precipitated substrate was removed by centrifugation at 15,000 × g for 30 min. The amount of digested substrate was determined by measuring the supernatant at an optical density (OD) at 440 nm (azoalbumin and azocasein) and OD at 280 nm (hemoglobin). One unit of enzyme activity was defined as the amount of enzyme needed to increase the A440 and A280 by 0.1 OD unit.
Enzyme purification.
Cells were removed by centrifugation at 5,000 × g for 15 min, and the supernatant was filtered through a 0.2-μm-pore-size membrane filter (Nalgene Co., Rochester, N.Y.), precipitated with ammonium sulfate (40 to 60%), and centrifuged at 15,000 × g for 30 min. It was then dialyzed against distilled water, and the dialysate was applied to a 1.6- by 15-cm column of DEAE-Sepharose fast-flow beads equilibrated with 20 mM Tris-HCl (pH 8.0) buffer. Bound protein was eluted with a stepwise gradient of 0.1, 0.15, 0.25, 0.5, 0.75, and 1 M NaCl. The eluted fractions were collected, and protein concentration and enzyme activity were determined. Fractions containing proteolytic activity were pooled, dialyzed against distilled water at 4°C, and lyophilized. The partially purified enzyme was further purified by gelatin affinity chromatography (0.8- by 5-cm column) and equilibrated with 20 mM Tris-Cl (pH 8.0), and buffer and bound protein were eluted with a linear gradient of up to 1 M NaCl.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the method of Laemmli (13) using 12% (wt/vol) polyacrylamide gels (Novex Co.). Proteins were stained with Coomassie brilliant blue R-250 and destained to visualize the bands. Protein standards were phosphorylase b (97 kDa), BSA (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (30 kDa), soybean trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa) (Bio-Rad Co., Richmond, Calif.).
Determination of isoelectric point.
Isoelectric focusing was carried out with pH 3 to 10 polyacrylamide gels along with the standard markers human carbonic anhydrase B (pI 6.55), bovine carbonic anhydrase B (pI 5.85), β-lactaglobulin A (pI 5.2), soybean trypsin inhibitor (pI 4.55), glucose oxidase mannitol (pI 4.15), and methyl red (pI 3.75) (Pharmacia Biotech, Uppsala, Sweden).
Determination of optimal pH.
The optimal pH of the purified enzyme was determined in various buffers (pH range, 4.0 to 9.0). Ten microliters of enzyme solution in 290 μl of 0.1 M sodium acetate buffers (pH 4.0 to 5.5), 0.1 M phosphate buffers (pH 6.0 to 7.5), 0.1 M Tris-HCl buffers (pH 7 to 8.5), and 0.1 M glycine-OH buffers (pH 9.0) was incubated for 16 h at 37°C with 100 μl of 1% azoalbumin. Enzyme activity was measured at 440 nm with a spectrophotometer.
Determination of optimal temperature.
To determine the optimal temperature, 10 μl of enzyme solution in 290 μl of 0.1 M sodium phosphate buffer (pH 7.5) was incubated for 16 h at temperatures from 4 to 70°C with 100 μl of azoalbumin.
Effect of proteinase inhibitors.
The purified enzyme was preincubated at 37°C for 2 h in 0.1 M phosphate buffer (pH 7.5) with inhibitors, and 100 μl of substrate was added. The reaction mixtures were incubated at 37°C for 16 h, and enzyme activity was measured. The inhibitors used in this study were leupeptin (100 μM), iodoacetic acid (1 mM), diisopropylfluorophosphate (DFP) (100 μM), phenylmethylsulfonyl fluoride (PMSF) (1 mM), transepoxysuccinyl-l-leucylamide-(4-guanidino)butane (E-64) (10 μM), N-α-p-tosyl-l-lysine-chloromethyl ketone (TLCK) (100 μM), N-tosyl-l-phenylalanine-chloromethyl ketone (TPCK) (100 μM), pepstatin A (1 μM), and EDTA (5 mM). All inhibitors were purchased from Sigma.
Degradation of natural substrates.
Hemoglobin (bovine), β-casein (from bovine milk), and gamma globulin (from bovine plasma) were purchased from Sigma. They were dissolved in 0.1 M sodium phosphate buffer (pH 7.5) at a concentration of 5 mg/ml and incubated with purified enzyme for various times. SDS-PAGE was performed to measure degradation as described previously.
RESULTS
Strain selection and culture condition.
Of the 12 C. neoformans isolates, isolate NHPY24 showed the largest clear zone and was used for proteinase purification. It was cultured in 0.1% polypeptone and 1% BSA-YCB broth medium at 37°C for 7 days. Proteinase activity was measured with 1% azoalbumin as the substrate. The crude enzyme solution was pooled for proteinase purification.
Proteinase activity assay.
One percent azoalbumin (100% degradation) was the best substrate compared to 1% azocasein (30% degradation) and 1% hemoglobin (42% degradation). One percent azoalbumin was used as substrate in each purification step.
Purification of enzyme.
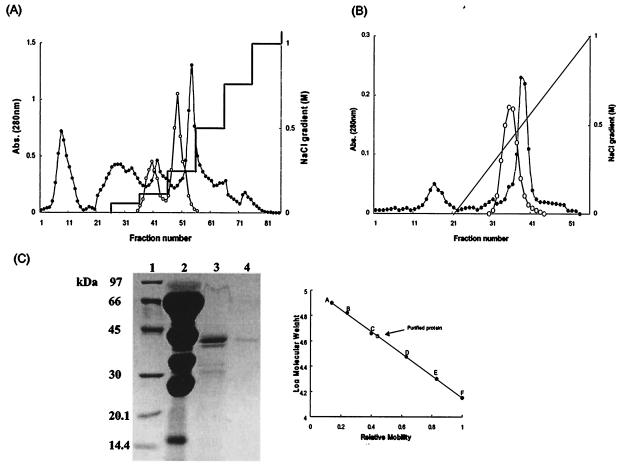
Two peaks of proteolytic activity (Fig. 1A) were generated by the first ion-exchange column. The three fractions of the 0.25 M NaCl gradient with highest proteolytic activity were pooled, dialyzed, concentrated, and applied to the gelatin affinity column (Fig. 1B). The two fractions from this with the highest proteolytic activity were pooled, dialyzed, and concentrated for further study.
FIG. 1.
Proteinase purification and SDS-PAGE. (A) First DEAE-Sepharose column chromatograph. The column was equilibrated with 20 mM Tris-HCl (pH 8.0) buffer, and bound proteins were eluted with a step gradient of 0.1, 0.15, 0.25, 0.5, 0.75, and 1.0 M NaCl. (B) Second gelatin affinity chromatograph of the active fractions from the first DEAE column eluted with a linear gradient of NaCl. The protein concentration was measured with a spectrophotometer at 280 nm (•), and proteolytic activity, with azoalbumin as the substrate, was measured at 440 nm absorbance (○). (C) SDS-12% PAGE of C. neoformans extracellular proteinase. Lane 1, standard SDS-PAGE markers; lane 2, crude extracts (40 to 60% ammonium sulfate precipitate); lane 3, fractions with proteolytic activity from the first DEAE-Sepharose chromatograph; lane 4, fractions with proteolytic activity from the second gelatin affinity chromatograph. SDS-PAGE markers: A, phosphorylase B (97 kDa); B, BSA (66 kDa); C, ovalbumin (45 kDa); D, carbonic anhydrase (30 kDa); E, soybean trypsin inhibitor (20.1 kDa); F, α-lactalbumin (14.4 kDa). The molecular mass of the purified enzyme was determined from its mobility relative to the protein standards.
SDS-PAGE.
Concentrated solutions obtained at each purification step were investigated by electrophoresis to assess their purity. The active fractions produced by gelatin affinity chromatography yielded just one band, which was of 43kDa (Fig. 1C).
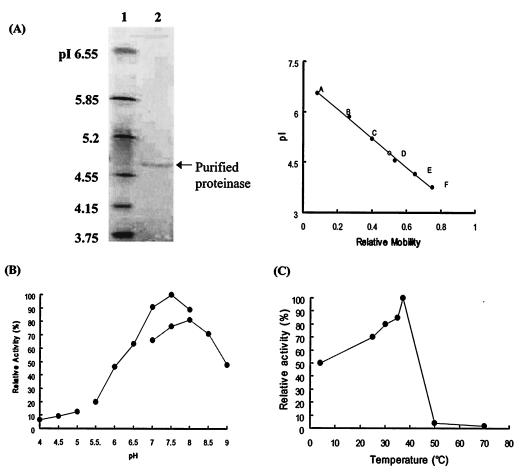
Optimal pH, temperature, and pI of the purified proteinase.
The purified enzyme had a pI of 4.77 (Fig. 2A) and a maximum activity in 0.1 M phosphate buffer (pH 7.5). The activity increased linearly from pH 6.0 to 7.5, and 80% of the total enzyme activity was manifested between pH 7.0 and 8.0 (Fig. 2B). It had a narrow temperature optimum with a maximum at 37°C, and it was almost completely inactive at 50 to 70°C (Fig. 2C).
FIG. 2.
Characterization of the enzyme purified from C. neoformans culture filtrates. (A) Isoelectric focusing and pI determination. The isoelectric focusing gel range was pI 3 to 10. Lane 1, standard pI markers; lane 2, purified enzyme. The pI value of the purified enzyme was determined by the mobility of the purified enzyme relative to the standard pI markers. pI markers: A, human carbonic anhydrase B (pI 6.55); B, bovine carbonic anhydrase B (pI 5.85); C, β-lactaglobuin A (pI 5.2); D, soybean trypsin inhibitor (pI 4.55); E, glucose oxidase mannitol (pI 4.15); F, methyl red (dye) (pI 3.75). (B) pH dependence of the purified enzyme. The purified enzyme was incubated with azoalbumin in various pH buffer solutions. The maximal activity, seen in 0.1 M phosphate buffer (pH 7.5), is taken as 100%. Enzyme activity increased linearly from pH 6.0 to 7.5, and 80% of the total activity was exhibited between pH 7.0 and 8.0. (C) Temperature dependence of the purified proteinase. Ten microliters of purified enzyme was incubated in 290 μl of 0.1 M sodium phosphate buffer (pH 7.5) and 100 μl of azoalbumin substrate solution at 4, 25, 30, 35, 37, 50, and 70°C for 16 h. The proteolytic activity remaining was measured at OD440 by spectrophotometer.
Effect of proteinase inhibitors.
The purified enzyme was inhibited by serine proteinase-specific inhibitors such as PMSF and DFP and general inhibitors such as TLCK and TPCK. It was not inhibited by cysteine proteinase inhibitor (E-64, leupeptin, and iodoacetic acid) or metalloproteinase inhibitor (EDTA) and aspartic proteinase inhibitor (pepstatin A) (Table 1).
TABLE 1.
Effects of various inhibitors on proteolytic activity of purified enzymea
| Inhibitor (concn) | Relative activity (%) |
|---|---|
| None (control) | 100 |
| E-64 (10 μM) | 94 |
| Leupeptin (100 μM) | 107 |
| Iodoacetic acid (1 mM) | 80 |
| TLCK (100 μM) | 27 |
| TPCK (100 μM) | 17 |
| PMSF (1 mM) | 0.5 |
| DFP (100 μM) | 8 |
| EDTA (10 mM) | 90 |
| Pepstatin A (1 μM) | 94 |
The inhibitors were tested at their maximum effective concentrations. Activity against azoalbumin in the absence of inhibitors was taken as 100% (control).
Natural substrate degradation.
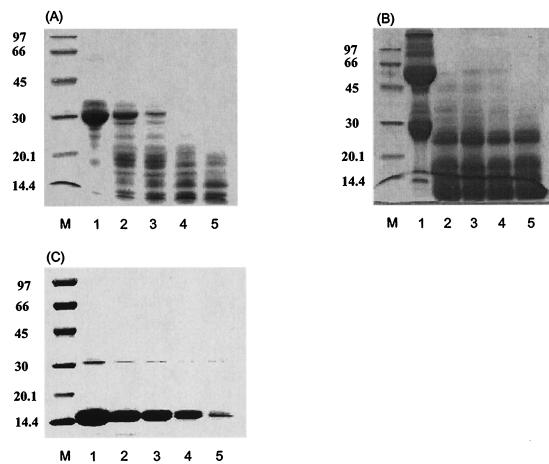
The purified enzyme progressively degraded β-casein, gamma globulin, and hemoglobin. β-Casein was rapidly degraded, and the other substrates apparently degraded in about 24 h (Fig. 3).
FIG. 3.
Time-dependent degradation of natural substrates. Substrates dissolved in 0.1 M sodium phosphate buffer (pH 7.5) at a concentration of 5 mg/ml were incubated with purified enzyme. Results are shown for the degradation of gamma globulin (A), β-casein (B), and hemoglobin (C). Lanes: M, standard marker; 1, gamma globulin (A), β-casein (B), and hemoglobin (C) controls; 2, 6 h of reaction; 3, 12 h of reaction; 4, 24 h of reaction; 5, 48 h of reaction.
DISCUSSION
Proteins secreted by fungal pathogens could be involved in their invasive process and might be useful in vaccine design (4). The identification and characterization of the C. neoformans extracellular protein is important because proteinases, esterases, and lipases are associated with virulence in other pathogens (5, 10, 11, 12, 14, 20, 22). C. neoformans is usually considered to be nonproteolytic (21), but a few studies have focused interest on its proteolytic activity (2, 16). Extracellular proteinase activity in C. neoformans was first reported in 1972 (16). In the present study, we collected the extracellular proteinase in YCB medium supplemented with 1% BSA and 0.1% polypeptone. Among the 12 C. neoformans clinical isolates, NHPY24 showed the largest clearing zone in agar supplemented with BSA as a substrate. Chen et al. characterized cryptococcal extracellular proteolytic activity in vitro as a serine proteinase and found it associated with proteins of approximately 200, 100, and 50 kDa (3), In this study, the purified proteolytic enzyme had a molecular mass of 43 kDa as determined by SDS-PAGE. Because the purified enzyme was inhibited by serine proteinase inhibitors such as PMSF and DFP, we concluded that it was a serine proteinase. Goodley and Hamilton succeeded in isolating an extracellular 200 kDa proteinase from the culture filtrate of C. neoformans and showed it to be a Ca2+- and Mg2+-dependent serine proteinase with an optimal pH at 7.5 to 8.5 (Abstr. 2nd Int. Conf. Cryptococcus Cryptococcosis). The purified proteinase in this study had almost the same characteristics, but it differed in pH optimum and molecular mass. The purified proteinase is thus a new C. neoformans proteinase. Müller and Sathi (16) first demonstrated the ability of C. neoformans to degrade or split 2 of a total of 13 human plasma proteins tested in their immunoelectrophoretic study. Cryptococcal proteinases degrade several host proteins such as elastin, collagen, fibrin, fibrinogen, complement factors, and immunoglobulins (2, 3, 16), suggesting that they may be important in tissue disruption and the perturbation of host immunity. The purified proteinase from isolate NHPY24 degraded hemoglobin, β-casein, and gamma globulin. It could be involved in tissue disruption and C. neoformans penetration, but its function needs further investigation.
REFERENCES
- 1.Aoke, S., S. Ito-Kuwa. K. Nakamura, J. Kato, K. Ninomiya, and V. Vidotto. 1994. Extracellular proteolytic activity of Cryptococcus neoformans. Mycopathologia 128:143-150. [DOI] [PubMed] [Google Scholar]
- 2.Brueske, C. 1986. Proteolytic activity of a clinical isolate of Cryptococcus neoformans. J. Clin. Microbiol. 23:631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen. L.-C., E. S. Blank, and A. Casadevall. 1996. Extracellular proteinase activity of Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 3:570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen. L.-C., L. A. Pirofski, and A. Casadevall. 1997. Extracellular proteins of Cryptococcus neoformans and host antibody response. Infect. Immun. 65:2599-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Commenil, P., L. Belingheri, and M. S. Dehorter. 1995. Purification and properties of an extracellular lipase from the fungus Botrytis cinerea. Lipids 30:351-356. [DOI] [PubMed] [Google Scholar]
- 6.Currie, B. P., and A. Casadevall. 1994. Estimation of the prevalence of cryptococcal infection among patients infected with the human immunodeficiency virus in New York City. Clin. Infect. Dis. 19:1029-1033. [DOI] [PubMed] [Google Scholar]
- 7.David, G., S. C. Lee, and A. Casadevall. 1994. Pathogenesis of pulmonary Cryptococcus neoformans infection in the rat. Infect. Immun. 62:4755-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilton. A. J., and J. Goodley. 1993. Purification of the 115-kilodalton exoantigen of Cryptococcus neoformans and its recognition by immune sera. J. Clin. Microbiol. 31:335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamilton, A. J., L. Jeavons, P. Hobby, and R. J. Hay. 1992. A 34- to 38-kilodalton Cryptococcus neoformans glycoprotein produced as an exoantigen bearing a glycosylated species-specific epitope. Infect. Immun. 60:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito, A., A. Mukaiyama, Y. Itoh, K. Nagase, I. B. Thogersen, J. J. Enghild, Y. Sasaguri, and Y. Mori. 1996. Degradation of interleukin 1beta by matrix metalloproteinases. J. Biol. Chem. 271:14657-14660. [DOI] [PubMed] [Google Scholar]
- 11.Koning, B., K.-E. Jaeger, A. E. Sage, M. L. Vasil, and W. Konig. 1996. Role of Pseudomonas aeruginosa lipase in inflammatory mediator release from human inflammatory effector cells (platelets, granulocytes, and monocytes). Infect. Immun. 64:3252-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon-Chung, K. J., I. Polacheck, and T. J. Popkin. 1982. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J. Bacteriol. 150:1414-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Maeda, H., and T. Yamamoto. 1996. Pathogenic mechanisms induced by microbial proteases in microbial infections. Biol. Chem. Hoppe-Seyler 377:217-226. [DOI] [PubMed] [Google Scholar]
- 15.Morrison, C. J., S. F. Hurst, S. L. Brag, R. J. Kuyhendall, H. Diaz, D. W. McLaughlin, and E. Reiss. 1993. Purification and characterization of the extracellular aspartyl proteinase of Candida albicans: removal of extraneous proteins and cell wall mannoprotein and evidence for lack of glycosylation. J. Gen. Microbiol. 139:1177-1186. [DOI] [PubMed] [Google Scholar]
- 16.Müller, H. E., and K. K. Sathi. 1972. Proteolytic activity of Cryptococcus neoformans against human plasma proteins. Med. Microbiol. Immunol. 158:129-134. [DOI] [PubMed] [Google Scholar]
- 17.Negi, M., R. Tsuboi, T. Matsui, and H. Ogawa. 1984. Isolation and characterization of proteinase from Candida albicans: substrate specificity. J. Investig. Dermatol. 83:32-36. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes, J. J. C., I. Polacheck, and K. J. Kwon-Chung. 1982. Phenoloxidase activity and virulence in isogenic strains of Cryptococcus neoformans. Infect. Immun. 36:1175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross, A., and I. E. P. Taylor. 1981. Extracellular glycoprotein from virulent and avirulent Cryptococcus species. Infect. Immun. 31:911-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakata, Y., T. Akaike, M. Suga, S. Ijiri, M. Ando, and H. Maeda. 1996. Bradykinin generation triggered by Pseudomonas proteases facilitates invasion of the systemic circulation by Pseudomonas aeruginosa. Microbiol. Immunol. 40:415-423. [DOI] [PubMed] [Google Scholar]
- 21.Staib, F. 1965. Serum-proteins as nitrogen source for yeastlike fungi. Sabouraudia 4:187-193. [DOI] [PubMed] [Google Scholar]
- 22.Tsuboi, R., H. Komatsuzaki, and H. Ogawa. 1996. Induction of an extracellular esterase from Candida albicans and some of its properties. Infect. Immun. 64:2936-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner, S. H., R. Cherniak, and E. Reiss. 1984. Fractionation and characterization of galactoxylomannan from Cryptococcus neoformans. Carbohydr. Res. 125:343-349. [DOI] [PubMed] [Google Scholar]
- 24.Vartivarian, S. E., G. H. Reyes, E. S. Jacobson, P. G. James, R. Cherniak, V. R. Mumaw, and M. J. Tingier. 1989. Localization of mannoprotein in Cryptococcus neoformans. J. Bacteriol. 171:6850-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]