Abstract
The susceptibility of human immunodeficiency virus type 2 (HIV-2) to protease inhibitors (PI) is largely unknown. We studied HIV-2 protease genes from 21 HIV-2-infected patients who were exposed or not exposed to PI. The aim of this study was (i) to characterize the polymorphism of HIV-2 protease in the absence of drug, (ii) to know whether the HIV-2 protease gene naturally harbors HIV-1 drug resistance codons, and (iii) to identify mutations emerging under PI-selective pressure. Sixty-five HIV-2 RNA or proviral DNA samples were directly sequenced from the plasma or peripheral blood mononuclear cells of 8 patients who had received PI and 13 patients who had never received any antiretroviral. In untreated patients, the highest amino acid variability in HIV-2 protease was observed at positions 14, 40, 43, 46, 65 and 70, and seven codons (10V, 32I, 36I, 46I, 47V, 71V, and 73A) associated with drug resistance in HIV-1 were highly prevalent. In addition, at six positions (positions 7, 46, 62, 71, 90, and 99), the amino acid variability or the amino acid frequencies or both differed significantly in PI-treated and untreated patients, suggesting that mutations 7K→R, 46V→I, 62V→A/T, 71V→I, 90L→M and 99L→F were occurring under PI-selective pressure. At these positions, at least one sample simultaneously harbored both wild-type and mutated codons, while substitutions at positions 62, 71, 90, and 99 were confirmed in a longitudinal analysis. Moreover, the presence of codons 46I and 99F in the absence of drug in HIV-2 subtype B proteases may reflect natural resistance to PI. In conclusion, the present study revealed that HIV-2 strains harbor specific patterns of natural polymorphism and resistance.
The human immunodeficiency virus type 2 (HIV-2) was first identified in 1986 in West Africa (10). In this area, the prevalence of HIV-2 infections varies from 1 to 10% or more depending on the country (18, 22, 31, 45). The majority of cases identified outside West Africa have been in European countries, mostly in Portugal, where HIV-2 infections account for 10 to 13% of HIV infections (36). It is estimated that approximately 1% of HIV-infected patients in France could be infected with HIV-2. In the Marseilles area, HIV-2 is responsible for approximately 0.6% of cases of HIV infections and 0.4% of cases of AIDS. Seven HIV-2 subtypes, A through G, have been identified to date (8, 17, 27, 38, 46), but subtype A is the most common worldwide (17, 46) and in France (11). Although both HIV-1 and HIV-2 can cause AIDS and have similar clinical and biological characteristics (9, 19), major differences can be highlighted: plasma HIV RNA titers are lower in HIV-2-infected patients than in HIV-1-infected patients (3, 4, 12, 30, 35), individuals infected with HIV-2 have a slower disease progression (2, 24, 37, 44), and HIV-2 shows lower transmissibility (1, 2, 23, 24, 44) compared with HIV-1. These factors explain at least in part why HIV-2 is less disseminated worldwide than HIV-1 (5, 13, 26). In developed countries, some HIV-2-infected individuals have been treated with antiretroviral agents. However, until now, no large-scale clinical trial has been conducted and systematic quantitation of HIV-2 RNA levels in plasma has not been available in clinical practice. Moreover, in areas where HIV-2 infection is endemic, access to antiretroviral drugs is limited. Thus, little information is available on the clinical efficiency of antiretroviral drugs in HIV-2-infected patients and on HIV-2 drug resistance mutations (34). Although protease inhibitors (PI) have contributed to the significant reduction of morbidity and mortality due to HIV-1 infection in developed countries (25, 42), their efficacy against HIV-2 has been poorly documented. Moreover, the few studies related to PI resistance of HIV-2 are controversial: HIV-2 isolates appear to be sensitive in vitro to most PI according to some authors (40, 41), whereas they could be intrinsically resistant to at least some PI according to others (G. Collin, D. Descamps, C. Apetrei, F. Damond, S. Souquiere, I. Loussert-Ajaka, F. Simon, and F. Brun-Vezinet, Abstr. 2nd Int. Workshop HIV Drug Resist. Treat. Strategies, abstr. 48, 1998; J. Goncalves, S. Coellio, F. Antunes, and J. Monitz-Pereira, Abstr. 11th Int. Workshop HIV Drug Resist. Treat. Strategies, abstr. 32, 2002). To date, only one study has reported the presence of genotypic changes in the protease gene of HIV-2 which could be drug resistance-associated mutations (34). The present study was done with both untreated and PI-treated patients and had three objectives: (i) to characterize the polymorphism of the HIV-2 protease in the absence of PI; (ii) to know whether natural polymorphisms are found at homologous positions in the HIV-2 and HIV-1 protease and whether amino acids that confer resistance to PI in HIV-1 are found in HIV-2; and (iii) to identify mutations emerging under PI selective pressure.
MATERIALS AND METHODS
Patients and sequences.
Our cohort included 31 HIV-2-infected patients being treated at different hospitals in Marseilles and the surrounding area. Twenty-one HIV-2 protease sequences were available from 13 patients who had never undergone antiretroviral therapy, and 35 sequences were available from 8 patients receiving PI-containing regimens. For three PI-naive patients and five PI-treated patients, 11 and 32 sequences were available, respectively, from sequential samples. For one PI-treated patient, a sequence was also available before initiation of antiretroviral therapy. Nine sequences from three patients treated with nucleosidic inhibitors of reverse transcriptase (RT) were also analyzed. In all, 65 sequences were studied. They included 27 DNA and 38 RNA sequences. Both DNA and RNA sequences were analyzed when available for the same sample. However, for the statistical analysis, DNA sequences were excluded to avoid considering the same sample twice.
Nucleic acid extraction and purification.
Whole blood was collected in tubes containing EDTA. Plasma was aliquoted after a centrifugation step and stored at −80°C until it was processed. Viral RNA was concentrated from 500 μl of plasma by ultracentrifugation for 1 h at 17,000 × g and then extracted using the QIAamp Viral RNA Kit (Qiagen, Courtaboeuf, France). It was eluted in 50 μl of elution buffer. Peripheral blood mononuclear cells (PBMC) were separated from blood samples collected in EDTA by Ficoll-Hypaque centrifugation (Eurobio, Les Ullis, France). Aliquots containing 1 × 106 to 5 × 106 PBMC measured by cell count were frozen as dry pellets at −80°C until they were processed. The PBMC pellets were thawed, and total DNA was extracted using a QIAamp DNA minikit (Qiagen). Prepared RNA and DNA were directly analyzed or stored at −80°C.
Nucleic acid RT-PCR or PCR amplification. (i) RNA amplification.
A 10-μl sample of extracted viral RNA was retrotranscribed and amplified using SuperScript One-Step RT-PCR with Platinium Taq (Invitrogen Life Technologies, Carlsbad, Calif.) using 50 pmol of the outer primers (forward: H2Mp1, 5′-GGGAAAGAAGCCCCGCAACTTC, 1,859; reverse: H2Mp2, 5′-GGGATCCATGTCACTTGCCA, 3,612; the primer location was defined in reference to HIV-2 ROD [GenBank accession number M15390]). Protease and RT genomic regions were amplified in the same PCR product of 1,753 bp. This amplification was carried out in a 50-μl reaction mixture under the conditions recommended by the manufacturer. RT-PCR involved a retrotranscription step at 45°C for 30 min, followed by a polymerase activation-initial denaturation step at 94°C for 2 min, 40 amplification cycles (for 30 s at 94°C, 45 s at 56°C, and 2 min at 72°C), and then an elongation step at 72°C for 10 min. Nested PCR was performed using 1 to 10 μl of the previous amplification product, 50 pmol of inner primers (forward: H2Mp3, 5′-ACTTACTGCACCTCGAGCA, 2,020 bp; reverse: H2Mp4, 5′-CCCAAATGACTAGTGCTTCTT, 3,527 bp), 0.2 mM each deoxynucleoside triphosphate, 1X polymerase buffer (Roche, Mannheim, Germany), and 2.5 U of enzyme in a final volume of 50 μl to obtain a genomic fragment of 1,507 bp. PCR conditions were as follows: denaturation at 94°C for 2 min followed by 40 cycles (for 30 s at 94°C, 45 s at 58°C, and 2 min at 72°C), and then incubation at 72°C for 5 min. The PCR products were analyzed in a 1.5% agarose gel with ethidium bromide.
(ii) Proviral DNA amplification.
Procedure and primers were identical to those described above for amplification of RNA, except for the reverse transcription step. A 10-μl volume of extracted HIV-2 DNA was initially used.
DNA and RNA pol analysis.
The resulting PCR-amplified DNA fragments were purified using Multiscreen PCR (Millipore, Molshein, France) as specified by the manufacturer. The PCR product was used as the DNA template for nucleotide sequencing analysis of the HIV-2 protease-RT coding region with eight primers (forward: H2Mp3; H2Mp6, 5′-AAAAGAGATCTGTGCAAAAATGG, 2,482 bp; H2Mp9, 5′-GGATGATATCTTAATAGCTAG, 2,932 bp; reverse: H2Mp5, 5′-AGTTTTGGTCCATCTTTCCC, 2,441 bp; H2Mp7, 5′-GGGTATTATAAGGATTAGTTGG, 2,555 bp; H2Mp8, 5′-AATTGCTGGTGATCCCTTC, 2,857 bp; H2Mp10, 5′-GATGTCATTGACTGTCC, 3,151 bp; H2Mp4). Cycle sequencing of both strands was performed on the GeneAmp PCR system 9600 instrument (Applied Biosystems, Branchburg, N.J.) with the Big Dye Terminator cycle-sequencing kit (Applied Biosystems). Excess dye-labeled terminators were removed from the extension products on Sephadex G-50 Superfine on NAHVN 4550 plates (Millipore), and the purified products were sequenced on an ABI Prism 3100 genetic analyzer (Applied Biosystems). The nucleotide sequences of the protease-RT genes were aligned and translated with the Auto Assembler and Sequence Navigator software programs (Applied Biosystems) by using the sequences of HIV-2 ROD and HIV-1 HXB2 (GenBank accession numbers M15390 and K03455, respectively), as references.
Variability study.
We created the ASVARAP program (http://ifr48.free.fr/recherche/jeu_cadre/jeu_rickettsie.html), which works with a Microsoft Excel file and automatically analyzes, reveals, and graphically represents amino acid variability in a set of sequences. We generated alignments in Genetic Data Environment format from our sets of sequences with ClustalX v.1.8 (39). Our program calculated the proportion of sequences that harbored an amino acid at a given position that was not the most frequently found in the studied set of sequences.
Statistical analysis.
Chi-square or Fisher tests were used to assess the significance of differences between sets of sequences.
RESULTS
Patients and HIV-2 protease sequences.
The main epidemiological, immunological, and clinical features of the 31 patients are summarized in Table 1. Twenty-six individuals had been infected several years beforehand, and five were newly diagnosed during the previous year. Twenty-eight patients have been followed up until now, two were lost to follow-up, and one died. The mean length of follow-up was 48 months (range, 6 to 139 months). Twenty-six individuals were from or had epidemiological links with sub-Saharan Africa, four were French without epidemiological links with areas of high HIV-2 endemicity, and one case was not documented. The means of HIV-2 transmission were heterosexual contact in twenty-six cases, heterosexual contact plus intravenous drug use or transfusion in two cases, transfusion in one case, and unknown in two cases. Twenty-two patients were at stage A, three were at stage B, and six were at stage C according to the Centers for Disease Control and Prevention classification (1993). Sequencing and phylogenetic analysis could be carried out for 25 patients and indicated that they were infected by HIV-2 subtype A (n = 21) or B (n = 4). Thirteen patients received antiretroviral therapy, and 16 were never treated. Two cases were not documented. The median duration of treatment was 36 months. Among the treated patients, 11 received PI in association with RT inhibitors and were treated for a median of 28 months (range, 2 to 60 months). The median CD4 cell count at the beginning of follow-up was 452 cells/mm3. The lowest CD4 cell count reached during follow-up ranged from 4 to 1,192 cells/mm3, with a median of 243 cells/mm3.
TABLE 1.
Epidemiological, clinical, and immunological characteristics of the 31 HIV-2-infected patients
| Patient | Age (yr) | Gendera | Geographic origin (eventual epidemiological link with area of high endemicity) | Transmission group | Disease stage | Duration of follow-up (mos) | First CD4+ cell count available during follow- up (cells/mm3) | HIV-2 subtype (pol gene) | Antiretroviral therapy | PI (when administered) |
|---|---|---|---|---|---|---|---|---|---|---|
| MRT-1 | 38 | F | Algeria | Heterosexual | B | 71 | 210 | A | Yes | NFV |
| MRT-2 | 31 | M | Ivory Coast | Heterosexual | A | 9 | 37 | B | No | |
| MRT-3 | 46 | F | Guinea-Bissau | Heterosexual | A | 65 | 616 | NAc | No | |
| MRT-4 | 40 | F | France | Heterosexual | A | 34 | 445 | A | No | |
| MRT-5 | 35 | F | France (Congo) | Heterosexual | A | 22 | 676 | A | No | |
| MRT-6 | 35 | F | France (subsaharan Africa) | Heterosexual | A | 20 | 599 | NA | No | |
| MRT-7 | 47 | M | France (subsaharan Africa) | Heterosexual | A | 61 | 451 | A | Yes | RTV, NFV |
| MRT-8 | 52 | F | Ivory Coast | Transfusion | A | 119 | 308 | A | Yes | SQV, RTV, NFV, IDV |
| MRT-9 | 46 | F | Senegal | Heterosexual | A | 21 | 404 | A | No | |
| MRT-10 | 51 | M | Senegal | Heterosexual | A | 16 | 213 | NA | Yes | |
| MRT-11 | 40 | M | France | IVDUb, heterosexual | A | 54 | 1,883 | A | No | |
| MRT-12 | 38 | F | Burkina Faso | Heterosexual | A | 75 | 200 | A | ND | ND |
| MRT-13 | 27 | F | Senegal | Heterosexual | A | 9 | 580 | A | No | |
| MRT-14 | 41 | M | Guinea-Bissau | Unknown | A | 6 | 453 | A | No | |
| MRT-15 | 61 | F | No data | Heterosexual | A | 60 | 498 | A | No | |
| MRT-16 | 61 | M | Guinea-Bissau, Ivory Coast, Burkina Faso | Heterosexual | C | 12 | 62 | B | Yes | NFV |
| MRT-17 | 37 | M | Ivory Coast, Burkina Faso | Heterosexual | A | 133 | 464 | B | No | |
| MRT-18 | 50 | M | Central African Republic | Heterosexual | A | 11 | 1,019 | NA | No | |
| MRT-19 | 40 | M | Burkina Faso | Heterosexual | A | 8 | 578 | A | No | |
| MRT-20 | 47 | M | Guinea-Bissau | Heterosexual | C | 105 | 805 | A | Yes | IDV, NFV |
| MRT-21 | 40 | M | Senegal | Heterosexual | A | 57 | 1204 | A | No | |
| MRT-22 | 57 | M | Senegal | Heterosexual | A | 55 | 126 | A | Yes | NFV, LPV/r |
| MRT-23 | 47 | M | Guinea-Bissau | Heterosexual | C | 75 | 375 | A | ND | ND |
| MRT-24 | 41 | F | Guadeloupe | Heterosexual, transfusion | A | 38 | 156 | A | Yes | RTV, IDV, NFV |
| MRT-25 | 45 | F | France (Cape Verde islands) | Heterosexual | B | 52 | 69 | A | Yes | IDV, NFV, LPV/r, APV, RTV |
| MRT-26 | 56 | F | Guinea-Bissau | Heterosexual | A | 130 | 763 | A | No | |
| MRT-27 | 37 | F | Ghaana | Heterosexual | C | 20 | 136 | NA | Yes | IDV, LPV/r |
| MRT-28 | 72 | F | Senegal | Heterosexual | A | 21 | 507 | A | No | |
| MRT-29 | 45 | M | Senegal | Heterosexual | C | 12 | 10 | A | Yes | NFV |
| MRT-30 | 40 | F | Burkina Faso | Heterosexual | B | 139 | 600 | NA | Yes | |
| MRT-31 | 47 | M | Burkina Faso | Unknown | C (died) | 45 | NDd | B | Yes | NFV |
F, female; M, male.
IVDU, intravenous drug use.
NA, not available.
ND, no data.
Finally, 65 sequences of the HIV-2 protease from 21 patients were studied. For analysis of natural polymorphism and natural resistance, we used 21 sequences from 13 of the 16 patients who had never received any antiretroviral therapy. For analysis of amino acid changes selected under PI selective pressure, we used 35 sequences from 8 of the 11 patients treated with PI.
Natural polymorphism in HIV-2 protease.
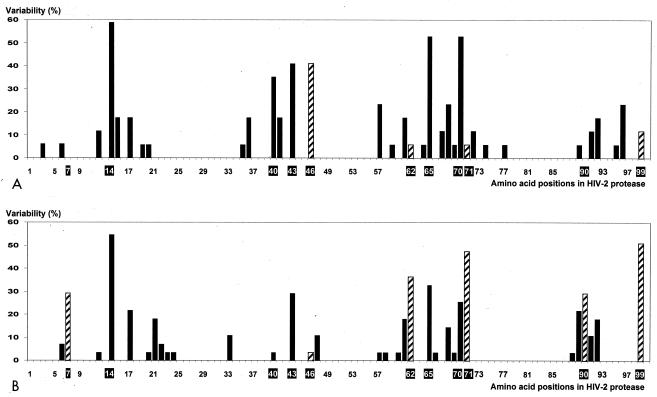
The sequences obtained from patients who had never been treated were used for analysis of natural polymorphism. We considered that an amino acid position was polymorphic when at least two different amino acids were found. Amino acid variability at a given position was established by calculating the percentage of sequences harboring an amino acid that was different from the most frequent one at this position in the set of sequences. Twenty-one sequences from 13 untreated patients were analyzed (Fig. 1A). We found that 34 (34%) of the 99 amino acid positions of HIV-2 protease varied spontaneously. As shown in Fig. 1A, the highest amino acid variability was found at positions 14, 40, 43, 46, 65, and 70 of HIV-2 protease, where more than 30% of the sequences were variable. Position 40 harbored six different amino acids. Interestingly, three areas from positions 25 to 32, 48 to 56, and 78 to 87 were free of mutations in all sequences from untreated patients.
FIG. 1.
Amino acid variability at each site of HIV-2 protease for untreated individuals (A) and patients treated with protease inhibitor-containing regimen (B). The variability was considered to be the percentage of sequences that contained, at a given position, an amino acid that was not the most frequently found in the studied set of sequences. Amino acid positions of interest (highly variable or potentially associated with resistance to protease inhibitors) are in bold type. Hatched bars indicate positions thought to be associated with resistance to protease inhibitors.
Analysis of positions that are classically associated with HIV-1 drug resistance.
In the second step of our analysis, 21 positions associated with HIV-1 drug resistance according to the International AIDS Society (http://www.iasusa.org/resistance_mutations/resistance.pdf) were compared with the same positions in HIV-2 protease sequences from naive patients. We found that the amino acids mostly or always found at seven positions in sequences of the HIV-2 protease corresponded to those conferring resistance in HIV-1: 10V, 32I, 36I, 46I, 47V, 71V and 73A (Table 2).
TABLE 2.
Frequency of amino acids in HIV-2 protease, from untreated patients, at positions associated with PI resistance in HIV-1
| Virus | HIV-2 protease amino acid at positiona:
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 20 | 24 | 30 | 32 | 33 | 36 | 46 | 47 | 48 | 50 | 53 | 54 | 63 | 71 | 73 | 77 | 82 | 84 | 88 | 90 | |
| Wild-type HIV-1b | L | K | L | D | V | L | M | M | I | G | I | F | I | L | A | G | V | V | I | N | L |
| Drug-resistant HIV-1c | F, I, R, V | M, R | I | N | I | F | I | I, L | V | V | V, L | L | L, M, V | P | V, T | S, A | I | A, F, T, S | V | S, D | M |
| HIV-2 subtype A (con- sensus sequence)d | V | V | L | D | I | V | I (V) | I (V) | V | G | I | F | I | E | V | A | T | I | I | N | L |
| HIV-2 subtype B (con- sensus sequence)d | V | V | L | D | I | V | I | I | V | G | I | F | I | E | V | A | T | I | I | N | L |
| HIV-2 strains from untreated patientc | V (100) | V (94), I (6) | L (100) | D (100) | I (100) | V (100) | I (82), V (12), V/I (6) | I (59), V (35), T (6) | V (100) | G (100) | I (100) | F (100) | I (100) | E (100) | V (94), I (6) | A (100) | T (100) | I (100) | I (100) | N (100) | L (100) |
Below each amino acid position is the list of amino acids that are observed and in parentheses is the percentage of sequences concerned. The predominant amino acid for each position is given first. Amino acids that are the most frequently found or always present in HIV-2 strains and are associated with drug resistance in HIV-1 are in bold type.
Wild-type sequence from the reference HIV-1 strain HXB2 (GenBank accession number K03455).
According to the International AIDS Society (IAS; http://www.iasusa.org/resistance_mutations/resistance.pdf).
Available at http://hiv-web.lanl.gov/content/hiv-db/ALIGN_CURRENT/ALIGN-INDEX.html; nonpredominant amino acids are indicated in parentheses.
Identification of PI resistance mutations.
In the present study, mutations were considered to be selected under a PI-containing antiretroviral regimen based on the presence of at least two of the following criteria: (i) a significantly higher amino acid variability at a given position in PI-treated than in naive patients; (ii) a statistically significant difference between the frequency of amino acids at a given position in naive and PI-treated patients; (iii) progressive selection of mutated codons observed in longitudinal studies under selective pressure in PI-treated patients, and (iv) simultaneous presence as a mixture of codons considered as wild-type and mutated in at least one sequence. A significantly higher amino acid variability was found at positions 7, 62, 71, 90, and 99 in PI-treated patients than in treatment-naive patients (P < 0.05): 30 versus 0%, 37 versus 6%, 48 versus 6%, 30 versus 0%, and 52 versus 12%, respectively (Fig. 1). Moreover, the frequencies of amino acids 46I, 62A/T, 71I, 90M, and 99F were significantly higher in the PI-treated patients than in the untreated patients (P < 0.05) (Table 3). An arginine was found at position 7 in 0% of sequences from untreated patients and 18.5% of sequences from PI-treated patients, but the difference was not statistically significant (P = 0.07). The presence of mixtures including both wild-type and mutated viral populations at positions 7, 46, 62, 71, 90, and 99 was observed in at least one sequence obtained from a single blood sample. Longitudinal analysis of five PI-treated individuals showed the progressive selection of mutations 62V→A/T, 71V→I, 90L→M, and 99L→F. Finally, these results led us to speculate that the substitutions 7K→R, 46V→I, 62V→A/T, 71V→I, 90L→M, and 99L→F were occurring under selective pressure by PI. Interestingly, the mutated amino acids found in HIV-2 sequences at positions 46 and 90 are major codons of resistance to PI in HIV-1 protease.
TABLE 3.
Frequency of amino acids in HIV-2 protease, from untreated and PI-containing-regimen-treated patients, at positions suspected of being associated with resistance to PI
| Virus | HIV-2 protease amino acid at positiona:
|
|||||
|---|---|---|---|---|---|---|
| 7 | 46 | 62 | 71 | 90 | 99 | |
| Wild-type HIV-1b | Q | M | I | A | L | F |
| HIV-2 subtype A (consensus sequence)c | K (N) | I (V) | V (I) | V | L | L |
| HIV-2 subtype B (consensus sequence)c | R | I | V | V | L | F |
| HIV-2 strains from patients with no antiretroviral therapy | K (100) | I (59), V (35), T (6) | V (94), I (6) | V (94), I (6) | L (100) | L (88), F (12) |
| HIV-2 strains from patients receiving PI-containing anti- retroviral therapy | K (70), R (11), K/R (7), K/Q (4), K/E (4), Q (4) | I (96), V/I (4) | V (63), A (19), T (11), V/A (7) | I (52), V (41), V/I (7) | L (70), M (26), L/M (4) | F (48), L (48), L/F (4) |
Below each amino acid position is the list of amino acids that are observed, and in parentheses is the percentage of sequences concerned. The predominant amino acid for each position is given first. Amino acids found to be selected by PI-containing antiretroviral therapy are in bold type.
Wild-type sequence from the reference HIV-1 strain HXB2 (GenBank accession number K03455).
Available at http://hiv-web.lanl.gov/content/hiv-db/ALIGN_CURRENT/ALIGN-INDEX.html; nonpredominant amino acids are indicated in parentheses.
HIV-2 subtype specificities.
In our study, the HIV-2 strains obtained from antiretroviral-naive patients were classified as belonging to subtype A (n = 11) or B (n = 2) based on pol sequence analysis (Table 1). The amino acid homology between consensus protease sequences of subtypes A and B was 82%. We identified several codons which could represent a specific subtype signature. For example, in subtype A sequences, positions 12, 67, and 91 harbored a threonine, a leucine, and a threonine, respectively, whereas subtype B sequences had a glutamine or a lysine, a valine, and an asparagine. In addition, all of the subtype B strains available in our study and in the Los Alamos HIV sequence database (http://hiv-web.lanl.gov/content/hiv-db/ALIGN_CURRENT/ALIGN-INDEX.html) harbored an isoleucine at codon 46. Interestingly, at position 99, all HIV-2 subtype B sequences from antiretroviral-naive and PI-treated patients harbored a phenylalanine previously found to emerge under a PI-containing regimen.
Associations or exclusions of mutations.
We studied the potential associations or exclusions of amino acids at each position in HIV-2 protease in all of the available HIV-2 protease sequences. Amino acid 90M (observed in 11 of 65 sequences) was rarely associated with amino acid 7R (in one sequence), 62A or 62T (in one sequence), or 99F (in four cases). In contrast, amino acid 7R, found in eight sequences only from PI-treated patients, was always associated with amino acid 46I and in most cases was associated with amino acid 62A or 62T (in six sequences), amino acid 71I (in seven sequences), or amino acid 99F (in six sequences).
DISCUSSION
We analyzed 65 HIV-2 protease sequences from 21 HIV-2-infected patients. Although they have similar structures (40), HIV-2 and HIV-1 proteases are approximately 50% divergent in their amino acid compositions (21). Moreover, differences in conformation (32) and interaction parameters with substrates or PI have been described (15, 29, 43). It is important to know if natural polymorphism and mutations which confer resistance to PI in HIV-1 are found at homologous positions in the HIV-2 protease. Natural polymorphism patterns are radically different in HIV-2 and HIV-1 proteases. In our study, highly polymorphic positions of HIV-2 protease from antiretroviral-naive patients (positions 14, 40, 43, 46, 65, and 70) were generally highly conserved in HIV-1 strains. The highest levels of natural polymorphism in HIV-1 were observed at positions 35, 37, 41, 62, 63, 64, 77, and 93 (7, 20). In addition to rough differences in amino acid composition between HIV-2 and HIV-1 (21), our findings indicate that natural polymorphisms occur at specific and different positions in HIV-2 and HIV-1 sequences. For example, position 63, which is highly polymorphic in HIV-1 strains (7, 20), harbored only a glutamic acid in HIV-2. The extent to which polymorphism influences or predisposes to the evolution of drug resistance remains to be determined (7, 16, 28). In contrast, we observed that the three regions from positions 25 to 32, including the active site of HIV protease, 48 to 56, corresponding to the flap region, and 78 to 87 were mostly conserved (6). Interestingly, the last three regions were equally conserved in HIV-1 protease and harbored the same amino acids at most positions (7, 20, 21), strongly suggesting a common structural backbone for both enzymes. In the sequences from antiretroviral-naive HIV-2-infected patients, we found HIV-1 drug resistance-associated codons in most cases (amino acids 36I, 46I, and 71V) or in all cases (amino acids 10V, 32I, 47V, and 73A). Of the HIV-2 sequences from untreated patients, 59% harbored an isoleucine at position 46, corresponding in HIV-1 to a major drug resistance mutation to indinavir (IDV) and to a minor drug resistance mutation to ritonavir (RTV), nelfinavir (NFV), amprenavir (APV), lopinavir plus ritonavir (LPV/r), and atazanavir (ATV). These data are in agreement with those of other studies, suggesting a naturally reduced susceptibility to these drugs even in untreated individuals (34; D. Pienazek, M. Rayfield, J. Nkengasong, W. Heneine, V. Soriano, C. Zeh, S. M. Agwale, C. Wambebe, L. Odama, T. Folks, and M. L. Kalish, Abstr. 8th Conf. Retroviruses Opportunistic Infect., abstr. 461, 2001). Recently, Pienazek et al. studied 75 HIV-2 protease sequences (68 and 32% belonging to subtypes A and B, respectively) and found that all of them harbored at least four positions associated with PI resistance in HIV-1 at amino acids 10, 32, 36, 46, 47, 71, or 77, with 89% harboring 46I (D. Pieniazek, M. Rayfield, D. Hu, J. Nkengasong, V. Soriano, W. Heneine, C. Zeh, S. Agwale, C. Wambebe, L. Odama, and M. L. Kalish, Abstr. 10th Conf. Retroviruses Opportunistic Infect. abstr. 629, 2003). Interestingly, Collin et al. observed a lower phenotypic susceptibility to RTV of HIV-2 strains compared with HIV-1 strains in drug-naive patients (Collin et al., Abstr. 2nd Int. Workshop HIV Drug Resist. Treat. Strategies, 1998). As a matter of fact, the concept of natural drug resistance has already been suggested for HIV-1 group O isolates with respect to nonnucleosidic RT inhibitors: a tyrosine-to-cysteine substitution at codon 181 of the RT gene was associated with high-level phenotypic resistance (14). Recently, a structural study suggested that the presence of an isoleucine at position 181 of HIV-2 RT could contribute to the inherent drug resistance of HIV-2 to nonnucleosidic RT inhibitors (33).
Genotypic studies of the development of in vivo drug resistance in HIV-2-infected patients are rare (34; D. Descamps, F. Damond, S. Matheron, I. Farfara, S. Lastere, P. Campa, P. Foiny, S. Pueyo, G. Chene, F. Brun-Vezinet and the ANRS French HIV-2 cohort, Abstr. 11th Int. Workshop HIV Drug Resist. Treat. Strategies, abstr. 138, 2002), and no trial of antiretroviral therapy in HIV-2-infected patients has been reported to date. In contrast to HIV-1 infection, we lack data providing accurate information about the impact of primary or secondary protease mutations on both viral fitness and the level of drug resistance in HIV-2 infection. In the present study, all eight patients undergoing antiretroviral treatment received NFV whereas the other PI were given sequentially: IDV (four patients), RTV (four patients), LPV/r (two patients), saquinavir (SQV) (one patient), and APV (one patient) (Table 1). In agreement with other studies (J. N. Telles, D. Descamps, S. Matheron, G. Collin, A. Leprêtre, F. Damond, F. Simon, F. Brun-Vezinet and the ANRS French HIV-2 Cohort, Abstr. 3rd Int. Workshop Drug Resist. Treat. Strategies, abstr. 54, 1999; M. Schutten, M. Van der Ende, B. Van der Hogen, et al., Abstr. 7th Eur. Conf. Clin. Aspects Treat. HIV Infect. abstr. 1237, 1999; C. Adje, R. Cheingsong, J. G. Garcia-Lerma, T. Roels, T. Chorba, D. R. Adams, R. A. Otten, R. Respess, J. Nkengasong, and W. Heneine, Abstr. 5th Int. Workshop HIV Drug Resist. Treat. Strategies, abstr. 57, 2001; Descamps, et al., Abstr. 11th Int. Workshop HIV Drug Resist. Treat. Strategies, 2002), we found substitutions associated with PI resistance at positions 46 (V→I), 71 (V→I), or 90 (L→M) in HIV-2 infected patients receiving IDV-, SQV-, NFV-, or RTV-containing regimens. In contrast, we did not find the previously reported PI resistance-associated mutations at positions 54 (I→L or M), 82 (I→M or F) or 84 (I→V) (Descamps et al., Abstr. 11th Int. Workshop HIV Drug Resist. Treat. Strategies, 2002; Goncalves et al., Abstr. 11th Int. Workshop HIV Drug Resist. Treat. Strategies, 2002; Schutten, et al., Abstr. 7th Eur. Conf. Clin. Aspects Treat. HIV Infect., 1999). Nor did we find mutations T12P, G17Q, R72A, or M76V, conferring phenotypic resistance to IDV and SQV as reported in a recent study (Goncalves et al., Abstr. 11th Int. Workshop HIV Drug Resist. Treat. Strategies, 2002). In our study, the K7R mutation was found in the patients treated with LPV/r, RTV, or SQV and in most cases seemed to be associated with an isoleucine at positions 46 and 71, an alanine or a threonine at position 62, and a phenylalanine at position 99. The L99F mutation was selected by NFV-, LPV/r-, SQV-, or IDV-containing regimens, suggesting a relatively broad cross-resistance spectrum. In addition, our data, although limited by the number of the samples studied, suggest that a natural resistance to PI could be subtype dependent since HIV-2 strains of subtype B from PI-naive patients display both V46I and L99F mutations. The clinical significance of the substitutions that we described as being associated with PI selective pressure warrants further investigation and confirmation by both phenotypic and site-directed mutagenesis studies.
In conclusion, this genotypic study of HIV-2 protease revealed that despite an amino acid identity of approximately 50% between HIV-2 and HIV-1 proteases, HIV-2 strains harbor specific hot spots of natural polymorphism and resistance profile (both natural and drug selected). On the one hand, we found amino acids associated with PI resistance in HIV-1 at 7 positions in HIV-2 protease which could be associated with natural resistance of HIV-2 to PI. On the other hand, we described six amino acid substitutions that seemed to emerge under selective pressure by PI. Two of these last substitutions are major PI resistance mutations in HIV-1 (V46I and L90M), but we described three new amino acid substitutions at positions 7 (K→R), 62 (V→A/T), and 99 (L→F) selected under PI pressure and not present in HIV-1. In contrast, as in HIV-1 protease, we identified three conserved areas in HIV-2 protease which could represent critical regions for the preservation of a functional and stable structure.
REFERENCES
- 1.Adjorlolo-Johnson, G., K. M. De Cock, E. Ekpini, K. M. Vetter, T. Sibailly, K. Brattegaard, D. Yavo, R. Doorly, J. P. Whitaker, and L. Kestens. 1994. Prospective comparison of mother-to-child transmission of HIV-1 and HIV-2 in Abidjan, Ivory Coast. JAMA 272:462-466. [PubMed] [Google Scholar]
- 2.Ancelle, R., O. Bletry, A. C. Baglin, F. Brun-Vezinet, M. A. Rey, and P. Godeau. 1987. Long incubation period for HIV-2 infection. Lancet i:688-689. [DOI] [PubMed]
- 3.Andersson, S., H. Norrgren, Z. da Silva, A. Biague, S. Bamba, S. Kwok, C. Christopherson, G. Biberfeld, and J. Albert. 2000. Plasma viral load in HIV-1 and HIV-2 singly and dually infected individuals in Guinea-Bissau, West Africa: significantly lower plasma virus set point in HIV-2 infection than in HIV-1 infection. Arch. Intern. Med. 160:3286-3293. [DOI] [PubMed] [Google Scholar]
- 4.Berry, N., K. Ariyoshi, S. Jaffar, S. Sabally, T. Corrah, R. Tedder, and H. Whittle. 1998. Low peripheral blood viral HIV-2 RNA in individuals with high CD4 percentage differentiates HIV-2 from HIV-1 infection. J. Hum. Virol. 1:457-468. [PubMed] [Google Scholar]
- 5.Bock, P. J., and D. M. Markovitz. 2001. Infection with HIV-2. AIDS 15:S35-S45. [DOI] [PubMed] [Google Scholar]
- 6.Boden, D., and M. Markowitz. 1998. Resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 42:2775-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bossi, P., M. Mouroux, A. Yvon, F. Bricaire, H. Agut, J. M. Huraux, C. Katlama, and V. Calvez. 1999. Polymorphism of the human immunodeficiency virus type 1 (HIV-1) protease gene and response of HIV-1-infected patients to a protease inhibitor. J. Clin. Microbiol. 37:2910-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Z., A. Luckay, D. L. Sodora, P. Telfer, P. Reed, A. Gettie, J. M. Kanu, R. F. Sadek, J. Yee, D. D. Ho, L. Zhang, and P. A. Marx. 1997. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J. Virol. 71:3953-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clavel, F. 1987. HIV-2, the West African AIDS virus. AIDS 1:135-140. [PubMed] [Google Scholar]
- 10.Clavel, F., D. Guetard, F. Brun-Vezinet, S. Chamaret, M. A. Rey, M. O. Santos-Ferreira, A. G. Laurent, C. Dauguet, C. Katlama, and C. Rouzioux. 1986. Isolation of a new human retrovirus from West African patients with AIDS. Science 233:343-346. [DOI] [PubMed] [Google Scholar]
- 11.Damond, F., C. Apetrei, D. L. Robertson, S. Souquiere, A. Lepretre, S. Matheron, J. C. Plantier, F. Brun-Vezinet, and F. Simon. 2001. Variability of human immunodeficiency virus type 2 (HIV-2) infecting patients living in france. Virology 280:19-30. [DOI] [PubMed] [Google Scholar]
- 12.Damond, F., M. Gueudin, S. Pueyo, I. Farfara, D. L. Robertson, D. Descamps, G. Chene, S. Matheron, P. Campa, F. Brun-Vezinet, and F. Simon. 2002. Plasma RNA viral load in human immunodeficiency virus type 2 subtype A and subtype B infections. J. Clin. Microbiol. 40:3654-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Cock, K. M., G. Adjorlolo, E. Ekpini, T. Sibailly, J. Kouadio, M. Maran, K. Brattegaard, K. M. Vetter, R. Doorly, and H. D. Gayle. 1993. Epidemiology and transmission of HIV-2. Why there is no HIV-2 pandemic. JAMA 270:2083-2086. [DOI] [PubMed] [Google Scholar]
- 14.Descamps, D., G. Collin, F. Letourneur, C. Apetrei, F. Damond, I. Loussert-Ajaka, F. Simon, S. Saragosti, and F. Brun-Vezinet. 1997. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J. Virol. 71:8893-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan, N., K. B. Rank, J. W. Leone, R. L. Heinrikson, C. A. Bannow, C. W. Smith, D. B. Evans, S. M. Poppe, W. G. Tarpley, and D. J. Rothrock. 1995. The differential processing of homodimers of reverse transcriptases from human immunodeficiency viruses type 1 and 2 is a consequence of the distinct specificities of the viral proteases. J. Biol. Chem. 270:13573-13579. [PubMed] [Google Scholar]
- 16.Frater, J., D. Dunn, J. N. Weber, and M. O. McClure. 2002. Association between secondary mutations in human immunodeficiency virus type 1 protease and therapeutic outcome. J. Infect. Dis. 185:1376. [DOI] [PubMed] [Google Scholar]
- 17.Gao, F., L. Yue, D. L. Robertson, S. C. Hill, H. Hui, R. J. Biggar, A. E. Neequaye, T. M. Whelan, D. D. Ho, and G. M. Shaw. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghys, P. D., K. Fransen, M. O. Diallo, V. Ettiegne-Traore, I. M. Coulibaly, K. M. Yeboue, M. L. Kalish, C. Maurice, J. P. Whitaker, A. E. Greenberg, and M. Laga. 1997. The associations between cervicovaginal HIV shedding, sexually transmitted diseases and immunosuppression in female sex workers in Abidjan, Cote d'Ivoire. AIDS 11:F85-F93. [DOI] [PubMed] [Google Scholar]
- 19.Kanki, P. 1991. Biological features of HIV-2. An update. AIDS Clin. Rev. 1991:17-38. [PubMed]
- 20.Kozal, M. J., N. Shah, N. Shen, R. Yang, R. Fucini, T. C. Merigan, D. D. Richman, D. Morris, E. Hubbell, M. Chee, and T. R. Gingeras. 1996. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat. Med. 2:753-759. [DOI] [PubMed] [Google Scholar]
- 21.Kuiken, C., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber (ed.). HIV sequence compendium 2001. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.M.
- 22.Langley, C. L., E. Benga-De, C. W. Critchlow, I. Ndoye, M. D. Mbengue-Ly, J. Kuypers, G. Woto-Gaye, S. Mboup, C. Bergeron, K. K. Holmes, and N. B. Kiviat. 1996. HIV-1, HIV-2, human papillomavirus infection and cervical neoplasia in high-risk African women. AIDS 10:413-417. [DOI] [PubMed] [Google Scholar]
- 23.Marlink, R. 1996. Lessons from the second AIDS virus, HIV-2. AIDS 10:689-699. [DOI] [PubMed] [Google Scholar]
- 24.Marlink, R., P. Kanki, I. Thior, K. Travers, G. Eisen, T. Siby, I. Traore, C. C. Hsieh, M. C. Dia, and E. H. Gueye. 1994. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science 265:1587-1590. [DOI] [PubMed] [Google Scholar]
- 25.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, S. D. Holmberg, and HIV Outpatient Study Investigators. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 26.Peeters, M. 2001. The genetic variability of HIV-1 and its implications. Transfus. Clin. Biol. 8:222-225. [DOI] [PubMed] [Google Scholar]
- 27.Peeters, M., B. Koumare, C. Mulanga, C. Brengues, B. Mounirou, F. Bougoudogo, S. Ravel, F. Bibollet-Ruche, and E. Delaporte. 1998. Genetic subtypes of HIV type 1 and HIV type 2 strains in commercial sex workers from Bamako, Mali. AIDS Res. Hum. Retroviruses 14:51-58. [DOI] [PubMed] [Google Scholar]
- 28.Perno, C. F., A. Cozzi-Lepri, C. Balotta, F. Forbici, M. Violin, A. Bertoli, G. Facchi, P. Pezzotti, G. Cadeo, G. Tositti, S. Pasquinucci, S. Pauluzzi, A. Scalzini, B. Salassa, A. Vincenti, A. N. Phillips, F. Dianzani, A. Appice, G. Angarano, L. Monno, G. Ippolito, M. Moroni, and A. d' Arminio Monforte. 2001. Secondary mutations in the protease region of human immunodeficiency virus and virologic failure in drug-naive patients treated with protease inhibitor-based therapy. J. Infect. Dis. 184:983-991. [DOI] [PubMed] [Google Scholar]
- 29.Phylip, L. H., A. D. Richards, J. Kay, J. Kovalinka, P. Strop, I. Blaha, J. Velek, V. Kostka, A. J. Ritchie, and A. V. Broadhurst. 1990. Hydrolysis of synthetic chromogenic substrates by HIV-1 and HIV-2 proteinases. Biochem. Biophys. Res. Commun. 171:439-444. [DOI] [PubMed] [Google Scholar]
- 30.Popper, S. J., A. D. Sarr, K. U. Travers, A. Gueye-Ndiaye, S. Mboup, M. E. Essex, and P. J. Kanki. 1999. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J. Infect. Dis. 180:1116-1121. [DOI] [PubMed] [Google Scholar]
- 31.Poulsen, A. G., B. Kvinesdal, P. Aaby, K. Molbak, K. Frederiksen, F. Dias, and E. Lauritzen. 1989. Prevalence of and mortality from human immunodeficiency virus type 2 in Bissau, West Africa. Lancet i:827-831. [DOI] [PubMed]
- 32.Priestle, J. P., A. Fassler, J. Rosel, M. Tintelnot-Blomley, P. Strop, and M. G. Grutter. 1995. Comparative analysis of the X-ray structures of HIV-1 and HIV-2 proteases in complex with CGP 53820, a novel pseudosymmetric inhibitor. Structure 3:381-389. [DOI] [PubMed] [Google Scholar]
- 33.Ren, J., L. E. Bird, P. P. Chamberlain, G. B. Stewart-Jones, D. I. Stuart, and D. K. Stammers. 2002. Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors. Proc. Natl. Acad. Sci. USA 99:14410-14415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodes, B., A. Holguin, V. Soriano, M. Dourana, K. Mansinho, F. Antunes, and J. Gonzalez-Lahoz. 2000. Emergence of drug resistance mutations in human immunodeficiency virus type 2-infected subjects undergoing antiretroviral therapy. J. Clin. Microbiol. 38:1370-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon, F., S. Matheron, C. Tamalet, I. Loussert-Ajaka, S. Bartczak, J. M. Pepin, C. Dhiver, E. Gamba, C. Elbim, and J. A. Gastaut. 1993. Cellular and plasma viral load in patients infected with HIV-2. AIDS 7:1411-1417. [DOI] [PubMed] [Google Scholar]
- 36.Soriano, V., P. Gomes, W. Heneine, A. Holguin, M. Doruana, R. Antunes, K. Mansinho, W. M. Switzer, C. Araujo, V. Shanmugam, H. Lourenco, J. Gonzalez-Lahoz, and F. Antunes. 2000. Human immunodeficiency virus type 2 (HIV-2) in Portugal: clinical spectrum, circulating subtypes, virus isolation, and plasma viral load. J. Med. Virol. 61:111-116. [PubMed] [Google Scholar]
- 37.Switzer, W. M., S. Wiktor, V. Soriano, A. Silva-Graca, K. Mansinho, I. M. Coulibaly, E. Ekpini, A. E. Greenberg, T. M. Folks, and W. Heneine. 1998. Evidence of Nef truncation in human immunodeficiency virus type 2 infection. J. Infect. Dis. 177:65-71. [DOI] [PubMed] [Google Scholar]
- 38.Takehisa, J., M. Osei-Kwasi, N. K. Ayisi, O. Hishida, T. Miura, T. Igarashi, J. Brandful, W. Ampofo, V. B. Netty, M. Mensah, M. Yamashita, E. Ido, and M. Hayami. 1997. Phylogenetic analysis of HIV type 2 in Ghana and intrasubtype recombination in HIV type 2. AIDS Res. Hum. Retroviruses 13:621-623. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomasselli, A. G., J. O. Hui, T. K. Sawyer, D. J. Staples, C. Bannow, I. M. Reardon, W. J. Howe, D. L. DeCamp, C. S. Craik, and R. L. Heinrikson. 1990. Specificity and inhibition of proteases from human immunodeficiency viruses 1 and 2. J. Biol. Chem. 265:14675-14683. [PubMed] [Google Scholar]
- 41.Vacca, J. P., B. D. Dorsey, W. A. Schleif, R. B. Levin, S. L. McDaniel, P. L. Darke, J. Zugay, J. C. Quintero, O. M. Blahy, and E. Roth. 1994. L-735,524: an orally bioavailable human immunodeficiency virus type 1 protease inhibitor. Proc. Natl. Acad. Sci. USA 91:4096-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vittinghoff, E., S. Scheer, P. O'Malley, G. Colfax, S. D. Holmberg, and S. P. Buchbinder. 1999. Combination antiretroviral therapy and recent declines in AIDS incidence and mortality. J. Infect. Dis. 179:717-720. [DOI] [PubMed] [Google Scholar]
- 43.Weber, J., P. Majer, J. Litera, J. Urban, M. Soucek, J. Vondrasek, J. Konvalinka, P. Novek, J. Sedlacek, P. Strop, H. G. Krausslich, and I. Pichova. 1997. Potency comparison of peptidomimetic inhibitors against HIV-1 and HIV-2 proteinases: design of equipotent lead compounds. Arch. Biochem. Biophys. 341:62-69. [DOI] [PubMed] [Google Scholar]
- 44.Whittle, H., A. Egboga, J. Todd, T. Corrah, A. Wilkins, E. Demba, G. Morgan, M. Rolfe, N. Berry, and R. Tedder. 1992. Clinical and laboratory predictors of survival in Gambian patients with symptomatic HIV-1 or HIV-2 infection. AIDS 6:685-689. [DOI] [PubMed] [Google Scholar]
- 45.Wilkins, A., D. Ricard, J. Todd, H. Whittle, F. Dias, and A. Paulo Da Silva. 1993. The epidemiology of HIV infection in a rural area of Guinea-Bissau. AIDS 7:1119-1122. [DOI] [PubMed] [Google Scholar]
- 46.Yamaguchi, J., S. G. Devare, and C. A. Brennan. 2000. Identification of a new HIV-2 subtype based on phylogenetic analysis of full-length genomic sequence. AIDS Res. Hum. Retroviruses 16:925-930. [DOI] [PubMed] [Google Scholar]